Abstract
Extensive gene expression during meiosis is a hallmark of spermatogenesis. Although it was generally accepted that RNA transcription ceases during meiosis, recent observations suggest that some transcription occurs in postmeiosis. To further resolve this issue, we provide direct evidence for the de novo transcription of RNA during the postmeiotic phases. These results strengthen the newly emerging notion that postmeiotic transcription is dynamic and integral to the overall process of spermatogenesis.
SPERMATOGENESIS is a fundamental developmental process producing male sex cells, the spermatozoa. In Drosophila melanogaster, this process occurs in three phases: a diploid mitotic phase, which increases cell numbers and size; a second, diploid meiotic phase characterized by intense transcriptional activity and important structural changes; and a final postmeiotic haploid phase of sperm morphogenesis and maturation. Early experiments using autoradiography demonstrated RNA transcription during mitosis and early meiosis but no transcriptional activity during postmeiosis (Olivieri and Olivieri 1965). On the basis of this and other similar studies, it has generally been accepted that the postmeiotic phase is devoid of transcriptional activity and led to the prevailing notion that proteins required later during the postmeiotic stages were translated from mRNAs produced during meiosis and stored in the cytoplasm (Schäfer et al. 1995). Two contemporary studies have challenged this view, one using a targeted gene expression approach demonstrating mRNA accumulation in postmeiotic spermatids (Barreau et al. 2008) and our recent microarray analysis estimating substantial genome-wide expression during postmeiosis (Vibranovski et al. 2009). This latter study demonstrated that 20–30% of testis genes transcribed are over expressed during postmeiosis in comparison to meiosis. However, both studies failed to provide clear-cut direct evidence for postmeiotic transcription: both studies measured, either qualitatively or quantitatively, mRNA in postmeiotic cells but did not provide direct evidence for the production of nascent RNA. In addition, in the first study, only a small subset of genes was analyzed and the second study measured bulk mRNA levels from dissected tissues and therefore expression at the cellular level remains unclear. Here, we directly visualized RNA transcripts in intact testes using 5-bromouridine (BrU) and we describe their cellular and subcellular distributions during spermatogenesis (Figure 1).
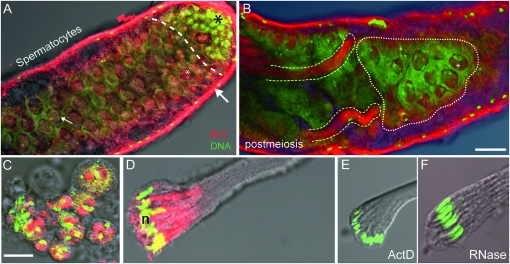
Figure 1.—
Transcription in the Drosophila testis visualized following BrU incorporation. Overlay confocal fluorescent/light microscopic images of the apical and middle regions of intact testes (upper) and individual cyst cells (lower) showing localized BrU labeling (red) and both DNA and RNA (green). (A) Apical regions showing hub and stem cell divisions and spermatogonia (black asterisk) distinct from the adjacent area that predominantly contains spermatocytes (dashed line). A strong localized BrU signal in presumptive nucleoli (white asterisk) were observed as bright red dots in nuclei of spermatocytes near the apical end and more diffuse BrU signals were observed in enlarged spermatocyte nuclei located more posteriorly (e.g., small white arrow). The bulk of BrU incorporation is observed in somatic sheath cells (large white arrow). (B) Distal midregion view of testis showing spermatocytes (white stippling) and postmeiotic spermatid bundles (yellow stippling). Diffuse BrU incorporation in spermatocyte nuclei (asterisk) and inside spermatid bundles are clearly visible. (C) Enhanced views of BrU incorporation in isolated spermatocytes and (D) elongating spermatid bundles provided clear evidence for de novo RNA transcription in postmeiotic phases. Similarly staged postmeiotic spermatid bundles treated with either (E) actinomycin D (ActD) or (F) Rnase treatment resulted in a nearly complete loss of BrU signal, providing further confirmation that the BrU signal was a consequence of RNA transcription. Methods: For these, and images shown in Figure S1, the following procedures and reagents were used (modified from Chang et al. 2000; Ohtsu et al. 2008): For BrU incorporation, testes dissected in TB1 (testis buffer 1) and treated with a mixture of BrU (100 mM) (Sigma- Aldrich) and DOTAP (0.2 μg/μl) (Roche Molecular Bio) for either 1 hr (intact testes), 30 min (meiotic cells), or 20 min (postmeiotic cells). DOTAP is a liposomal transfection reagent for the highly efficient transfection of negatively charged molecules such as RNA into eukaryotic cells. Cells were then washed three times for 5 min each with TB1. Cells were fixed and processed for indirect immunofluorescence: stained with a primary anti-bromo-deoxyuridine (anti BrdU) mouse monoclonal antibody [2 μg/μl, in BTP (PBS-T 0.1%, BSA0.5%)] (Roche Molecular Bio) for 1 hr, briefly washed three times prior to staining with a goat Cy5-cyanine anti-mouse IgG secondary antibody (Jackson Immuno Research) diluted 1/400 for 1 hr. Then, cells were imaged using a Zeiss LSM 510 confocal microscope (BrU, red; DNA, green). We note that similar results were obtained using BrUTP. Bar, 50 μm (upper); 10 μm (lower). Rnase treatment was performed using DNase-free RNase A solution at 1 mg/ml, in 2× SSC.
As expected, in intact testis, strong BrU signals were observed in somatic cells of the outer sheath, in presumptive nucleoli of primary spermatocytes (Figure 1A). We also observed a surprisingly strong BrU signal in developing spermatid bundles during postmeiosis (Figure 1B). Additionally, BrU incorporation in isolated spermatocytes (Figure 1C) and in isolated postmeiotic spermatid bundles (Figure 1D) routinely displayed strong BrU signal near spermatid nuclei. The latter result provides direct evidence for de novo RNA synthesis in postmeiosis. Those cells were isolated from an intact testis prior to addition of BrU and thus eliminating the possibility that intercellular transport of RNA molecules from surrounding somatic testis sheath cells in intact testes was responsible for the observed BrU signals in postmeiotic cells. The BrU signal was reduced in the presence of actinomycin D, a general inhibitor of RNA synthesis (Figure 1E), confirming that the BrU signal in these cells was dependent on RNA synthesis. In addition, a virtual complete loss of BrU signal in Rnase-treated postmeiotic spermatid bundles provided further confirmation that the BrU signal was a consequence of RNA presence (Figure 1F). Statistical analysis confirmed BrU signal inhibition by actinomycin D and Rnase compared to control signal (Fisher exact test, P ≤ 0.0005). Taken together, these results strongly support the conclusion that robust RNA transcription is prevalent in the postmeiotic phases.
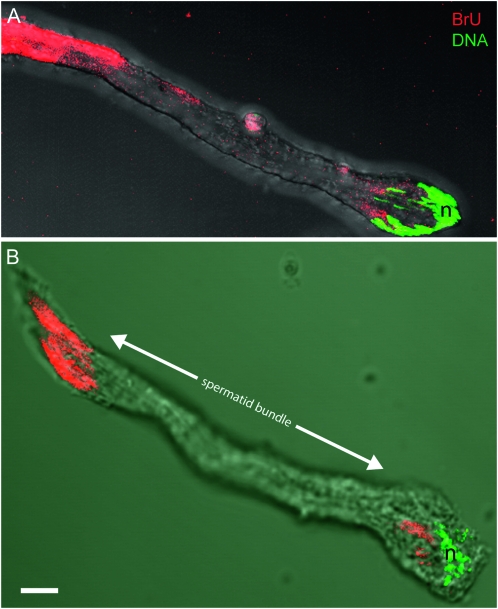
Confocal imaging and 3D reconstruction of testes demonstrated the subcellular localization of BrU signal within developing spermatocytes (supporting information, Figure S1, A and B). Additional 3D reconstructions of isolated elongating spermatid bundles suggested periodic phases (“waves”) of postmeiotic transcription where blocks of BrU signal were observed between intervening regions devoid of signal (Figure 2). We observed this general phenomenon in repeated independent experiments and, although our data do not fully explain the origin or the temporal dynamics of this process, they do raise the intriguing possibility that RNA is produced by active transcription in spermatid nuclei and then actively transported down the bundle.
Figure 2.—
Merged confocal and transmitted light images of isolated spermatid bundles labeled with BrU. (A) 3D reconstruction of the apical end of a single postmeiotic spermatid bundle (note, ∼10% of the entire bundle is shown). The characteristic grouping of nuclei (n) denotes the apical end of the bundle. BrU staining is apparent both in the apical region near nuclei and along the growing spermatid axonemes. Note the absence of BrU signal in the middle region (between arrowheads) of the bundle. (B) Full view early stage of an elongating spermatid showing BrU signal at both apical and distal ends. Bar 10 μm.
In summary, our work extends previous studies because the use of BrU incorporation in isolated spermatids provides direct evidence for the production of nascent RNA in postmeiotic cells. Therefore, it further strengthens the newly emerging notion that postmeiotic transcription is dynamic and integral to the overall process of spermatogenesis in D. melanogaster.
In addition to obvious potential functions during the latter stages of sperm maturation, are there other consequences for RNA products of postmeiotic transcription? One intriguing possibility is that a subset of these RNAs is packaged into mature sperm and delivered to the egg at fertilization. Indeed, microarray studies indicate robust mRNA signals in purified Drosophila (personal communication, S. Russell, S. Dorus and T. L. Karr) and mammalian sperm (Krawetz 2005). Our results suggest that transcripts observed in mature sperm might arise from all three major phases of spermatogenesis including the postmeiotic phase. Finally, the postmeiotic transcriptional dynamics suggested by our results further strengthen the notion that haploid gamete gene expression may have important consequences for the evolution of sperm competition and sperm cooperation by generating variation in and among sibling spermatozoa (Parker and Begon 1993; Immler 2008).
Acknowledgments
We thank Melina Hale and Ru Yi Teow for helpful assistance with the confocal microscope. We also thank Michael B. Eisen for suggesting experiments. This work was supported by a National Science Foundation CAREER award (MCB0238168) and National Institutes of Health (NIH) grants (R01GM065429-01A1 and R01GM078070-01A1) (to M.L.), and an NIH American Recovery and Reinvestment Act (ARRA) supplement award (R01GM0780-0351) to M.L., T.L.K., and H.L. M.D.V. was supported by a Pew Latin America fellowship. H.F.L.'s research was supported by Booth School of Business, University of Chicago. T.L.K. was supported by Biodesign Institute, Arizona State University.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.118919/DC1.
References
- Barreau, C., E. Benson, E. Gudmannsdottir, F. Newton and H. White-Cooper, 2008. Post-meiotic transcription in Drosophila testes. Development 135 1897–1902. [DOI] [PubMed] [Google Scholar]
- Chang, W. Y., N. A. Winegarden, J. P. Paraiso, M. L. Stevens and J. T. Westwood, 2000. Visualization of nascent transcripts on Drosophila polytene chromosomes using BrUTP incorporation. Biotechniques 29 934–936. [DOI] [PubMed] [Google Scholar]
- Immler, S., 2008. Sperm competition and sperm cooperation: the potential role of diploid and haploid expression. Reproduction 135 275–283. [DOI] [PubMed] [Google Scholar]
- Krawetz, S. A., 2005. Paternal contribution: new insights and future challenges. Nat. Rev. Genet. 6 633–642. [DOI] [PubMed] [Google Scholar]
- Ohtsu, M., M. Kawate, M. Fukuoka, W. Gunji, F. Hanaoka, et al., 2008. Novel DNA microarray system for analysis of nascent mRNAs. DNA Res. 15 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri, G., and A. Olivieri, 1965. Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster. Mutat. Res. 2 366–380. [DOI] [PubMed] [Google Scholar]
- Parker, G. A., and M. E. Begon, 1993. Sperm competition games: sperm size and number under gametic control. Proc. Biol. Sci. 253 255–262. [DOI] [PubMed] [Google Scholar]
- Schäfer, M., K. Nayernia, W. Engel and U. Schäfer, 1995. Translational control in spermatogenesis. Dev. Biol. 172 344–352. [DOI] [PubMed] [Google Scholar]
- Vibranovski, M. D., H. F. Lopes, T. L. Karr and M. Long, 2009. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5 e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]