Abstract
The ability to identify genetic markers in nonmodel systems has allowed geneticists to construct linkage maps for a diversity of species, and the sex-determining locus is often among the first to be mapped. Sex determination is an important area of study in developmental and evolutionary biology, as well as ecology. Its importance for organisms might suggest that sex determination is highly conserved. However, genetic studies have shown that sex determination mechanisms, and the genes involved, are surprisingly labile. We review studies using genetic mapping and phylogenetic inferences, which can help reveal evolutionary pattern within this lability and potentially identify the changes that have occurred among different sex determination systems. We define some of the terminology, particularly where confusion arises in writing about such a diverse range of organisms, and highlight some major differences between plants and animals, and some important similarities. We stress the importance of studying taxa suitable for testing hypotheses, and the need for phylogenetic studies directed to taxa where the patterns of changes can be most reliably inferred, if the ultimate goal of testing hypotheses regarding the selective forces that have led to changes in such an essential trait is to become feasible.
THE ever-increasing accessibility of genetic markers is allowing sex-determining regions to be genetically mapped in a growing number of nonmodel organisms. There are several reasons for studying sex determination. In animals, gonadal differences are often accompanied by striking somatic secondary sexual dimorphisms, which are interesting in an evolutionary context (Shine 1989; Badyaev 2002). In plants, females and males often differ in flower morphology and abundance (Dawson and Geber 1998), and, although sex differences are often minor outside the flowers (or inflorescences), they do exist (Dawson and Geber 1998; Eppley and Wenk 2001). The genetic control of these phenotypes is a fundamental biological process, and studying sex determination pathways is important in animal developmental biology (Adams and McLaren 2002; Pinyopich et al. 2003), including genetic pathway evolution (Wilkins 1995; Williams and Carroll 2009).
Until recently, sex determination was generally studied by testing for genetic control vs. partial or complete environmental influences. Genetic systems were examined cytologically to determine the level of heteromorphism between the sex chromosomes and to identify whether females or males are heterogametic (see the comprehensive review in Bull 1983). Male heterogametic systems, referred to as XY, were also tested to identify whether the Y chromosome carries a male-determining gene, as in almost all therian mammals and most dioecious plants so far studied, or whether sex is determined through X–autosome balance, as in Drosophila and Caenorhabditis elegans (Haag 2005). For female heterogametic (ZW) species, analogous tests were used to identify how femaleness is determined.
Until recently, sex-determining genes and regions could be genetically mapped in only a few model species, but now that molecular genetic markers can be developed in nonmodel species, new information is becoming available about how genetic sex determination (GSD) mechanisms have changed during evolutionary history. It has long been known from genetic mapping in model systems, including mammals, and (more recently) birds, that sex chromosomes often have large nonrecombining regions (Bull 1983; Charlesworth 1991; Charlesworth et al. 2005). However, in other organisms, nonrecombining regions are not always large and may sometimes be absent. The evolution of sexual reproduction and recombination have been the focus of many years of discussion in evolutionary biology (Otto 2009), and studies of sex chromosomes are important for understanding why recombination is often lost, and elucidating the evolutionary consequences of recombination suppression (Charlesworth 1996; Otto and Barton 1997; Barton and Charlesworth 1998). The adaptation of genes on the sex chromosomes is also interesting, because this location affects the outcome of sex-specific selection pressures (Rice 1984; Charlesworth et al. 1987; Vicoso and Charlesworth 2006; Mank 2009a). Finally, the mechanism of sex determination can affect sex ratios (West and Sheldon 2002; Dorken and Pannell 2008; West 2009) and is therefore significant in evolutionary ecology.
Sex determination is also relevant in applied biology. In many domesticated animals, one sex may be of greatest economic interest to farmers and breeders. Modern meat production is largely based on males, including industrial production of chicken, cattle, and many fish, whereas females are the sex required for milk (cattle) and egg (chicken) production. Similarly, a few crop plants are dioecious, and, in some of these, the crop is produced by females (e.g., grapes, dates, and papaya), while in other species the sexes differ in characteristics such as fiber or chemical content. Because immature birds, fish, and plants have no obvious phenotypic sex differences, maximizing agricultural returns often requires genetically sexing juveniles. Mapping sex determination is an important first step toward identifying the sex-determining genes or finding other sex-specific markers to develop molecular sexing methods.
In this review, we first summarize recent developments in genetic mapping of sex determination, concentrating on nonmodel plants and animals with genetic sex determination. We show how this information can be useful for understanding the evolution of sex determination and sex chromosomes and identify some important unanswered questions.
SOME IMPORTANT CONCEPTS AND TERMINOLOGY
As we will describe, genetic results have revealed changes in both the mechanism of sex determination and in the location of preexisting sex-determining genes. Given our wide taxonomic scope, it is important to note, first, that the terms X and Y chromosomes (or Z and W) do not necessarily mean a chromosome pair carrying the genes giving the earliest sex-determining signal to the developmental system. In some systems, the sex chromosomes and autosomes jointly determine sex, such as the X–autosome balance systems of Drosophila, C. elegans, and the plant Rumex (Navajas-Perez et al. 2005). Cases such as these probably evolved from ancestors whose sex chromosomes did carry such genes. Mechanisms of sex determination often vary among closely related taxa, and there are several hypotheses to explain transitions between different systems (e.g., Bull 1983; van Doorn and Kirkpatrick 2007; Vuilleumier et al. 2007; Kozielska et al. 2010), but here we focus on new data about empirically demonstrated transitions, for which genetic mapping results are particularly illuminating.
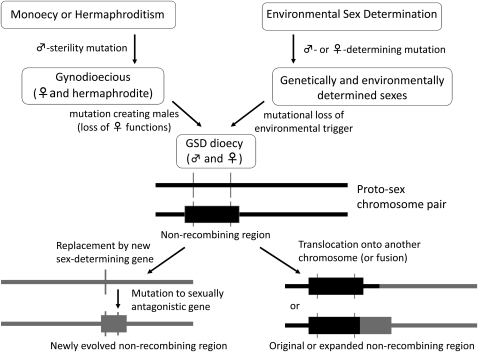
Second, unlike the well-known sex chromosomes of therian mammals and birds, sex-determining chromosomes in some species lack large nonrecombining regions (Bull 1983). This situation can arise in two ways (Figure 1). Sometimes a sex-determining region has not yet evolved a nonrecombining region; this is often referred to as a proto sex chromosome. (See glossary of terms in Table 1.) Alternatively, a recent replacement of the sex-determining locus can produce a homomorphic chromosome pair with single-gene sex determination, and also no nonrecombining region. Replacement of an existing sex-determining gene by a new one will often lead to production of homozygotes for the original sex-determining locus, except when the change involves a fully dominant male-determining gene. When the original sex-determining locus lies within a large sex-specific nonrecombining region, such as a genetically degenerated Y chromosome that lacks many genes carried by the X, such YY individuals will often be homozygous lethal, preventing fixation of the new gene. However, such replacements could occur in plant and fish species where homozygotes for the sex-determining locus are viable. Such recently arisen sex-determining chromosomes are thus also called proto sex chromosomes. If a nonrecombining sex-determining region, such as a dominant male-determining region of a Y chromosome, moves to a new genomic location (e.g., in a chromosome fusion), the original nonrecombining region may be retained. If not, the new sex-determining chromosome may evolve suppressed recombination, either immediately through the chromosome rearrangement itself, or through a later decrease in crossing over.
Figure 1.—
Potential changes during the evolution of sex-determining regions from different possible starting states and possible changes in their subsequent evolution. In the evolution of dioecy from hermaphroditism, females are assumed to evolve first through a recessive male-sterility mutation. Starting from ESD, females might arise by loss of the ability to respond to an environmental trigger for male development. Males in either pathway then arise by a linked mutation causing failure to develop female structures, resulting in proto-sex chromosomes. Before recombination suppresssion has become extensive and genetic degeneration of one sex chromosome has occurred, the linkage group associated with sex determination can change by the replacement of a new sex-determining gene or via the transpositation of the existing sex-determining region to a new autosome. Translocations between the sex chromosomes and autosomes may also occur, and may involve either proto-sex chromosomes or degenerated ones.
TABLE 1.
Glossary of terms
| Term | Definition |
|---|---|
| Dioecy | plant species with separate male and female individuals (see gonochorism in animals, below). |
| Environmental sex determination (ESD) | sex-determining system where maleness or femaleness is induced by an environmental cue during development, such as the temperature control of sex in alligators and crocodiles. |
| Genetic sex determination (GSD) | the situation when alleles or alternative types of a particular linkage group (such as a Y or W chromosome) influence the probability of developing as a male or a female. |
| Genetic degeneration | the absence (or lack of function) of Y-linked alleles (in male-heterogametic systems), or W-linked ones (in female-heterogametic species) of many genes carried by the homologous sex chromosome. |
| Gonochorism | the term in animals referring to species with separate males and females (see dioecy in plants, above). |
| Hermaphroditism | in animals, species where individuals have both male and female gonads at the same time (simultaneous hermaphroditism), or an ovotestis, or where an individual may change sex (sequential hermaphroditism) during its life cycle. In plants, hermaphroditism denotes species whose flowers have both male and female parts. |
| Heterogamety | sex chromosome system in which males or females have heteromorphic sex chromosomes. In male heterogamety (including most mammals, Drosophila, and dioecious species of the plant Silene), males are XY and females XX, and in female heterogamety, including in birds and Lepidoptera, females are ZW, and males ZZ. Heterogamety is also sometimes used when the sex chromosomes are homomorphic (nonheteromorphic) to indicate which sex is heterozygous for the sex-determining region. |
| Monoecy | plant species with separate male and female flowers on the same individual plant. |
| Neo-sex chromosomes | systems where a sex chromosome has undergone a fusion or translocation with a formerly autosomal chromosome. |
| Proto-sex chromosomes | systems where a chromosome carries a newly arisen sex-determining gene or a newly evolved sex-determining region, but recombination suppression has not yet evolved and therefore there is no sex chromosome heteromorphism. |
| Sex chromosome heteromorphism | the degree to which the X and Y chromosomes (or Z and W chromosomes) differ from one another. This condition gives rise to either male or female heterogamety. |
| Subdioecy | presence of hermaphrodites as well as male and female individuals, found in some plant species populations. |
Given these varied possibilities, we use the term “sex-determining chromosome” for homomorphic situations, and reserve “sex chromosome” for situations when a large chromosome region is nonrecombining and there is a genetically degenerate Y or W chromosome. When reviewing plants, we follow convention and use the word “gender” to indicate maleness and femaleness of diploid individuals in species with separate sexes (dioecy) and also to haploid gametophytes of species where these produce only male or female gametes. For diploid plants, we also use the conventional terms “ovules and pollen” for the haploid stage. For gonochoristic animals, we use the word sex to refer to whether an individual is male or female, and refer to their gametes as sperm and eggs, respectively. There should be no confusion with the alternative meaning “sexual reproduction” (as opposed to asexual).
GENETIC MAPPING OF SEX-DETERMINING REGIONS
Table 2 summarizes plant and animal studies that have mapped the genomic region or regions associated with GSD. The term GSD is somewhat vague, and here we use it to mean that the alleles or alternative types of a particular linkage group (such as a Y or W chromosome) influence the probability of developing as a male or a female. Genetic control of sex need not be complete—in many organisms, environmental conditions, experimental treatments, or the genetic background, can affect the phenotypic sex of individuals. Our table therefore excludes species thought to have wholly environmental sex determination (ESD), but includes species where genetic influences have been demonstrated, even if environmental influences also affect or modify individuals' sex. The table also distinguishes between species with heteromorphic sex chromosomes (male or female heterogamety) and those where one sex is heterozygous for a sex-determining region, but without overt heteromorphism (in which case we specify whether females or males are the heterozygous sex).
TABLE 2.
Genetically mapped regions of sex determination in nonmodel plants and animals
| Species | Type of markers | Type of GSD | Comments |
|---|---|---|---|
| Plants | |||
| Non-flowering plants | |||
| Ceratodon purpureus (moss) | AFLPs (McDaniel et al. 2007) | Males heterozygous | |
| Marchantia polymorpha (liverwort) | Various (Okada et al. 2001) | Male heterogametic | |
| Dicotyledons | |||
| Rosid clade | |||
| Vitis (grape vine, Rhamnales) | SSRs, SSCPs, morphological traits (Marguerit et al. 2009) | Subdioeciousa | Three alleles for a single gene appears to confer maleness, femaleness, or dioecy |
| Carica papaya (papaya, Brassicales) | RAPDs, AFLPs, SSRs (Sondur et al. 1996; Ma et al. 2004; Chen et al. 2007a) | Male heterogametic | Male-determining Y |
| Basal Eudicots | |||
| Silene latifolia (Caryophyllales) | SSCPs, genic SNP markers (Guttman and Charlesworth 1998; Bergero et al. 2007) | Male heterogametic | Male-determining Y, at least three genes influencing male fertility present on Y chromosome |
| Spinicia oleracea (spinach, Caryophyllales) | AFLPs and SSRs (Khattak et al. 2006) | Males heterozygous | |
| Rumex species (sorrel and dock, Caryophyllales) | Cytogenetics only (Bartkowiak 1971; Singh and Smith 1971) | Male heterogametic (XY or XY1Y2) | Male-determining Y– or X–autosome balance (Navajas-Perez et al. 2005) |
| Salix viminalis, Populus (willow, poplar and aspen, Malpighiales) | AFLPs (Semerikov et al. 2003); SSRs (Yin et al. 2008) | Females heterozygous | |
| Cannabis sativa (Urticales) | AFLPs (Peil et al. 2003) | Male heterogametic | |
| Humulus lupulus (hops, Rosales) | RAPD (Polley et al. 1997); RAPDs, AFLPs, SSRs (Seefelder et al. 2000) | Male heterogametic | |
| Fragaria virginiana (wild strawberry, Rosales) | SSRs (Spigler et al. (2008) | Subdioecious a, females heterozygous | Two linked genes affecting sex determination |
| Asterid clade | |||
| Actinidia chinensis (kiwifruit, Ericales) | SSRs (Fraser et al. (2009) | Male heterozygous | |
| Monocotyledons | |||
| Asparagus officinalis (asparagus) | AFLPs (Reamon-Buttner et al. 1998;, Telgmann-Rauber et al. 2007) | Males heterozygous | Sex chromosomes may be related to Silene and Carica sex chromosomes (Telgmann-Rauber et al. 2007) |
| Dioscorea tokoro (wild yarm) | AFLPs (Terauchi and Kahl 1999) | Males heterozygous | |
| Animals | |||
| Invertebrates | |||
| Schistosoma mansoni (blood fluke) | SSRs (Criscione et al. 2009); FISH (Taguchi et al. 2007) | Female heterogametic | |
| Haliotis discus (Pacific abalone) | AFLPs, RAPDs, and SSRs (Liu et al. 2006) | Male heterogametic | |
| Insects | |||
| Anopheles gambiae (mosquito) | BACs (Krzywinski et al. 2004) | Male heterogametic | |
| Culex tarsalis (mosquito) | SSRs (Venkatesan et al. 2009) | Males heterozygous | |
| Apis mellifera (honey bee) | RAPDs (Hunt and Page 1994); SSRs and SNPs (Solignac et al. 2004) | Haplodiploid | Complementary sex determination |
| Bracon (wasp) | RAPDs (Holloway et al. 2000) | Haplodiploid | |
| Bombus terrestis (buff-tailed bumblebee) | RAPDs and microsatellites (Gadau et al. 2001) | Haplodiploid | |
| Mayetiola destructor (Hessian fly) | RAPDs (Formusoh et al. 1996) | Male heterogametic | Postzygotic chromosome elimination (Benatti et al. 2009) |
| Bombyx mori (silkworm) | RAPDs (Abe et al. 1998) | Female heterogametic | W chromosome is homologous to wild ancestor, B. mandarina. |
| Allenemobius socius (ground cricket) | RAPDs and allozymes (Chu and Howard 1998) | Male heterogametic | |
| Leptinotarsa decemlineata (Colorado potato beetle) | AFLPs and anonymous codominant markers (Hawthorne 2001) | Male heterogametic (X0) | |
| Crustaceans | |||
| Penaeus japonicus (kurua prawn) | AFLPs (Li et al. 2003) | Female heterogametic | |
| Litopenaeus vannamei (Pacific white shrimp) | AFLPs and SSRs (Zhang et al. 2007) | Female heterogametic | |
| Fish | |||
| Catfishes (Siluriformes) | |||
| Ictalurus punctatus (channel catfish) | SSRs (Waldbieser et al. 2001) | Males heterozygous | |
| Flatfishes (Pleuronectiforms) | |||
| Paralichthys olivaceus (Japanese flounder) | SSRs and AFLPs (Coimbra et al. 2003) | Male heterozygous | |
| Scophthalmus maximus (turbot) | SNPs (Martinez et al. 2009) | Male heterozygous | |
| Cynoglossus semilaevis (sole) | AFLP (Chen et al. 2007a); BAC screening (Shao et al. 2010) | Female heterogametic | |
| Pufferfish (Tetraodontiformes) | |||
| Takifugu rubripes (fugu) | Genetic markers (Kikuchi et al. 2007) | Males heterozygous | |
| Sticklebacks (Gasterosteiformes) | |||
| Pungitius pungitius (ninespine stickleback) | SSRs (Shapiro et al. 2009); SSRs and FISH (Ross et al. 2009) | Male heterogametic | Sex determination is in different location from other stickleback species. |
| Gasterosteus aculeatus | SSRs (Peichel et al. 2004) | Male heterogametic | G. wheatlandi males are X1X2Y. |
| (threespine stickleback), G. wheatlandi (black-spotted stickleback) | SSRs and FISH (Ross et al. 2009) | ||
| Apeltes quadracus (fourspine stickelback) | SSRs and FISH (Ross et al. 2009) | Females heterogametic | |
| Salmon (Salmoniformes) | |||
| Salvelinus alpinus (Arctic charr), S. namaycush (Atlantic salmon) | SSRs and AFLPs (Woram et al. 2003, 2004) | Males heterozygous | Y chromosome is not orthologous to other salmonids. |
| Salmo salar (Atlantic salmon), S. trutta (brown trout) | SSRs and AFLPs (Woram et al. 2003); SSRs and allozymes (Gharbi et al. 2006) | Males heterozygous | Y chromosome is not orthologous to other salmonids. |
| Oncorhynchus mykiss, O. kisutch, O. clarki | SSRs and AFLPs (McClelland and Naish 2008; Woram et al. 2003); various (Nichols et al. 2003); SSRs and FISH (Alfaqih et al. 2008; Rexroad et al. 2008) | Males heterozygous | |
| Cichlids (Perciformes) | |||
| Oreochromis karongae, O. aureus, O. tanganicae | SSRs and FISH (Cnaani and Kocher 2008; Cnaani et al. 2008) | Females heterogametic | Females heterozygous in some O. aureus populations (Lee et al. 2004) |
| Oreochromis niloticus (Nile tilapia) | SSRs and FISH (Cnaani et al. (2008) | Male heterozygous | Only in some populations (Lee et al. 2003) |
| Tilapia mariae (spotted tilapia) | SSRs and FISH (Cnaani et al. 2008) | Female heterogametic | |
| Tilapia zillii (red-bellied tilapia) | SSRs and FISH (Cnaani et al. 2008) | Male heterozygous | |
| Aulonocara baenschi, Metriaclima barlowi, M. phaeos, M. zebra, Pseudotropheus polit | SSRs (Ser et al. 2010) | Males heterozygous | Heterozygosity is associated with Lake Malawi cichlid linkage group 7 |
| Labeotropheus trewavasae, Metriaclima callainos, M. fainzilberi | SSRs (Ser et al. 2010) | Females heterozygous | Heterozygosity is associated with Lake Malawi cichlid linkage group 5 |
| Metriaclima pyrsonatus | SSRs (Ser et al. 2010) | Both male and female heterozygosity | Both Lake Malawi cichlid linkage groups 5 and 7 present in populations |
| Rice fish (Beloniformes) | |||
| Oryzias latipes | Chromosome walking, FISH (Matsuda et al. 2002); SSRs (Kimura et al. 2006) | Males heterozygous | Sex determined by DMY, a recent duplicate of DMRT1 |
| Oryzias javanicus, O. hubbsi | SNPs and FISH (Takehana et al. 2008) | Females heterogametic | O. javanicus W chromosome is not homologous to O. hubbsi |
| Oryzias luzonensis, O. dancena, O. minutillus | SNPs and FISH (Takehana et al. 2007; Nagai et al. 2008) | Males heterozygous | Y chromosomes of O. luzonensis, O. dancena, and O. minutillus are not homologous |
| Killifish and livebearers (Cyprinodontiformes) | |||
| Nothobranchius furzeri (African killifish) | SSRs (Valenzano et al. 2009) | Males heterozygous | |
| Poecilia reticulata (guppy) | SNPs (Tripathi et al. 2009a) | Male heterogametic | |
| Xiphophorus | Allozymes and morphological markers (Morizot et al. 1991) | Male heterogametic | W chromosome present in some X. maculatus populations (Kallman 1984) |
| Amphibians | |||
| Hyla arborea (European tree frog) | SSRs (Berset-Brändli et al. 2006) | Male heterozygous | Males are achiasmate |
| Ambystoma mexicanus (tiger salamander) | SNPs (Smith and Voss 2009) | Female heterozygous | |
| Rana rugosa (Japanese frog) | Phylogenetic inference from sex-linked gene (Miura et al. 1998) and FISH (Uno et al. 2008) | Female and male heterogametic populations | |
| Reptiles | |||
| Dragon lizards (Agamidae) | |||
| Pogona vitticeps, P. barbata | FISH (Ezaz et al. 2005) chromosome walking (Quinn et al. 2010) | Female heterogametic | Z and W are microchromosomes |
| Amphibolurus nobbi | FISH (Ezaz et al. 2009) | Female heterogametic | ZW sex chromosomes probably orthologous to those in Pogona |
| Ctenophorus fordi | FISH (Ezaz et al. 2009) | Female heterogametic | ZW sex chromosomes are not orthologous to those in Pogona and Amphibolurus |
Heteromorphic sex chromosomes are termed either male or female heterogamety (XY and ZW systems, respectively), and sex-determining chromosomes without marked heteromorphism are termed male or female heterozygosity. AFLP, amplified fragment length polymorphism; BAC, bacterial artificial chromosome; FISH, florescent in situ hybridization; RFLP, random amplified length polymorphism; RAPD, random amplified length polymorphism; SNP, single nucleotide polymorphism; SSCP, single strand conformation polymorphism; SSR, simple sequence repeat, also known as microsatellites.
Hermaphrodites present as well as males and females.
In plants, “sex determination” is sometimes used to mean control of the floral organs involved in pollen and ovule production, as well as the developmental genetics of sterility mutants, such as gynoecious melons, without normal male functions (Martin et al. 2009). Here, however, we maintain the usage that prevails in animal genetics and use this term only for genes affecting whether the individual plant develops as a male or a female. We will therefore not consider cytoplasmic male sterility in plants (e.g., Fishman and Willis 2006; Chase 2007). We also exclude fungal mating-type genes (Fraser et al. 2004), because these species are not dioecious and have no gamete dimorphism resembling sperm and eggs. For the same reason, we excluded mating-type regions of isogamous algae (Ferris et al. 2002).
Most of the mapping studies are based on linkage analysis of genetic markers typed in families, but FISH studies have also been very valuable, and genome sequencing is likely to become increasingly important. Most mapping so far has not identified the sex-determining genes (these are still known in only a few well-studied species), but has located the region(s) controlling sex determination (Holloway et al. 2000; Hawthorne 2001; Lee et al. 2004; Solignac et al. 2004; Telgmann-Rauber et al. 2007; Spigler et al. 2008).
Finally, recombination rates can be estimated separately in the male and the female parents of families (Hedrick 2007; Mank 2009b). In some species, recombination is absent, or nearly absent, throughout the genome in the heterogametic sex (Huxley 1928; Berset-Brändli et al. 2008). More often, recombination is suppressed in one sex in limited genomic regions (Naruse et al. 2000; Chen et al. 2007b; Yin et al. 2008), including the familiar situations in organisms such as mammals with no crossing over across most of the X and Y chromosomes in males. Recombination suppression in the region surrounding the sex-determining locus is a first step in the evolution of heteromorphic sex chromosomes (Figure 1).
Sex determination in plants and genetic mapping of sex-determining loci:
We review data from plants first, because many dioecious plants probably evolved recently from hermaphrodite ancestors, and this recent origin is likely to underlie some major differences in the genetics of sex determination between plants vs. those animals with much older sex chromosomes, such as mammals and birds. In plants, as in animals, hermaphroditism or environmental sex determination (e.g., Talamali et al. 2003) are possible ancestral states from which dioecy could have evolved. Dioecy in plants may also evolve from monoecy, where separate male and female flowers exist on the same individual (Renner and Ricklefs 1995; Dorken and Barrett 2004). In any of these ancestral states, all the genetic information required for the production of both anthers and pistils is present in each individual. The evolution of GSD must therefore involve mutations causing male and female sterility.
Dioecy in plants probably often evolved from hermaphroditism or monoecy by an initial recessive male-sterility mutation (which guaranteed outcrossing and resulted in a polymorphic population with females and hermaphrodites, see Figure 1), followed by one or more mutations increasing male fertility, creating males (or male-biased hermaphrodites) but reducing female functions (Charlesworth and Charlesworth 1978). When these later male-defining mutations occur in a linked region on the homologous chromosome to that containing the male-sterility allele, this creates a proto-Y chromosome (Westergaard 1958; Charlesworth and Charlesworth 1978). Because the initial male-sterility mutation is often recessive (homozygous individuals develop as females), the model predicts that females will be the homozygous sex and that the later female-suppressing mutations must be at least partially dominant. This model therefore predicts a predominance of male heterogamety in recently evolved dioecious species (Charlesworth and Charlesworth 1978), suggesting that female heterogamety may derive from male heterogametic ancestors.
Comprehensive reviews of dioecious plants indeed find that males are mostly the heterozygous sex (Westergaard 1958; Bull 1983). Among the best studied dioecious taxa, male heterogamety has evolved twice in the genus Silene (Mrackova et al. 2008), twice more in the Hawaiian genus Schiedia in the same family, Caryophyllaceae (Sakai et al. 2006), and twice yet again in the genus Rumex in another family (Navajas-Perez et al. 2005). In all these cases, dioecy probably evolved within the past 10–15 million years (dioecy in Silene is estimated to have evolved more recently than in Rumex, and dioecious Schiedia evolved more recently still).
Female heterogamety has been demonstrated in the genus Cotula and other members of the daisy family (Lloyd 1975; Bull 1983), and in Populus (Yin et al. 2008). In these taxa, dioecy may be older (Populus is in the family Salicaceae, whose members are almost all dioecious, suggesting a considerable age for dioecy), and male heterogamety could exist in species not yet studied. In the subdioecious plant, Fragaria virginiana (with hermaphrodites as well as males and females) females are heterozygous. However, males and females appear to have evolved very recently; hermaphrodites are also present, and recombinants occur between the sex-determining genes (Spigler et al. 2008). This may be an unexpected instance of de novo evolution of female heterogamety, and fully dioecious Fragaria species, if they exist, may have evolved recombination suppression in the sex-determining region.
In several plants with male-heterozygous systems, suppressed recombination is limited to small regions of the sex-determining chromosomes; sometimes, no markers fully linked to the sex-determining region have been discovered, even with dense mapping (Table 2). Possibly these species evolved dioecy recently, and there has simply not been enough time to evolve suppressed recombination across the entire sex-determining chromosome pair. Alternatively, evolution of suppressed recombination across most of the chromosome pair may not be inevitable (see our later discussion of the evolution of recombination suppression below).
The evolution of dioecy has been dated in only a handful of plant taxa, leaving it currently unclear whether all plant sex-determining chromosomes without large nonrecombining regions are young, and systems with heteromorphism older; there are no current mapping results for old angiosperm sex chromosomes. Phylogenetic studies suggest that dioecy evolved in the common ancestor of the families Begoniaceae, Cucurbitaceae, Datiscaceae, and Tetrameleaceae, probably from hermaphroditism (Zhang et al. 2006), and it has probably been lost many times in the first two large families (Mabberley 1997). This suggests considerable age, consistent with known XY heteromorphic sex chromosomes in dioecious Cucurbitaceae (Sinha et al. 2007). By mapping genes to these chromosomes, it may be possible to estimate the chromosomes' age using sequence divergence between X–Y ortholog pairs and to determine whether the same genomic region is involved in sex determination in all these taxa.
The ages of more plant sex chromosomes should be estimated, including those of species with undifferentiated potentially proto-X–Y pairs, such as papaya and the moss Ceratodon purpureus (see Table 2). It will therefore be important to sequence genes in the nonrecombining regions. In papaya, many sex-linked nongenic markers (mostly AFLPs) were found in a physically small nonrecombining sex chromosome region (Liu et al. 2004), which suggests high divergence of the male-specific region. However, the four papaya X–Y gene pairs so far studied all have synonymous site divergence <10%, suggesting that recombination stopped quite recently in this part of the chromosome pair (Yu et al. 2008). Genes from other locations in the papaya nonrecombining region are, however, needed to test whether the entire male-specific region evolved suppressed recombination recently.
In many plants, the genetic basis remains unclear, including stinging nettles Urtica dioica (Glawe and Jong 2009), Byronia dioica (Correns 1928), and Antennaria dioica (von Ubisch 1936), but males are again probably the heterozygous sex and females homozygous (reviewed in Westergaard 1958). These cases may involve recently evolved dioecy, breakdown of dioecy, or plants whose gender is subject to environmental influences. More phylogenetic and genetic studies (including using genetic markers) should clarify these situations.
Despite their fairly recent origins, some modifications of XY systems have occurred in several plants. XY1Y2 neo-sex chromosome systems have arisen in Humulus japonicus (Kihara and Hirayoshi 1932; Kim et al. 2008), and Silene diclinis (Howell et al. 2009), and twice independently in Rumex (Smith 1963, 1969; Navajas-Perez et al. 2005), and Viscum fischeri has a multiple Y system (Barlow and Wiens 1976). These situations probably evolved by X–autosome translocations (Figure 1; changing the ancestrally autosomal element into a chromosome that segregates from the new arm of the X, denoted by Y2, while Y1 denotes the ancestral Y), although fission of the Y cannot yet be ruled out in Rumex; genetic maps using genic markers might be able to establish whether the Y chromosomes still carry functional genes (as in Silene; see for instance Marais et al. 2008), in which case their gene content could be compared and this possibility tested.
Within the genus Rumex, an X–autosome balance sex-determining mechanism has also evolved, apparently from a male-determining Y mechanism retained in most species (Navajas-Perez et al. 2005). It is less clear whether the few known plants with female heterogamety (ZW systems) evolved from XY systems, vs. de novo evolution of female heterogamety (see above). It will be interesting to discover whether taxa with known ZW dioecious plants, have relatives with male heterogamety. If so, phylogenetic studies like those in Silene (Desfeux et al. 1996) and Rumex (Navajas-Perez et al. 2005) might reveal transitions between XY and ZW sex chromosomal systems.
Overall, the results from plants indicate that dioecy has evolved many times, sometimes quite recently, through major loss-of-function mutations producing unisexual females and males, usually with a male-determining “Y” chromosome. Plant sex-determining regions are probably often small, consistent with viability of homozygotes for the Y chromosome in some species (Westergaard 1958). However, few species have been genetically mapped, and the physical sizes of the regions are mostly unknown, because finding multiple sex-linked markers can imply either a large physical size or a large divergence time. More phylogenetic studies are needed to determine ancestral vs. derived states and to estimate the ages of more sex-determining chromosomes.
Genetic mapping of sex-determining loci in animals:
Sex chromosome evolution in animals has mainly been studied using therian mammals (Potrzebowski et al. 2008), Drosophila (Koerich et al. 2008), and birds (Mank and Ellegren 2007). Within each of these taxa, GSD arose just once, long ago (Matsubara et al. 2006; Veyrunes et al. 2008). Despite originating separately, recent linkage maps of the therian mammal X and bird Z chromosomes have revealed that recombination between the sex chromosomes was initially suppressed over a limited region of the chromosome pair, and later, in both lineages, extended across most of the recombining “pseudoautosomal” genome regions, resulting in “evolutionary strata” (Lahn and Page 1999; Skaletsky et al. 2003; Ross et al. 2005; Nam and Ellegren 2008; see Figure 2). Similar strata are found in the plant S. latifolia (Bergero et al. 2007). In birds, heteromorphism between the Z and W varies greatly (Mank and Ellegren 2007); the few ratite birds so far studied have only slight heteromorphism, implying a small region of restricted recombination (Tsuda et al. 2007), whereas, in more recently derived bird lineages (Neognathae—all modern birds except ratites and tinamous), the W chromosome is much smaller than the Z (Solari 1993). In situ hybridization studies show that the nonrecombining part of the W chromosome in Neognathae has lost most genes that were present on the ancestral autosome (Nishida-Umehara et al. 2007; Nanda et al. 2008). Similar results have been found in snakes (Matsubara et al. 2006).
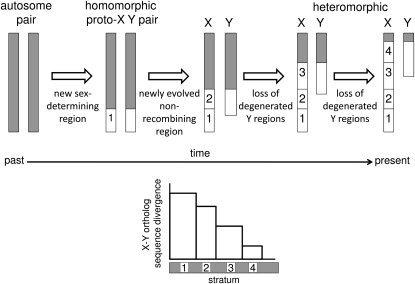
Figure 2.—
Evolutionary strata are regions where recombination ceased at different times during sex chromosome evolution, as recombination suppression (open bars) spreads gradually over the length of the sex chromosomes. Regions where recombination ceased first have greater sequence divergence between X–Y or Z–W orthologous pairs of genes than regions where recombination ceased recently. In this illustration of an X–Y pair, sex-determining genes evolved in the initially recombining region labeled 1, and recombination ceased first in this region, creating stratum 1. This led to genetic degeneration of this stratum in the Y chromosome, and the evolution of chromosome heteromorphism. The process was repeated when recombination ceased in stratum 2, then stratum 3. The cessation of recombination in stratum 4 leaves only a small “pseudoautosomal region” on the X and Y chromosomes where recombination still occurs.
Compared with other animal clades, the sex chromosomes of mammals and birds are highly conserved, apart from addition of some repeated copies of testis-expressed genes to the Z and X chromosomes (Mueller et al. 2008; Ellott et al. 2010). Most therian mammals have XY GSD, and the X has changed very little since the addition, ∼166 million years ago, of the q arm of the current placental X to the ancestral therian X, which remains the marsupial X (Waters et al. 2001). The gene content and gene order of the therian X chromosome have changed little (Murphy et al. 1999; Raudsepp et al. 2004; Delgado et al. 2009), apart from duplications of genes onto the X and movements of some genes off the X (Potrzebowski et al. 2009), and formation of neo-sex chromosomes in some species (reviewed in Bakloushinskaya 2009). The avian Z chromosome gene content and order are even more conserved (Dawson et al. 2007; Backström et al. 2008). As the existence of strata of different divergence levels between the sex chromosomes is the main new addition to our knowledge about the evolution of these chromosomes in mammals and birds to have emerged from genetic maps, and vertebrate sex determination was recently reviewed by Graves and Peichel (2010), the remainder of this review concentrates on recent genetic results on the genetic control of sex in nonmodel invertebrates and from fish, amphibians, and reptiles.
Invertebrates:
GSD is found in many invertebrate taxa, and male heterogamety is widespread, although female heterogamety is documented in schistosomes (Platyhelminthes) (Taguchi et al. 2007; Criscione et al. 2009), the lepidoptera (Abe et al. 1998), and some crustaceans (Li et al. 2003; Zhang et al. 2007). Many species have X0 systems, including some nematodes and insects, and these most likely evolved from XY GSD (Shearman 2002), after genetic degeneration of the Y chromosome and movement of genes essential for male functions to other chromosomes. Such gene movement has been documented in Drosophila pseudoobscura (Carvalho and Clark 2005; Koerich et al. 2008; Larracuente et al. 2010). X0 species, such as C. elegans (Gladden et al. 2007), must also have a dosage-sensitive sex determination system, where the different amounts of one or more X chromosome transcripts between the sexes provide a “counting mechanism” relative to autosomal elements. It will be interesting to identify sex-determining mechanisms in other nematodes, and study their sex chromosomes, which may have undergone neo-sex chromosome formation and other changes (Streit 2008).
In insects, chromosome fusions and translocations, involving both the X (McAllister and Charlesworth 1999) and the Y (MacKnight 1939; Bachtrog and Charlesworth 2002) have occurred many times, creating nonrecombining neo-sex chromosome systems in species such as Drosophila, where recombination does not occur in the heterogametic sex. Neo-sex chromosomes have also evolved by fusions in X0 systems of grasshoppers (White 1973). Unlike Drosophila, many such species have crossing over in both sexes, and the added arm can continue to recombine, though rearrangements probably often decrease the chiasma frequency (reviewed in Marti and Bidau 1995; Charlesworth and Wall 1998). Some termite species have multiple translocations involving the sex chromosomes, with chiasmata largely in terminal regions (Luycx 1981). It is unclear whether such rearrangements were selected because it is advantageous to move genes to the sex chromosomes, and estimates of the relative rates of such events for both sex chromosomes and autosomes have not yet established whether the sex chromosomes are disproportionately involved in translocations (Charlesworth et al. 1987).
The prevalence of male-heterogametic GSD suggests that it was probably the ancestral state when separate sexes were first established in these insects. However, the long evolutionary time since the insect ancestor makes inferring the ancestral state difficult. Even when two taxa both have XY systems, they may not have evolved from a single ancestral XY system—a new GSD system could have re-evolved in a lineage after being lost. For instance, the mosquito Anopheles gambiae resembles Drosophila in having a large region in which the X and Y do not recombine. However, these species diverged ∼250 MYA, and genome sequencing shows that the ancestral Drosophila X (the chromosome arm homologous to the D. melanogaster X, or the pre-fusion portion of the D. pseudoobscura X), carries few genes present on the X chromosome of A. gambiae, and, conversely, most A. gambiae X chromosome genes are scattered on various Drosophila autosomes (Zdobnov et al. 2002). Even among mosquitoes, the sex-determining genes may have been replaced. Aedes mosquitoes (like other Culicinae) have no sex chromosome heteromorphism, and so far no nonrecombining region has been found in the Culex tarsalis sex-determining chromosome (Presgraves and Orr 1998; Venkatesan et al. 2009). However, one arm of the A. aegypti sex-determining chromosome is probably homologous with the D. melanogaster X (Nene et al. 2007).
In several Dipteran insects, sex-determining genes are known to move their chromosomal locations without major chromosomal rearrangements. Some housefly populations have an XY system, and it is unknown whether the different genomic locations of sex-determining regions in other populations represent gene movements or replacements by new sex-determining genes (reviewed in Hediger et al. 2010). In Megaselia scalaris, there are no heteromorphic sex chromosomes (Traut et al. 1999), and a male-determining factor may have moved around the genome within a transposable element (Traut and Willhoeft 1990). This species may allow tests of whether a nonrecombining sex chromosome-like region has evolved from a single-gene system, but it is currently unclear whether there is indeed an associated nonrecombining region (Traut and Wollert 1998). Information about the existence of a nonrecombining region should emerge from new approaches for obtaining genetic markers and from genome sequencing (Rasmussen and Noor 2009).
Although some of these changes could be due to movements of existing genes, sex-determining systems and genes differ among insects. Replacement of a sex-determining gene by another gene, on the same or a different chromosome, may be involved in the transitions between XY and ZW systems (for example, in the hessian fly, Mayetiola destructor, a new sex-determining gene competing with the ancestral system of male heterogamety seems to be present on a neo-W chromosome; Benatti et al. 2009), or from a male-determining Y to an X–autosome balance or dosage-sensitive system. If sex-determining genes can be identified, it should become possible to distinguish such new sex-determining genes from movements of existing sex-determining genes.
Replacement of the sex-determining genes by a new gene controlling maleness or femaleness in insects often leaves the downstream parts of the pathway largely unchanged (Wilkins 1995; Graham et al. 2003; Pane et al. 2005; Siegal and Baker 2005; Cho et al. 2007). An example is the major change in the mechanism of sex determination represented by the haplodiploid system in Hymenopteran insects (reviewed by Bull 1983), where females lay either fertilized eggs that generally develop as females, or unfertilized ones that develop as haploid males.
Even among different Hymenoptera, the systems differ. In the wasp Nasonia vitripennis, sex is determined through imprinting of the N. vitripennis transformer (Nvtra) locus (transformer is a gene already known to act in the Drosophila sex-determining pathway; see Marin and Baker 1998). The maternal Nvtra copy is silenced in developing embryos, and without transcription from this locus, individuals develop as male. Unfertilized eggs (with only a maternal Nvtra copy) therefore develop as males, while fertilized eggs (expressing the paternal copy) develop as females (Verhulst et al. 2010). In bees, however, the signal to switch the developmental system to male or female involves “complementary sex determination” (csd), with a polymorphic locus, such that most diploid zygotes are heterozygous, and possession of different alleles serves as a signal that controls female development, while haploid eggs (or homozygous ones produced by inbred pairings) develop as males.
Even among different bees, one sex-determining gene has been replaced by another; in different species, the functional csd gene differs (confusingly, since the notation csd is used for the different genes). The gene with the csd function in honeybees (Apis mellifera) was found to be a duplicated and changed form of a preexisting gene, fem. The fem gene is present in many non-Apis Hymenopteran species, including a bumblebee (Bombus), a stingless bee, Melipona, and even N. vitripennis, and is involved in a downstream step of sex determination, but not in the csd function (Hasselmann et al. 2008a). fem apparently controls the female–male switch through expression of alternative RNA transcripts in a system reminiscent of Drosophila sex determination (see Marin and Baker 1998; Siegal and Baker 2005). However, in honeybees and their close relatives A. cerana and A. dorsata, which diverged in the last 10 million years, fem is duplicated. The duplicate gene has taken over the csd function from an unknown gene that presumably had this function in an ancestor, while fem maintains the female developmental fate (Hasselmann et al. 2008a,b; Gempe et al. 2009).
Fish:
The fish ancestor is thought to have had separate males and females (Smith 1975), as simultaneous hermaphroditism is rare in the clade (Devlin and Nagahama 2002) and confined to the tips of the fish phylogeny, suggesting recent origins (Mank et al. 2006). However, determining whether the fish ancestor had GSD or ESD is difficult, because sex-determining mechanisms are known in only a small fraction of fish, and sex determination has undergone many changes (Figure 3 and reviewed in Volff et al. 2007), and the phylogeny of many higher-level fish taxa is unresolved (Mank et al. 2006; Setiamarga et al. 2009). In some clades, dioecy may have evolved from sequential hermaphroditism, which is widespread in fish, including the closest relatives of the androdioecious goby, Lythrypnus dalli (Drilling and Grober 2005), which could be in transition to or from dioecy.
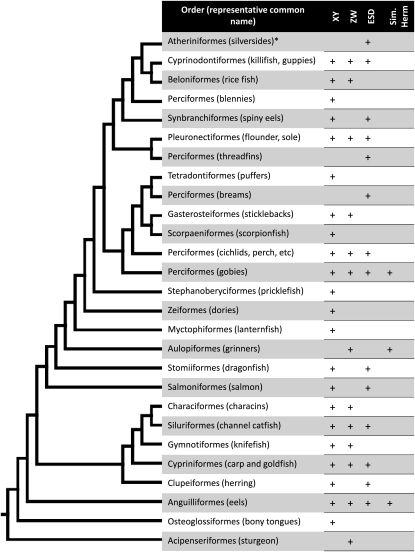
Figure 3.—
Sex determination in the actinopterygiian fishes (adapted from Mank et al. 2005, 2006). Shown is the supertree topology for the clade, presenting only the orders where sex determination is known for at least some members. Divergence dates of teleost clades are still contested and are therefore not indicated in the figure; however, it is helpful to note several nodes dated with a recently estimated molecular clock (Setiamarga et al. 2009). These include the origins of the Tetraodontiformes (80 MYA), the Beloniformes (115 MYA), the Salmoniformes (135 MYA), the Cypriniformes (190 MYA), and the Anguilliformes (220 MYA) and the divergence of the Gasterosteiformes from the Scorpaeniformes (150 MYA). Under environmental sex determination (ESD), we include sequential hermaphrodites where size plays a role in determining sex, as well as gonochorist species, where the environment can influence sex determination during development, and taxa with sequential hermaphrodites are noted separately (sim. herm.). Male heterogametic species are denoted by XY, even when there is no known chromosome heteromorphism, and female heterogametic species are similarly denoted by ZW. Sex determination in silversides has been shown to have a genetic component, although it is not known whether males or females are the heterozygous sex (Conover and Kynard 1981).
Thus, although fish are not ideal organisms for studying the initial evolution of separate sexes, several taxa are excellent for studying modifications of GSD (Mank et al. 2006), and some may allow studies of the evolution of GSD from initial states other than simultaneous hermaphroditism. A particularly interesting situation is when there is single-gene control after a change in the sex-determining system. Although sex chromosome heteromorphism is known in some fish (see Table 2), many species have homomorphic sex-determining chromosomes and could have single sex-determining genes (and should, strictly, not be called XY or ZW systems). It is currently unclear whether such new sex-determining genes can trigger the evolution of a nonrecombining sex chromosome-like region, representing a different origin for sex chromosomes from that in plants. If cases of young and heteromorphic sex chromosomes are found, they will be ideal for testing what has driven recombination suppression.
Many fish have large clutch sizes that facilitate linkage mapping, and genetic mapping in fish has revealed several new cases of GSD. For instance, genetic mapping in catfish (order Siluriformes) found a sex-determining chromosome with a small nonrecombining region, which is heterozygous in males (Waldbieser et al. 2001). In the hirame, a flounder (order Pleuronectiformes), males are heterozygous (Coimbra et al. 2003), but temperature also influences sex development (Yamamoto 1999). Another pleuronectiform, the half-smooth tongued sole, has female heterogamety, and many genes present only in females have been identified through a BAC library screen, suggesting W linkage (Shao et al. 2010). Among fish whose genomes have been sequenced, the pufferfish (Takifugu rubripes, order Tetraodontiformes) has a male-heterozygous region (Kikuchi et al. 2007). In the zebrafish (Danio rerio, Cypriniformes), sex is determined by a combination of genetic and environmental influences (Siegfried and Nusslein-Volhard 2008), while among its relatives, the goldfish has an XY system of GSD (Zhang and Wu 1985) and Leporinus (Characiformes) has a strongly heteromorphic ZW system, with largely heterochromatic W chromosomes, which have probably undergone W–autosome translocations (Parise-Maltempi et al. 2007).
Many of these findings are from distantly related isolated species, among which it is impossible to recognize large syntenic blocks or, until the sex-determining loci are identified, determine gene homology. Comparative mapping in fish is further complicated by the fact that several whole genome duplication events have occurred within the clade (Meyer and Schartl 1999; Christoffels et al. 2004; Mank and Avise 2006). However, five clades have provided information about changes in sex-determining loci and could potentially lead to estimates of turnover rates.
In sticklebacks, genetic mapping has been important for evolutionary studies of adaptation and has led to the development of a panel of molecular genetic markers (Peichel et al. 2001; Shapiro et al. 2009). Their use in different species in the family Gasterosteidae has revealed male heterogametic GSD in both Gasterosteus aculeatus and Pungitius pungitius. Both have sex chromosome heteromorphism, suggesting a large nonrecombining region (Ross and Peichel 2008; Shapiro et al. 2009). The X1X2Y GSD system in G. wheatlandi (the sister species to G. aculeatus) is probably derived by a Y–autosome fusion involving an XY pair in a common ancestor (Ross et al. 2009); surprisingly, this fused the linkage groups corresponding to the X chromosomes in G. aculeatus (LG19) and the more distant relative, P. pungitius (LG12). It is uncertain whether the ancestor of G. aculeatus and P. pungitius had an XY system (implying replacement or movement of the male-determining gene in one species) or whether one of these lineages evolved this independently from a different ancestral state. A more distant relative, Apeltes quadracus, has female heterogamety with visible chromosome heteromorphism. In Culaea inconstans (sister to P. pungitius), there is no evidence of heteromorphic sex chromosomes or GSD, although markers linked to a sex-determining gene may be found once more markers and chromosomes are mapped in this species (Ross et al. 2009).
The salmon family are somewhat similar. Early cytological studies showed clearly differentiated male heterogametic sex chromosomes in several species (Thorgaard 1977), and later genetic mapping in Oncorhynchus, Salvelinus, and Salmo confirmed male heterozygous GSD in most species, though with only slightly differentiated sex chromosomes and some environmental influence (Nichols et al. 2003; Woram et al. 2004; Gharbi et al. 2006; Alfaqih et al. 2008). However, like the sticklebacks, the sex-determining chromosomes in different species do not have the same gene content and probably did not evolve from a common single ancestral XY pair. Few genes are currently mapped, so the chromosomal homologies in the different species, and the evolutionary history of the changes, are not yet certain. Genetic mapping with microsatellite loci is starting to indicate some chromosomal homologies, even between distantly related fish (Rexroad et al. 2008). However, these homologies have not yet been shown to extend to entire chromosomes having the same gene content in distant relatives, and fish chromosomes appear to have been extensively rearranged (Naruse et al. 2004) and it is unlikely that entire arms will be recognizable as they are in Drosophila and Hymenoptera. The X and Y chromosomes may have evolved de novo four or more times from different autosomes within the Salmonid family, or a sex-determining region may have moved to an autosome several times, creating a new, homomorphic sex-determining chromosome. It is also possible that a new sex-determining gene may have replaced a preexisting one in some species, and heteromorphism evolved later in some cases. A final alternative is that the recent whole genome duplication in this family created duplicate sex-determining genes, and different copies were lost in different lineages. Finer mapping could reveal syntenies and exclude or confirm these scenarios.
The medaka, Oryzias latipes (Cyprinodontiformes) is a clear case of a new sex-determining gene replacing an unknown preexisting one. Linkage mapping (Naruse et al. 2000) identified a sex-determining chromosome pair (9) containing a small (258 kb) region specific to males (Kondo et al. 2006). In this region, males are heterozygous, and there is no homology (and no recombination) with the homologous “X” chromosome (Kondo et al. 2006). Further genetic studies identified a duplicate of the DMRT1 gene in this region (Matsuda et al. 2002; Nanda et al. 2002). This duplicate “DMY” gene has superseded an ancestral gene previously controlling the maleness development pathway, which is presumably still involved in some congeneric rice fishes (Matsuda et al. 2002). In the region with homology between the X and Y that surrounds the sex-determining locus, recombination in males is lower than in females. Interestingly, when heterozygotes for the male-specific region are given a sex reversal treatment, making them female, they recombine at a rate like that of “XX” females (Kondo et al. 2001). The reduced recombination in males is therefore not directly caused by the insertion, but rather, depends on the sex phenotype. Sex-specific recombination rates have not been published for other genome regions, so it is unclear whether there are genome-wide differences in recombination.
Although DMRT1 homologs are involved in male determination in many animals, including Drosophila, C. elegans, mammals (Raymond et al. 2000; Haag 2005), and birds (Smith et al. 1999, 2009), the DMY gene is missing in several Oryzias species (Kondo et al. 2004; Tanaka et al. 2007), although these species retain the ancestral DMRT1 locus. Additional linkage mapping indicates independent changes to female heterogametic GSD in O. javanicus and O. hubbsi (Takehana et al. 2008).
A particularly interesting group of fish for mapping studies includes the guppy, Poecilia reticulata and Xiphophorus, in the same superorder, Atherinomorpha, as medaka (Figure 3). Classical genetic work in both species found an XY GSD system and demonstrated that many male color patterns are caused by polymorphisms in genes fully or partially Y linked (Winge 1927; Kallman 1970). However, the platyfish, Xiphophorus maculatus, also has a “W” chromosome carrying a female-determining gene. Xiphophorus species may be changing between female and male heterogamety, and in X. maculatus, like the frog Rana ragosa (see below), the X and W chromosomes are at least partially homologous (reviewed in Schultheis et al. 2009). The male color polymorphisms may be maintained by sexual antagonism, being advantageous to males, but disadvantageous to females (Fisher 1931; Kallman 1970; Rice 1984). These species therefore hold promise for testing the hypothesis that sexually antagonistic genes have driven selection for reduced X–Y recombination (see Figure 1 and below). Large numbers of SNP markers have been obtained for genetic mapping in guppies, using EST sequences (Tripathi et al. 2009a,b), and new progress can now occur in comparing the maps of these species' sex chromosomes (Morizot et al. 1991, 2001). No marker completely linked to the sex-determining locus has yet been found in either species, but part of the guppy sex chromosome may recombine less frequently in males than females (Tripathi et al. 2009a). The guppy sex chromosome is homologous with the O. luzonensis sex chromosome (Tanaka et al. 2007) and with medaka autosome 12 (Tripathi et al. 2009b), raising the possibility that the guppy can help to identify the sex-determining region in the ancestor of the medaka. Recent estimates of the age of these taxa are large, suggesting that changes in sex determination may not be rapid.
In tilapia and cichlid species (family Cichlidae, in the ancient and polyphyletic order Perciformes), there are again species with sex-determining (nonheteromorphic) chromosomes called X, Y, Z, and W, plus environmental influences complicating genetic analyses (Baroiller et al. 2009; Ser et al. 2010). Tilapia are farmed for food, and males have higher growth rates than females in commercial production (Beardsmore et al. 2001), therefore a method to sex fingerlings would be valuable. However, changes in the sex-determining region may frustrate efforts to develop a single method to sex multiple species unless the same gene turns out to be involved across the clade.
Within the tilapiine cichlids (genus Oreochromis), use of genetic markers has shown that females are heterogametic in O. karongae (Cnaani et al. 2008) and O. tanganicae (Cnaani and Kocher 2008), while in O. niloticus males are the heterozygous sex (Cnaani et al. 2008), and O. aureus has a polygenic sex-determination system (Lee et al. 2004). In the closely related genus Tilapia, one species, T. zillii, has GSD with heterozygous males, and the sex-determining chromosome is homologous to that in O. niloticus. In T. mariae, females are heterogametic, and the sex-determining chromosome is the same as that in O. karongae (LG3), but different from that in the male heterozygous species, LG1 (Cnaani et al. 2008). The genetic maps of Lake Malawi cichlids are almost perfectly colinear with those of tilapia cichlids (Ser et al. 2010). In Lake Malawi cichlids, female heterogamety may have evolved recently from male heterozygosity (Roberts et al. 2009; Ser et al. 2010), and a similar transition may have occurred independently in Oreochromis (Cnaani et al. 2008). It will be necessary to study more sets of closely related species to infer the rates and directions of changes and to determine whether these changes are due to transpositions of sex-determining genes or to the origin of new sex-determining genes, coopting the sex-determination pathway.
Amphibians and reptiles:
Most amphibians exhibit GSD and possess homomorphic sex-determining chromosomes (Eggert 2004). There have been many shifts between male and female heterogamety (Hillis and Green 1990), including among different populations of the Japanese frog, R. ragosa (Miura et al. 1998), where genetic mapping has shown that the X and W chromosomes are homologous (Uno et al. 2008). It is not yet understood how the change from one state to the other occurred, but the male heterozygous (XY) state is probably ancestral (Ogata et al. 2008). Genetic mapping in salamanders (Smith and Voss 2009) found that females are the heterozygous sex, and the sex-determining locus is not in a chromosome region with reduced recombination. In the European tree frog, Hyla arborea, males (the heterozygous sex) are nearly achiasmate (Berset-Brändli et al. 2008). The current genetic map is based on noncoding DNA so it is not yet known whether the male-specific region has degenerated or experienced gene loss.
In reptiles, temperature-based ESD predominates and was probably the ancestral state from which ZW GSD has repeatedly evolved, with few reversals (Janzen and Phillips 2006; Organ and Janes 2008). These inferences remain tentative, as there is no definitive reptile phylogeny (Olmo 2005; Pokorna and Kratochvil 2009). In the Australian dragon lizards, FISH mapping combined with phylogenetic analysis has demonstrated that female heterogametic sex determination evolved at least twice independently from temperature-sensitive ESD (Ezaz et al. 2005, 2009). Separate sexes and GSD in birds probably evolved independently of the change to GSD in reptiles, since the reptile DMRT1 homolog is autosomal (Kawai et al. 2007).
In animals, the transition from ESD to GSD may be more common than the reverse, although changes from GSD to ESD may have occurred in nematodes (Haag 2005). It has been suggested that ESD may be an “evolutionary trap” (Janzen and Phillips 2006; Pokorna and Kratochvil 2009) or that GSD may accelerate speciation, leading to most lineages having GSD, even with similar rates of transition (Organ et al. 2009). However, these alternative hypotheses are difficult to differentiate from each other, and no clear mechanism is known either to prevent reversion to ESD or promote speciation.
LESSONS FROM GENETIC MAPPING OF SEX-DETERMINING LOCI
Male and female heterogamety:
In clades with multiple types of GSD, including reptiles, fish, and plants, males are generally more often heterogametic or heterozygous for the sex-determining region than females (Westergaard 1958; Janzen and Phillips 2006; Mank et al. 2006). However, as explained above, in animals it is often unclear whether sex determination evolved from a situation without GSD, or by movement of a preexisting sex-determining gene, or by creation of a new one. The relative frequencies of male and female heterogamety will depend on the rates of such changes.
It has been argued that male heterogamety may evolve more readily, because, in animals, femaleness is probably often the developmentally “default state,” and maleness requires a positively male-determining gene (Raymond et al. 2000). With male heterogamety, the male-limitation of Y chromosomes (or Y-like regions) allows the male-determining gene to be restricted to males. Thus, female heterogametic GSD, where no genomic region has male-limited inheritance, may make it more difficult to specify male development. Known mechanisms of female heterogametic GSD indeed appear to be complex, with dosage-sensitive genes involved in male sex determination located on the Z chromosome of birds (Smith et al. 2009) and the lizard Pogona vitticeps (Quinn et al. 2007), and even more complicated interactions of the Z and W chromosomes in Xenopus frogs (Yoshimoto et al. 2008). However, femaleness is not always the default: in zebrafish, the germ line is required for ovary development, and in its absence the gonad develops as a testis (Siegfried and Nusslein-Volhard 2008).
Moreover, this argument can apply only to species whose GSD has evolved from a species that already has sex chromosomes, not to the initial stages of the evolution of separate sexes. Table 3 summarizes the types of selection that might be expected following the evolution of sex-determining regions from different ancestral states. In the initial evolutionary stage, when GSD evolved from hermaphroditism or ESD, the genes involved would not be confined to one sex. When GSD evolves from hermaphroditism in animals or monoecy in plants (diagrammed in Figure 1), there is, of course, no default sex. If females evolve first (see above), this predicts that males will generally be the heterozygous sex, because loss-of-function mutations creating females are probably generally recessive, and females would be homozygous.
TABLE 3.
Selective consequences that might be predicted when sex-determining regions evolve from the different potential starting states shown in Figure 1
| Possible starting states |
||||
|---|---|---|---|---|
| Hermaphroditism (no preexisting sex-specific development) | Sex-specific developmental functions already established |
|||
| Predicted effects | Monoecy | Environmental sex determination | Preexisting GSD system | |
| Initial genes changed | Major sterility mutations (involving at least two sex-determining genes) | Probably often major sterility mutations (involving at least two sex-determining genes) | New male or female determiners or major sterility mutations | New sex-determining gene, involving either loss- or gain- of-function mutations |
| Later changes in sex development and other pathways | Many other genes evolve sex-specific functions | Probably few changes | Probably few changes | Possibly several changes if sexual development is due to new genes, which interact with other pathways |
| Recombination suppression | Yes, initially in region with the sex-determining genes | Yes, initially in region with the sex-determining genes | Yes, initially in region with the sex-determining genes | Initially no, unless linked sexually antagonistic alleles arise in the region with the sex-determining genes |
However, GSD in animals seems often to have evolved from ESD, rather than from a simultaneously hermaphroditic state. In species with ESD, femaleness could, of course, be the default state, with an environmental trigger inducing maleness. This could change to GSD by the promoter controlling the expression of a male-determining gene becoming constitutive instead of inducible (Valenzuela and Shikano 2007; Valenzuela 2008); males would then be determined either genetically (without the environmental trigger) or environmentally. Under this scenario, maleness would most likely be dominant (assuming dominance of mutations causing constitutive expression; Lemos et al. 2008), and thus females would again be the homozygous sex. Unless there is a permanent environmental change (to the female-determining environment), evolution of full GSD would require a second genetic change, abolishing the response to the environmental trigger in individuals not carrying the constitutive maleness mutation, to create females in all environments (see Figure 1). Alternatively, GSD could arise by loss-of-function mutations that abolished responses to environmental cues determining male or female development (assuming that development would then switch to the other sex). Either sex could then be the heterozygous sex. In either scenario, during the initial stages before a Y chromosome or Y-like region evolves, the factor determining maleness could recombine with the female-determining one, yielding sterile individuals, so selection would presumably favor linkage of the two genes.
Suppression of recombination and sex chromosome heteromorphism:
All the organisms in Table 2 possess GSD, yet many of them lack visibly dimorphic sex chromosomes. This might suggest that, in contrast to the old and highly conserved sex chromosomes in birds, therian mammals, and Drosophila, most animal GSD systems and some of plants are quite young, and the sex chromosome pair have not yet diverged. In many animals, young homomorphic sex chromosomes may indicate a recent change in the location of the sex-determining gene (see Figure 1). However, the degree of sex chromosome heteromorphism is not a good indicator of age. First, chromosome rearrangements can produce heteromorphism, as in rainbow trout (Phillips et al. 2009), and even newly evolved sex chromosome systems could be heteromorphic. Furthermore, neither suppressed recombination nor heteromorphism always evolve in old-established sex chromosomes. Indeed, the homomorphic sex chromosomes in ratite birds and some reptiles clearly carry ancient GSD genes. To date, few sequences are available for comparing genes on XY or ZW pairs to estimate the time since recombination between these alleles ceased and thus a minimum age of a sex chromosome pair.
Moreover, in species with developmental plasticity in sexual differentiation, and when recombination suppression between the sex chromosomes depends on the phenotypic, rather than genotypic sex, as described above in the medaka (Kondo et al. 2001), occasional sex-reversed individuals may allow recombination between the Z and W or X and Y chromosomes. Old-established sex chromosomes could thus remain undifferentiated, apart from the sex-determining genes (Perrin 2009). This may be most common in poikilothermic vertebrates, where GSD is often accompanied by environmental influences, especially temperature, as in many fish, such as salmon (Devlin and Nagahama 2002) and platyfish (MacIntyre 1961). This could explain the prevalence of largely homomorphic sex chromosomes in fish (Arkhipchuk 1995). More studies of recombination between sex chromosomes in sex-reversed individuals are needed and also more studies of recombination rates in the two sexes.
The role of sexual antagonism in recombination suppression and GSD turnover:
Comparative mapping in closely related species has provided new details about the diversity of GSD systems, and similar types of changes are observed in very different taxa, from plants to mammals, including fusions and translocations creating neo-sex chromosomes, gene replacements (sometimes changing the identity of the sex-determining chromosome), and transitions between male and female heterogamety. Sexual antagonism may be involved in the evolution of some of this diversity in GSD, as close linkage with a sexually antagonistic gene on an autosome can create an advantage for a new sex-determining factor (Van Doorn and Kirkpatrick 2007). Anecdotal evidence exists for turnover of sex determination due to sexual conflict in cichlid (Roberts et al. 2009) and stickleback fish (Kitano et al. 2009). The evidence for considerable sex differences in gene expression in animals (Kopp et al. 2003; Morrow et al. 2008; Mainguy et al. 2009; Mank 2009a) suggests that sexual antagonism is possible at many genes. However, several other models for changes in GSD exist (Bull 1983; Vuilleumier et al. 2007; Kozielska et al. 2010), and it is difficult to test which model best fits the observed patterns.
It has often been suggested that the evolution of suppressed recombination between sex chromosomes may be due to sexual antagonism. Once a sex-determining region has evolved, mutations that are advantageous in males but disadvantageous in females are particularly likely to invade a population if they arise at loci linked to the male-determining region (Fisher 1931; Rice 1984). They can then sometimes remain polymorphic, with allele frequencies differing in the two sexes, which favors reduced recombination with the sex-determining region (Bull 1983; Rice 1987; Patten and Haig 2009). Such selection evidently acts when separate sexes are newly evolving and mutations at separate loci have opposite effects on male and female fertility (Charlesworth and Charlesworth 1978). After these initial mutations have become completely linked and behave as a single genetic locus, or when there is a single-locus sex-determining system because a new gene has replaced a previous system, sexually antagonistic alleles should start to accumulate at linked loci. It is currently unknown whether any new sex-determining gene that has replaced an existing one has been followed by evolution of a new nonrecombining region, but, if sexual antagonism is indeed important for evolution of nonrecombining sex chromosome regions, new sex-determining regions such as the medaka DMY region should also eventually do so. However, movement of an existing sex-determining region, creating a new sex-determining chromosome from a former autosome, may sometimes directly produce a region of suppressed recombination because of the chromosome rearrangement involved (see above).
Systems with greater levels of sexual antagonism may be the most likely to undergo progressive recombination suppression, as there are simply more sexually antagonistic loci, or loci with potentially sexually antagonistic alleles, near the sex-determining region. Highly heteromorphic sex chromosomes might therefore be most common in species with high levels of sexual conflict, and with sexual dimorphism, a likely indicator of sexual antagonism (Badyaev 2002). The male color polymorphisms in guppies and platyfish, mentioned above, are probably sexually antagonistic (Fisher 1931), as are the female coloration patterns in Lake Malawi cichlids, so these species hold promise for testing whether sexually antagonistic genes select for reduced recombination between the sex chromosomes. The scarcity of heteromorphic sex chromosomes in plants might, for instance, reflect low sexual dimorphism (Ashman 2003, 2005). However, it is not yet clear whether plant sex chromosomes evolved long enough ago for evolution of heteromorphic sex chromosomes to be expected.
Conservation in sex determination:
We have emphasized changes, but it is well established that some of the downstream components of the sex determination pathway in animals are retained over long evolutionary times, even across lineages with both male- and female-heterogametic GSD, as well as ESD. DMRT1 is involved in determining maleness in many animals, ranging from invertebrates to mammals (Ferguson-Smith 2007), but it is not always on the sex chromosomes. DMRT1 maps to the Z chromosome in birds, a duplicate is located on the W chromosome in Xenopus, and the gene is important in sex determination in both clades (Yoshimoto et al. 2008; Smith and Voss 2009, but see Kuroiwa 2009 and Zhao et al. 2010), whereas, in most male heterogametic species, DMRT1 is autosomal (Ferguson-Smith 2007), and is a downstream component in sex determination. The medaka fish is a notable exception where a duplicate of DMRT1 has assumed control of male development, rather as the honeybee csd gene has done in the very different haplodiploid sex determination system of Hymenoptera. In other cases, the earliest-acting controls have been lost from the sex determination pathway. In some mammals, the Y-linked gene SRY is the upstream regulator of DMRT1. SRY was probably recently recruited to the sex determination pathway, as it is restricted to therians (Williams and Carroll 2009), but the SRY gene is entirely absent in voles (Just et al. 1995; Chen et al. 2008).
In dioecious plants, very little is known about the molecular mechanisms of sex determination. Except in quite closely related plants, it is not possible to identify large regions of homologous genome, making it difficult to test whether the same chromosome carries sex-determining genes in different dioecious plants. However, it is likely that the sex-determining genes vary among taxa. Even within the genus Silene, there have been two independent origins of male heterogametic systems (Mrackova et al. 2008). It is estimated that large numbers of genes in plants can produce mutations with male or female sterility (Ohnishi 1985; Johnson-Brousseau and McCormick 2004; Peiffer et al. 2008). Different genes may therefore have undergone different sterility mutations in different dioecious taxa. One motivation for genetically mapping sex-determining regions is to identify the genes involved. In species with small sex-determining regions or single gene control, this may be possible, allowing the independent origins of sex determination to be tested, but it will remain very difficult when there is a large nonrecombining region.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
The rapid acceleration in development of genetic markers for nonmodel organisms has already made possible species- and clade-specific mapping of GSD in a variety of interesting plants and animals. Comparative genetic mapping of GSD in closely related clades can reveal whether apparently similar types of GSD are indeed due to shared ancestry and have the same sex-determining region, or whether changes have occurred in the location, and perhaps the identity, of the genes controlling sex determination. This can provide information about the directions of the changes that have occurred, a necessary preliminary to asking about the selective mechanisms of changes in sex determination and about how elements of the sex determination pathway can evolve while conserving the important functions.
Even without identifying the actual genes involved in sex determination, genetic linkage mapping of GSD in more taxa will reveal nonrecombining regions in species where recombination suppression has evolved. Linkage mapping can reveal X–Y and Z–W orthologs, which can then be used to date the divergence of the sex chromosomes and aid in understanding the age of GSD systems. This type of information on more and more diverse taxa will be needed to understand the evolutionary patterns and time course of events involved in the evolution of GSD and of sex chromosomes. Without such information, it will not be possible to understand the selective forces underlying these changes.
Acknowledgments
We thank Brian Charlesworth, Crispin Jordan, and Tobias Uller for helpful discussions, three anonymous reviewers for helpful comments, and we are grateful for funding from the Royal Society (to J.E.M.) and the Biotechnology and Biological Sciences Research Council (to D.C. and J.E.M.).
References
- Abe, H., M. Kanehara, T. Terada, F. Ohbayashi, T. Shimada et al., 1998. Identification of novel random amplified polymorphic DNAs (RAPDs) on the W chromosome of the domesticated silkworm, Bombyx mori, and the wild silkworm, B. mandarina, and their retrotransposable element-related nucleotide sequences. Genes Genet. Syst. 73 243–254. [DOI] [PubMed] [Google Scholar]
- Adams, I. R., and A. McLaren, 2002. Sexually dimorphic development of mouse primordial germ cells: switching from oogenesis to spermatogenesis. Development 129 1155–1164. [DOI] [PubMed] [Google Scholar]
- Alfaqih, M. A., R. B. Phillips, P. A. Wheeler and G. H. Thorgaard, 2008. The cutthroat trout Y chromosome is conserved with that of rainbow trout. Cytogenet. Genome Res. 121 255–259. [DOI] [PubMed] [Google Scholar]
- Arkhipchuk, V. V., 1995. Role of chromosomal and genome mutations in the evolution of bony fishes. Hydrobiol. J. 31 55–65. [Google Scholar]
- Ashman, T. L., 2003. Constraints on the evolution of males and sexual dimorphism: field estimates of genetic architecture of reproductive traits in three populations of gynodioecious Fragaria virginiana. Evolution 57 2012–2025. [DOI] [PubMed] [Google Scholar]
- Ashman, T. L., 2005. The limits of sexual dimorphism in vegetative traits in a gynodioecious plant. Am. Nat. 166 s5–s16. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D. and B. Charlesworth, 2002. Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416 323–326. [DOI] [PubMed] [Google Scholar]
- Backström, N., N. Karaiskou, E. H. Leder, L. Gustafsson, C. R. Primmer et al., 2008. A gene-based genetic linkage map of the collared flycatcher (Ficedula albicollis) reveals extensive synteny and gene-order conservation during 100 million years of avian evolution. Genetics 179 1479–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badyaev, A. V., 2002. Growing apart: an ontogenetic perspective on the evolution of sexual size dimorphism. Trends Ecol. Evol. 17 369–378. [Google Scholar]
- Bakloushinskaya, I., 2009. Evolution of sex determination in mammals. Biol. Bull. 36 167–174. [Google Scholar]
- Barlow, B. A., and D. Wiens, 1976. Translocation heterozygosity and sex ratio in Viscum fischeri. Heredity 37 27–40. [Google Scholar]
- Baroiller, J. F., H. D'Cotta, E. Bezault, S. Wessels and G. Hoerstgen-Schwark, 2009. Tilapia sex determination: where temperature and genetics meet. Comp. Biochem. Physiol. Part A 153 30–38. [DOI] [PubMed] [Google Scholar]
- Bartkowiak, E., 1971. Mechanisms of sex determination in Rumex hastatulus Baldw. Theor. Appl. Genet. 41 320–326. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and B. Charlesworth, 1998. Why sex and recombination? Science 281 1986–1990. [PubMed] [Google Scholar]
- Beardsmore, J. A., G. C. Mair and R. I. Lewis, 2001. Monosex male production in finfish as exemplified by tilapia: applications, problems and prospects. Aquaculture 197 282–301. [Google Scholar]
- Bellott, D., H. Skaletsky, T. Pyntikova, E. Mardis, T. Graves et al., 2010. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti, T. R., F. H. Valicente, R. Aggarwal, C. Zhao, J. G. Walling et al., 2009. A neo-sex-chromosome that drives post-zygotic sex determination in the hessian fly (Mayetiola destructor). Genetics 184 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero, R., A. Forrest, E. Kamau and D. Charlesworth, 2007. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset-Brändli, L., J. Jaquiery, S. Dubey and N. Perrin, 2006. A sex-specific marker reveals male heterogamety in European tree frogs. Mol. Biol. Evol. 23 1104–1106. [DOI] [PubMed] [Google Scholar]
- Berset-Brändli, L., J. Jaquiéry, T. Broquet, Y. Ulrich and N. Perrin, 2008. Extreme heterochiasmy and nascent sex chromosomes in European tree frogs. Proc. R. Soc. Lond. Ser. B 275 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J. J., 1983. Evolution of Sex Determining Mechanisms. Benjamin-Cummings, Menlo Park, CA.
- Carvalho, A. B., and A. G. Clark, 2005. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 307 108–110. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1991. The evolution of sex chromosomes. Science 251 1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1996. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6 149–162. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 1978. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112 975–997. [Google Scholar]
- Charlesworth, B., and J. D. Wall, 1998. Inbreeding, heterozygote advantage and the evolution of neo-X and neo-Y sex chromosomes. Proc. R. Soc. Lond. Ser. B 266 51–56. [Google Scholar]
- Charlesworth, B., J. A. Coyne and N. H. Barton, 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130 113–146. [Google Scholar]
- Charlesworth, D., B. Charlesworth and G. Marais, 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95 118–128. [DOI] [PubMed] [Google Scholar]
- Chase, C., 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23 81–90. [DOI] [PubMed] [Google Scholar]
- Chen, C. X., Q. Y. Yu, S. B. Hou, Y. J. Li, M. Eustice et al., 2007. b Construction of a sequence-tagged high-density genetic map of papaya for comparative structural and evolutionary genomics in brassicales. Genetics 177 2481–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. L., J. Li, S. P. Deng, Y. S. Tian, Q. Y. Wang et al., 2007. a Isolation of female-specific AFLP markers and molecular identification of genetic sex in half-smooth tongue sole (Cynoglossus semilaevis). Mar. Biotechnol. 9 273–280. [DOI] [PubMed] [Google Scholar]
- Chen, Y. Q., Y. W. Dong, X. J. Xiang, X. R. Zhang and B. C. Zhu, 2008. Sex determination of Microtus mandarinus mandarinus is independent of Sry gene. Mamm. Genome 19 61–68. [DOI] [PubMed] [Google Scholar]
- Cho, S., Z. Huang and J. Zhang, 2007. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels, A., E. G. L. Koh, J.-M. Chia, S. Brenner, S. Aparicio et al., 2004. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 21 1146–1151. [DOI] [PubMed] [Google Scholar]
- Chu, J. M., and D. J. Howard, 1998. Genetic linkage maps of the ground crickets Allonemobius fasciatus and Allonemobius socius using RAPD and allozyme markers. Genome 41 841–847. [Google Scholar]
- Cnaani, A., and T. D. Kocher, 2008. Sex-linked markers and microsatellite locus duplication in the cichlid species Oreochromis tanganicae. Biol. Lett. 4 700–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnaani, A., B. Y. Lee, N. Zilberman, C. Ozouf-Costaz, G. Hulata et al., 2008. Genetics of sex determination in tilapiine species. Sex. Dev. 2 43–54. [DOI] [PubMed] [Google Scholar]
- Coimbra, M. R. M., K. Kobayashi, S. Koretsugu, O. Hasegawa, E. Ohara et al., 2003. A genetic linkage map of the Japanese flounder, Paralichthys olivaceus. Aquaculture 220 203–218. [Google Scholar]
- Conover, D.O., and B.E. Kynard, 1981. Environmental sex determination: interaction of temperature and genotype in a fish. Science 213 577–579. [DOI] [PubMed] [Google Scholar]
- Correns, C., 1928. Bestimmung, Vererbung und Verteilung des Geschlechtes bei den höheren Pflanzen (Determination, inheritance and distribution of the sexes in the higher plants). Handbuch der Vererbungswissenschaft (Handbook of Genetics) 2 1–138. [Google Scholar]
- Criscione, C. D., C. L. L. Valentim, H. Hirai, P. T. LoVerde and T. J. C. Anderson, 2009. Genomic linkage map of the human blood fluke Schistosoma mansoni. Genome Biol. 10 R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, D. A., M. Åkesson, T. Burke, J. M. Pemberton, J. Slate et al., 2007. Gene order and recombination rate in homologous chromosome regions of the chicken and a passerine bird. Mol. Biol. Evol. 24 1537–1552. [DOI] [PubMed] [Google Scholar]
- Dawson, T. E., and M. A. Geber, 1998. Dimorphism in physiology and morphology, pp. 175–215 in Sexual Dimorphism in Flowering Plants, edited by M. A. Geber, T. E. Dawson and L. F. Delph. Springer-Verlag, Berlin.
- Delgado, C., P. D. Waters, C. Gilbert and J. A. M. Graves, 2009. Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years. Chromosome Res. 17 917–926. [DOI] [PubMed] [Google Scholar]
- Desfeux, C., S. Maurice, J. P. Henry, B. Lejeune and P. H. Gouyon, 1996. Evolution of reproductive systems in the genus Silene. Proc. R. Soc. Lond. Ser. B 263 409–414. [DOI] [PubMed] [Google Scholar]
- Devlin, R. H., and Y. Nagahama, 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208 191–364. [Google Scholar]
- Dorken, M. E., and S. C. G. Barrett, 2004. Sex determination and the evolution of dioecy from monoecy in Sagittaria latifolia (Alismataceae). Proc. R. Soc. Lond. Ser. B 271 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorken, M. E., and J. R. Pannell, 2008. Density-dependent regulation of sex ratio in an annual plant. Am. Nat. 171 824–830. [DOI] [PubMed] [Google Scholar]
- Drilling, C. C., and M. S. Grober, 2005. An initial description of alternative male reproductive phenotypes in the bluebanded goby, Lythrypnus dalli (Teleostei, Gobiidae). Environ. Biol. Fish. 72 361–372. [Google Scholar]
- Eggert, C., 2004. Sex determination: the amphibian models. Reprod. Nutr. Dev. 44 539–549. [DOI] [PubMed] [Google Scholar]
- Eppley, S. M., and E. H. Wenk, 2001. Reproductive biomass allocation in the dioecious perennial Acanthosicyos horrida. S. Afr. J. Bot. 67 10–14. [Google Scholar]
- Ezaz, T., A. E. Quinn, I. Miura, S. D. Sarre, A. Georges et al., 2005. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 13 763–776. [DOI] [PubMed] [Google Scholar]
- Ezaz, T., A. E. Quinn, S. D. Sarre, S. O'Meally, A. Georges et al., 2009. Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosome Res. 17 91–98. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith, M., 2007. The evolution of sex chromosomes and sex determination in vertebrates and the key role of DMRT1. Sex. Dev. 1 2–11. [DOI] [PubMed] [Google Scholar]
- Ferris, P. J., E. V. Armbrust and U. Goodenough, 2002. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1931. The evolution of dominance. Biol. Rev. 6 345–368. [Google Scholar]
- Fishman, L., and J. H. Willis, 2006. A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution 60 1372–1381. [DOI] [PubMed] [Google Scholar]
- Formusoh, E. S., J. H. Hatchett, W. C. Black and J. J. Stuart, 1996. Sex-linked inheritance of virulence against wheat resistance gene H9 in the Hessian fly (Diptera: Cecidomyiidae). Ann. Entomol. Soc. Am. 89 428–434. [Google Scholar]
- Fraser, J. A., S. Diezmann, R. L. Subaran, A. Allen, K. B. Lengeler et al., 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2 2243–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, L. G., G. K. Tsang, P. M. Datson, H. N. De Silva, C. F. Harvey et al., 2009. A gene-rich linkage map in the dioecious species Actinidia chinensis (kiwifruit) reveals putative X/Y sex-determining chromosomes. BMC Genomics 10 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadau, J., C. U. Gerloff, N. Kruger, H. Chan, P. Schmid-Hempel et al., 2001. A linkage analysis of sex determination in Bombus terrestris (L.) (Hymenoptera: Apidae). Heredity 87 234–242. [DOI] [PubMed] [Google Scholar]
- Gempe, T., M. Hasselmann, M. Schiøtt, G. Hause, M. Otte et al., 2009. Sex determination in honeybees: two separate mechanisms induce and maintain the female pathway. PLoS Biol. 7 e1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi, K., A. Gautier, R. G. Danzmann, S. Gharbi, T. Sakamoto et al., 2006. A linkage map for brown trout (Salmo trutta): chromosome homologies and comparative genome organization with other salmonid fish. Genetics 172 2405–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladden, J., B. Farboud and B. Meyer, 2007. Revisiting the X: a signal that specifies Caenorhabditis elegans sexual fate. Genetics 77 1639–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawe, G. A., and T. J. d. Jong, 2009. Complex sex determination in the stinging nettle Urtica dioica. Evol. Ecol. 23 635–649. [Google Scholar]
- Graham, P., J. Penn and P. Schedl, 2003. Masters change, slaves remain. Bioessays 25 1–4. [DOI] [PubMed] [Google Scholar]
- Graves, J., and C. Peichel, 2010. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 11 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman, D. S., and D. Charlesworth, 1998. An X-linked gene with a degenerate Y-linked homologue in a dioecious plant. Nature 393 263–266. [DOI] [PubMed] [Google Scholar]
- Haag, E. S., 2005. The evolution of nematode sex determination: C. elegans as a reference point for comparative biology, in WormBook, edited by The C. elegans Research Community. http://www.wormbook.org/chapters/www_evolutionsexdetermin/evolutionsexdetermin.html [DOI] [PMC free article] [PubMed]
- Hasselmann, M., T. Gempe, M. Schiøtt, C. Nunes-Silva, M. Otte et al., 2008. a Evidence for the evolutionary nascence of a novel sex determination pathway in honey bees. Nature 454 519–522. [DOI] [PubMed] [Google Scholar]
- Hasselmann, M., X. Vekemans, J. Pflugfelder, N. Koeniger, G. Koeniger et al., 2008. b Evidence for convergent nucleotide evolution and high allelic turnover rates at the complementary sex determiner gene of Western and Asian honeybees. Mol. Biol. Evol. 25 696–708. [DOI] [PubMed] [Google Scholar]
- Hawthorne, D. J., 2001. AFLP-based genetic linkage map of the Colorado potato beetle Leptinotarsa decemlineata: sex chromosomes and a pyrethroid-resistance candidate gene. Genetics 158 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger, M., C. Henggeler, N. Meier, R. Perez, G. Saccone et al., 2010. Molecular characterization of the key switch provides a basis for understanding the rapid divergence of the sex-determining pathway of the housefly. Genetics 184 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W., 2007. Sex: differences in mutation, recombination, selection, gene flow, and genetic drift. Evolution 61 2750–2771. [DOI] [PubMed] [Google Scholar]
- Hillis, D. M., and D. M. Green, 1990. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J. Evol. Biol. 3 49–64. [Google Scholar]
- Holloway, A. K., M. R. Strand, W. C. Black and M. F. Antolin, 2000. Linkage analysis of sex determination in Bracon sp near hebetor (Hymenoptera: Braconidae). Genetics 154 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, E. C., S. Armstrong and D. Filatov, 2009. Evolution of neo-sex chromosomes in Silene diclinis. Genetics 182 1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, G. J., and R. E. Page, 1994. Linkage analysis of sex determination in the honey bee (Apis mellifera). Mol. Gen. Genet. 244 512–518. [DOI] [PubMed] [Google Scholar]
- Huxley, J., 1928. Sexual differences in linkage in Gammarys chevruxi. J. Genet. 20 145–156. [Google Scholar]
- Janzen, F. J., and P. C. Phillips, 2006. Exploring the evolution of environmental sex determination, especially in reptiles. J. Evol. Biol. 19 1775–1784. [DOI] [PubMed] [Google Scholar]
- Johnson-Brousseau, S. A., and S. McCormick, 2004. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 39 761–775. [DOI] [PubMed] [Google Scholar]
- Just, W., W. Rau, W. Vogel, M. Akhverdian, K. Fredga et al., 1995. Absence of SRY in species of the vole Ellobius. Nat. Genet. 11 117–118. [DOI] [PubMed] [Google Scholar]
- Kallman, K. D., 1970. Sex determination and the restriction of sex-linked pigment patterns to the X and Y chromosomes of a Poeciliid fish, Xiphophorus maculatus, from the Belize and Sibun rivers of British Honduras. Zoologica 55 1–16. [Google Scholar]
- Kallman, K. D., 1984. A new look at sex determination in poeciliid fishes, pp. 95–171 in Genetics of Fishes, edited by B. J. Turner. Plenum Publishing, New York.
- Kawai, A., C. Nishida-Umehara, J. Ishijima, Y. Tsuda, H. Ota et al., 2007. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet. Genome Res. 117 92–102. [DOI] [PubMed] [Google Scholar]
- Khattak, J. Z. K., A. M. Torp and S. B. Andersen, 2006. A genetic linkage map of Spinacia oleracea and localization of a sex determination locus. Euphytica 148 311–318. [Google Scholar]
- Kihara, H., and J. Hirayoshi, 1932. Die Geschlechtschromosomen von Humulus japonicus Sieb. et. Zuce (Sex determination in Humulus japonicus Sieb. et. Zuce), pp. 248 in 8th Congress of the Japanese Association for the Advancement of Science, Plant Breeding Abstracts.
- Kikuchi, K., W. Kai, A. Hosokawa, N. Mizuno, H. Suetake et al., 2007. The sex-determining locus in the Tiger Pufferfish, Takifugu rubripes. Genetics 175 2039–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., C. Kim, J. Lee and J. Bang, 2008. Karyotype analysis and physical mapping using two rRNA genes in dioecious plant, Humulus japonicus Sieboid and Zucc. Genes Genomics 30 157–161. [Google Scholar]
- Kimura, T., K. Yoshida, A. Shimada, T. Jindo, H. M. M. Sakaizumi et al., 2006. Genetic linkage map of medaka with polymerase chain reaction length polymorphisms. Gene 363 24–31. [DOI] [PubMed] [Google Scholar]
- Kitano, J., J. A. Ross, S. Mori, M. Kume, F. C. Jones et al., 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461 1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich, L. B., X. Y. Wang, A. G. Clark and A. B. Carvalho, 2008. Low conservation of gene content in the Drosophila Y chromosome. Nature 456 949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, M., E. Nagao, H. Mitani and A. Shima, 2001. Differences in recombination frequencies during male and female meioses of the sex chromosomes of the medaka, Orzyias latipes. Genet. Res. 78 23–30. [DOI] [PubMed] [Google Scholar]
- Kondo, M., I. Nando, U. Hornung, M. Schmid and M. Schartl, 2004. Evolutionary origin of the medaka Y chromosome. Curr. Biol. 14 1664–1669. [DOI] [PubMed] [Google Scholar]
- Kondo, M., U. Hornung, I. Nanda, S. Imai, T. Sasaki et al., 2006. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 16 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp, A., R. M. Graze, S. Z. Xu, S. B. Carroll and S. V. Nuzhdin, 2003. Quantitative trait loci responsible for variation in sexually dimorphic traits in Drosophila melanogaster. Genetics 163 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielska, M., F. Weissing, L. W. Beukeboom and I. Pen, 2010. Segregation distortion and the evolution of sex determining mechanisms. Heredity 104 100–112. [DOI] [PubMed] [Google Scholar]
- Krzywinski, J., D. R. Nusskern, M. K. Kern and N. J. Besansky, 2004. Isolation and characterization of Y chromosome sequences from the African malaria mosquito Anopheles gambiae. Genetics 166 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa, A., 2009. No final answers yet on sex determination in birds. Nature 462 34. [DOI] [PubMed] [Google Scholar]
- Lahn, B.T., and D.C. Page, 1999. Four evolutionary strata on the human X chromosome. Science 286 964–967. [DOI] [PubMed] [Google Scholar]
- Larracuente, A. M., M. A. Noor and A. G. Clark, 2010. Translocation of Y-linked genes to the dot chromosome in Drosophila pseudoobscura. Mol. Biol. Evol. 27 1612–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B. Y., D. J. Penman and T. D. Kocher, 2003. Identification of a sex-determining region in Nile tilapia (Oreochromis niloticus) using bulked segregant analysis. Anim. Genet. 34 379–383. [DOI] [PubMed] [Google Scholar]
- Lee, B. Y., G. Hulata and T. D. Kocher, 2004. Two unlinked loci controlling the sex of blue tilapia (Oreochromis aureus). Heredity 92 543–549. [DOI] [PubMed] [Google Scholar]
- Lemos, B., L. O. Araripe, P. Fontanillas and D. L. Hartl, 2008. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc. Natl. Acad. Sci. USA 105 14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. T., K. Byrne, E. Miggiano, V. Whan, S. Moore et al., 2003. Genetic mapping of the kuruma prawn Penaeus japonicus using AFLP markers. Aquaculture 219 143–156. [Google Scholar]
- Liu, X. D., X. Liu, X. M. Guo, Q. K. Gao, H. G. Zhao et al., 2006. A preliminary genetic linkage map of the pacific abalone Haliotis discus hannai Ino. Mar. Biotechnol. 8 386–397. [DOI] [PubMed] [Google Scholar]
- Liu, Z., P. H. Moore, H. Ma, C. M. Ackerman, M. Ragiba et al., 2004. A primitive Y chromosome in Papaya marks the beginning of sex chromosome evolution. Nature 427 348–352. [DOI] [PubMed] [Google Scholar]
- Lloyd, D. G., 1975. Breeding systems in Cotula. III. Dioecious populations. New Phytol. 74 109–123. [Google Scholar]
- Luycx, P., 1981. A sex-linked esterase locus and translocation heterozygosity in a termite. Heredity 46 315–320. [Google Scholar]
- Ma, H., P. H. Moore, Z. Y. Liu, M. S. Kim, Q. Y. Yu et al., 2004. High-density linkage mapping revealed suppression of recombination at the sex determination locus in papaya. Genetics 166 419–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabberley, D., 1997. The Plant Book. Cambridge University Press, Cambridge, UK.
- MacIntyre, P., 1961. Spontaneous sex reversals of genotypic males in the Platyfish (Xiphophorus maculatus). Genetics 46 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKnight, R.H., 1939. The sex-determining mechanism of Drosophila miranda. Genetics 24 180–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainguy, J., S. D. Cote, M. Festa-Bianchet and D. W. Coltman, 2009. Father-offspring phenotypic correlations suggest intralocus sexual conflict for a fitness-linked trait in a wild sexually dimorphic mammal. Proc. R. Soc. Lond. Ser. B 276 4067–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank, J. E., 2009. a Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. Am. Nat. 173 141–150. [DOI] [PubMed] [Google Scholar]
- Mank, J. E., 2009. b The evolution of heterochiasmy: the role of sexual selection and sperm competition in determining sex-specific recombination rates in eutherian mammals. Genet. Res. 91 355–363. [DOI] [PubMed] [Google Scholar]
- Mank, J. E., and J. C. Avise, 2006. Phylogenetic conservation of chromosome numbers in actinopterygiian fishes. Genetica 127 321–327. [DOI] [PubMed] [Google Scholar]
- Mank, J. E., and H. Ellegren, 2007. Parallel divergence and degredation of the avian W sex chromosome. Trends Ecol. Evol. 22 389–391. [DOI] [PubMed] [Google Scholar]
- Mank, J. E., D. E. L. Promislow and J. C. Avise, 2005. Phylogenetic perspectives on the evolution of parental care in ray-finned fishes. Evolution 59 1570–1578. [PubMed] [Google Scholar]
- Mank, J. E., D. E. L. Promislow and J. C. Avise, 2006. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol. J. Linn. Soc. 87 83–93. [Google Scholar]
- Marais, G., M. Nicolas, R. Bergero, P. Chambrier, E. Kejnovsky et al., 2008. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr. Biol. 18 545–549. [DOI] [PubMed] [Google Scholar]
- Marguerit, E., C. Boury, A. Manicki, M. Donnart, G. Butterlin et al., 2009. Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor. Appl. Genet. 118 1261–1278. [DOI] [PubMed] [Google Scholar]
- Marin, J., and B. S. Baker, 1998. The evolutionary dynamics of sex determination. Science 281 1990–1994. [DOI] [PubMed] [Google Scholar]
- Marti, D., and C. Bidau, 1995. Male and female meiosis in a natural population of Dichroplus pratensis (Acrididae) polymorphic for Robertsonian translocations: a study of chiasma frequency and distribution. Hereditas 123 227–235. [Google Scholar]
- Martin, A., A. B. C. Troadec, M. Rajab, R. Fernandez, H. Morin et al., 2009. A transposon-induced epigenetic change leads to sex determination in melon. Nature 461 1135–1138. [DOI] [PubMed] [Google Scholar]
- Martinez, P., C. Bouza, M. Hermida, J. Fernandez, M. A. Toro et al., 2009. Identification of the major sex-determining region of turbot (Scophthalmus maximus). Genetics 183 1443–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K., H. Tarui, M. Toriba, K. Yamada, C. Nishida-Umehara et al., 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. USA 103 18190–18195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417 559–563. [DOI] [PubMed] [Google Scholar]
- McAllister, B. F., and B. Charlesworth, 1999. Reduced sequence variability on the neo-Y chromosome of Drosophila americana americana. Genetics 153 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland, E. K., and K. A. Naish, 2008. A genetic linkage map for coho salmon (Oncorhynchus kisutch). Anim. Genet. 39 169–179. [DOI] [PubMed] [Google Scholar]
- McDaniel, S. F., J. H. Willis and A. J. Shaw, 2007. A linkage map reveals a complex basis for segregation distortion in an interpopulation cross in the moss Ceratodon purpureus. Genetics 176 2489–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, A., and M. Schartl, 1999. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11 699–704. [DOI] [PubMed] [Google Scholar]
- Morizot, D. C., S. A. Slaugenhaupt, K. D. Kallman and A. Chakravarti, 1991. Genetic linkage map of fishes of the genus Xiphophorus (Teleostei, Poecilidae). Genetics 127 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizot, D. C., R. S. Nairn, P. Simhambhatla, L. Della Coletta, D. Trono et al., 2001. Xiphophorus genetic linkage map: beginnings of comparative gene mapping in fishes. Mar. Biotechnol. 3 S153–S161. [DOI] [PubMed] [Google Scholar]
- Morrow, E. H., A. D. Stewart and W. R. Rice, 2008. Assessing the extent of genome-wide intralocus sexual conflict via experimentally enforced gender-limited selection. J. Evol. Biol. 21 1046–1054. [DOI] [PubMed] [Google Scholar]
- Miura, I., H. Ohtani, M. Nakamura, Y. Ichikawa and K. Saitoh, 1998. The origin and differentiation of the heteromorphic sex chromosomes Z, W, X, and Y in the frog Rana rugosa, inferred from the sequences of a sex-linked gene, ADP/ATP translocase. Mol. Biol. Evol. 15 1612–1619. [DOI] [PubMed] [Google Scholar]
- Mrackova, M., M. Nicolas, R. Hobza, I. Negrutiu, F. Monéger et al., 2008. Independent origin of sex chromosomes in two species of the genus Silene. Genetics 179 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. L., S. K. Mahadevaiah, P. J. Park, P. E. Warburton, D. C. Page et al., 2008. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 40 794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W. J., S. Sun, Z.-Q. Chen, J. Pecon-Slattery and S. J. O'Brien, 1999. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 9 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, T., Y. Takehana, S. Hamaguchi and M. Sakaizumi, 2008. Identification of the sex-determining locus in the Thai medaka, Oryzias minutillus. Cytogenet. Genome Res. 121 137–142. [DOI] [PubMed] [Google Scholar]
- Nam, K., and H. Ellegren, 2008. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics 180 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler et al., 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, I., K. Schlegelmilch, T. Haaf, M. Schartl and M. Schmid, 2008. Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet. Genome Res. 122 150–156. [DOI] [PubMed] [Google Scholar]
- Naruse, K., S. Fukamachi, H. Mitani, M. Kondo, T. Matsuoka et al., 2000. A detailed linkage map of Medaka, Oryzias latipes: comparative genomics and genome evolution. Genetics 154 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse, K., M. Tanaka, K. Mita, A. Shima, J. Postlethwait et al., 2004. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 14 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navajas-Perez, R., R. de la Herran, G. L. Gonzalez, M. Jamilena, R. Lozano et al., 2005. The evolution of reproductive systems and sex-determining mechanisms within Rumex (Polygonaceae) inferred from nuclear and chloroplastidial sequence data. Mol. Biol. Evol. 22 1929–1939. [DOI] [PubMed] [Google Scholar]
- Nene, V., J. R. Wortman, D. Lawson, B. Haas, C. Kodira et al., 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, K. M., W. P. Young, R. G. Danzmann, B. D. Robison, C. Rexroad et al., 2003. A consolidated linkage map for rainbow trout (Oncorhynchus mykiss). Anim. Genet. 34 102–115. [DOI] [PubMed] [Google Scholar]
- Nishida-Umehara, C., Y. Tsuda, J. A. J Ishijima, A. Fujiwara, Y. Matsuda et al., 2007. The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res. 15 721–734. [DOI] [PubMed] [Google Scholar]
- Ogata, M., Y. Hasegawa, H. Ohtani, M. Mineyama and I. Miura, 2008. The ZZ/ZW sex-determining mechanism originated independently during the evolution of the frog, Rana rugosa. Heredity 100 92–99. [DOI] [PubMed] [Google Scholar]
- Ohnishi, O., 1985. Population genetics of cultivated buckwheat, Fagopyrum esculentum Moench. III. Frequency of sterility mutants in Japanese populations. Jpn. J. Genet. 60 391–404. [Google Scholar]
- Okada, S., T. Sone, M. Fujisawa, S. Nakayama, M. Takenaka et al., 2001. The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc. Natl. Acad. Sci. USA 98 9454–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo, E., 2005. Rate of chromosome changes and speciation in reptiles. Genetica 125 185–203. [DOI] [PubMed] [Google Scholar]
- Organ, C. L., and D. E. Janes, 2008. Evolution of sex chromosomes in Sauropods. Integr. Comp. Biol. 48 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ, C. L., D. E. Janes, A. Meade and M. Pagel, 2009. Genotypic sex determination enabled adaptive radiation of extinct marine reptiles. Nature 461 389–392. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., 2009. The evolutionary enigma of sex. Am. Nat. 174 S1–S14. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and N. H. Barton, 1997. The evolution of recombination: removing the limits to natural selection. Genetics 147 879–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane, A., A. D. Simone, G. Saccone and C. Polito, 2005. Evolutionary conservation of Ceratitis capitata transformer gene function. Genetics 171 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parise-Maltempi, P., C. Martins, C. Oliveira and F. Foresti, 2007. Identification of a new repetitive element in the sex chromosomes of Leporinus elongatus (Teleostei: Characiformes: Anostomidae): new insights into the sex chromosomes of Leporinus. Cytogenet. Genome Res. 116 218–223. [DOI] [PubMed] [Google Scholar]
- Patten, M. M., and D. Haig, 2009. Maintenance or loss of genetic variation under sexual and parental antagonism at a sex-linked locus. Evolution 63 2888–2895. [DOI] [PubMed] [Google Scholar]
- Peichel, C. L., K. S. Nereng, K. A. Ohgi, B. L. E. Cole, P. F. Colosimo et al., 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414 901–905. [DOI] [PubMed] [Google Scholar]
- Peichel, C. L., J. A. Ross, C. K. Matson, M. Dickson, J. Grimwood et al., 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14 1416–1424. [DOI] [PubMed] [Google Scholar]
- Peiffer, J., S. Kaushik, H. Sakai, M. Arteaga-Vazquez, N. Sanchez-Leon et al., 2008. A spatial dissection of the Arabidopsis floral transcriptome by MPSS. BMC Plant Biol. 8 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil, A., H. Flachowsky, E. Schumann and W. E. Weber, 2003. Sex-linked AFLP markers indicate a pseudoautosomal region in hemp (Cannabis sativa L.). Theor. Appl. Genet. 107 102–109. [DOI] [PubMed] [Google Scholar]
- Perrin, N., 2009. Sex reversal: a fountain of youth for sex chromosomes? Evolution 63 3043–3049. [DOI] [PubMed] [Google Scholar]
- Phillips, R., J. DeKoning, A. Ventura, K. Nichols, R. Drew et al., 2009. Recombination is suppressed over a large region of the rainbow trout Y chromosome. Anim. Genet. 40 925–932. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., G. S. Ditta, B. Savidge, S. J. Liljegren, E. Baumann et al., 2003. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. [DOI] [PubMed] [Google Scholar]
- Pokorna, M., and L. Kratochvil, 2009. Phylogeny of sex-determining mechanisms in squamate reptiles: Are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc. 156 168–183. [Google Scholar]
- Polley, A., E. Seigner and M. W. Ganal, 1997. Identification of sex in hop (Humulus lupulus) using molecular markers. Genome 40 357–361. [DOI] [PubMed] [Google Scholar]
- Potrzebowski, L., N. Vinckenbosch, A. C. Marques, F. Chalmel, B. Jegou et al., 2008. Chromosomal gene movements reflect the recent origin and biology of the therian sex chromosomes. PLoS Biol. 6 e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski, L., N. Vinckenbosch and H. Kaessmann, 2009. The emergence of new genes on the young therian X. Trends Genet. 26 1–4. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., and H. Orr, 1998. Haldane's rule in taxa lacking a hemizygous X. Science 282 952–954. [DOI] [PubMed] [Google Scholar]
- Quinn, A. E., A. Georges, S. D. Sarre, F. Guarino, T. Ezaz et al., 2007. Temperature sex reversal implied sex gene dosage in a reptile. Science 316 411. [DOI] [PubMed] [Google Scholar]
- Quinn, A. E., T. Ezaz, S. D. Sarre, J. A. Marshall Graves and A. Georges, 2010. Extension, single-locus conversion and physical mapping of sex chromosome sequences identify the Z microchromosome and pseudo-autosomal region in a dragon lizard, Pogona vitticeps. Heredity 104 410–417. [DOI] [PubMed] [Google Scholar]
- Rasmussen, D., and M. Noor, 2009. What can you do with 0.1x genome coverage? A case study based on a genome survey of the scuttle fly Megaselia scalaris (Phoridae). BMC Genomics 10 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp, T., E. Lee, S. Kata, C. Brinkmeyer, J. Mickelson et al., 2004. Exceptional conservation of horse-human gene order on X chromosome revealed by high-resolution radiation hybrid mapping. Proc. Natl. Acad. Sci. USA 101 2386–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C. S., M. W. Murphy, M. G. O'Sullivan, V. J. Bardwell and D. Zarkower, 2000. DMRT1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buttner, S. M., J. Schondelmaier and C. Jung, 1998. AFLP markers tightly linked to the sex locus in Asparagus officinalis L. Mol. Breed. 4 91–98. [Google Scholar]
- Renner, S. S., and R. E. Ricklefs, 1995. Dioecy and its correlates in the flowering plants. Am. J. Bot. 82 596–606. [Google Scholar]
- Rexroad, C. E., Y. Palti, S. A. Gahr and R. L. Vallejo, 2008. A second generation genetic map for rainbow trout (Oncorhynchus mykiss). BMC Genet. 9 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38 735–742. [DOI] [PubMed] [Google Scholar]
- Rice, W.R., 1987. Genetic hitchhiking and the evolution of reduced genetic activity of the human Y chromosome. Genetics 116 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R. B., J. R. Ser and T. D. Kocher, 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 362 998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J. A., and C. L. Peichel, 2008. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback. Genetics 179 2173–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J. A., J. R. Urton, J. Boland, M. D. Shapiro and C. L. Peichel, 2009. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 5 e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, M. T., D. V. Grafham, A. J. Coffey, S. Scherer, K. McLay et al., 2005. The DNA sequence of the human X chromosome. Nature 434 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, A. K., S. G. Weller, W. Wagner, M. Nepokroeff and T. M. Culley, 2006. Adaptive radiation and evolution of breeding systems in Schiedea (Caryophyllaceae), an endemic Hawaiian genus. Ann. Mo. Bot. Gard. 93 49–63. [Google Scholar]
- Schultheis, C., A. Bohne, M. Schartl, J. N. Volff and D. Galiana-Arnoux, 2009. Sex determination diversity and sex chromosome evolution in poeciliid fish. Sex. Dev. 3 68–77. [DOI] [PubMed] [Google Scholar]
- Seefelder, S., H. Ehrmaier, G. Schweizer and E. Seigner, 2000. Male and female genetic linkage map of hops, Humulus lupulus. Plant Breed. 119 249–255. [Google Scholar]
- Semerikov, V., U. Lagercrantz, V. Tsarouhas, A. Ronnberg-Wastljung, C. Alstrom-Rapaport et al., 2003. Genetic mapping of sex-linked markers in Salix viminalis L. Heredity 91 293–299. [DOI] [PubMed] [Google Scholar]
- Ser, J. R., R. B. Roberts and T. D. Kocher, 2010. Multiple interacting loci control sex determination in Lake Malawi cichild fish. Evolution 64 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiamarga, D. H. E., M. Miya, Y. Yamanoue, Y. Azuma, J. G. Inoue et al., 2009. Divergence time of the two regional medaka populations in Japan as a new time scale for compararative vertebrate genomics. Biol. Lett. 5 812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, C. W., S. L. Chen, C. F. Scheuring, J. Y. Xu, Z. H. Shan et al., 2010. Construction of two BAC libraries from half-smooth tongue sole Cynoglossus semilaevis and identification of clones containing candidate sex determining genes. Mar. Biotechnol. (in press) 10.1007/s10126–009–9242-x. [DOI] [PubMed]
- Shapiro, M. D., B. R. Summers, S. Balabhadra, J. T. Aldenhoven, A. L. Miller et al., 2009. The genetic architecture of skeletal convergence and sex determination in ninespine sticklebacks. Curr. Biol. 19 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman, D. C. A., 2002. The evolution of sex determination systems in Dipteran insects other than Drosophila. Genetica 116 25–43. [DOI] [PubMed] [Google Scholar]
- Shine, R., 1989. Ecological causes for the evolution of sexual dimorphism: a review of the evidence. Quart. Rev. Biol. 64 419–461. [DOI] [PubMed] [Google Scholar]
- Siegal, M. L., and B. S. Baker, 2005. Functional conservation and divergence of intersex, a gene required for female differentiation in Drosophila melanogaster. Dev. Genes Evol. 215 1–12. [DOI] [PubMed] [Google Scholar]
- Siegfried, K., and C. Nusslein-Volhard, 2008. Germ line control of female sex determination in zebrafish. Dev. Biol. 324 277–287. [DOI] [PubMed] [Google Scholar]
- Singh, R. B., and B. W. Smith, 1971. Mechanisms of sex determination in Rumex acetosella. Theor. Appl. Genet. 41 360. [DOI] [PubMed] [Google Scholar]
- Sinha, S., A. Guha, B. Sinha, R. Kumar and N. Banerjee, 2007. Average packing ratio and evolution of sex chromosomes in dioecious Coccinia indica and Trichosanthes dioica. Cytologia 72 369–372. [Google Scholar]
- Skaletsky, H., T. Kuroda-Kawaguchi, P. J. Minx, H. S. Cordum, L. Hillier et al., 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423 825–832. [DOI] [PubMed] [Google Scholar]
- Smith, B. W., 1963. The mechanism of sex determination in Rumex hastatulus. Genetics 48 1265–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, B. W., 1969. Evolution of sex-determining mechanisms in Rumex. Chromosomes Today 2 172–182. [Google Scholar]
- Smith, C., 1975. The evolution of hermaphroditism in fishes, pp. 295–310 in Intersexuality in the Animal Kingdom, edited by R. Reinboth. Springer-Verlag, Berlin.
- Smith, C. A., P. J. McClive, P. S. Western, K. J. Reed and A. H. Sinclair, 1999. Evolution: conservation of a sex-determining gene. Nature 402 601–602. [DOI] [PubMed] [Google Scholar]
- Smith, C. A., K. N. Roeszler, T. Ohnesorg, D. M. Cummins, P. G. Farlie et al., 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461 267–271. [DOI] [PubMed] [Google Scholar]
- Smith, J. J., and S. R. Voss, 2009. Amphibian sex determination: segregation and linkage analysis using members of the tiger salamander species complex (Ambystoma mexicanum and A.t. tigrinum). Heredity 102 542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari, A. J., 1993. Sex Chromosomes and Sex Determination in Vertebrates. CRC Press, Boca Raton, FL.
- Solignac, M., D. Vautrin, E. Baudry, F. Mougel, A. Loiseau et al., 2004. A microsatellite-based linkage map of the Honeybee, Apis mellifera L. Genetics 167 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondur, S. N., R. M. Manshardt and J. I. Stiles, 1996. A genetic linkage map of papaya based on randomly amplified polymorphic DNA markers. Theor. Appl. Genet. 93 547–553. [DOI] [PubMed] [Google Scholar]
- Spigler, R. B., K. S. Lewers, D. S. Main and T. L. Ashman, 2008. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity 101 507–517. [DOI] [PubMed] [Google Scholar]
- Streit, A., 2008. Reproduction in Strongyloides (Nematoda): a life between sex and parthenogenesis. Parasitology 135 285–294. [DOI] [PubMed] [Google Scholar]
- Taguchi, T., Y. Hirai, P. T. LoVerde, A. Tominaga and H. Hirai, 2007. DNA probes for identifying chromosomes 5, 6, and 7 of Schistoma mansoni. J. Parasitol. 93 724–726. [DOI] [PubMed] [Google Scholar]
- Takehana, Y., K. Naruse, S. Hamaguchi and M. Sakaizumi, 2007. Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma 116 463–470. [DOI] [PubMed] [Google Scholar]
- Takehana, Y., S. Hamaguchi and M. Sakaizumi, 2008. Different origins of ZZ/ZW sex chromosomes in closely related medaka fishes, Oryzias javanicus and O. hubbsi. Chromosome Res. 16 801–811. [DOI] [PubMed] [Google Scholar]
- Talamali, A., M. Bajji, A. L. Thomas, J. Kinet and P. Dutuit, 2003. Flower architecture and sex determination: How does Atriplex halimus play with floral morphogenesis and sex genes? New Phytol. 157 105–113. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Y. Takehana, K. Naruse, S. Hamaguchi and M. Sakaizumi, 2007. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene. Genetics 177 2075–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telgmann-Rauber, A., A. Jamsari, M. S. Kinney, J. C. Pires and C. Jung, 2007. Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. Mol. Genet. Genomics 278 221–234. [DOI] [PubMed] [Google Scholar]
- Terauchi, R., and G. Kahl, 1999. Mapping of the Dioscorea tokoro genome: AFLP markers linked to sex. Genome 42 752–762. [Google Scholar]
- Thorgaard, G. H., 1977. Heteromorphic sex chromosomes in male rainbow trout. Science 196 900–902. [DOI] [PubMed] [Google Scholar]
- Traut, W., and U. Willhoeft, 1990. A jumping sex determining factor in the fly Megaselia scalaris. Chromosoma 99 407–412. [Google Scholar]
- Traut, W., and B. Wollert, 1998. An X/Y DNA segment from an early stage of sex chromosome differentiation in the fly Megaselia scalaris. Genome 41 289–294. [DOI] [PubMed] [Google Scholar]
- Traut, W., K. Sahara, T. Otto and F. Marec, 1999. Molecular differentiation of sex chromosomes probed by comparative genomic hybridization. Chromosoma 108 173–180. [DOI] [PubMed] [Google Scholar]
- Tripathi, N., M. Hoffmann, D. Weigel and C. Dreyer, 2009. a Linkage analysis reveals the independent origin of Poeciliid sex chromosomes and a case of atypical sex inheritance in the guppy (Poecilia reticulata). Genetics 182 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, N., M. Hoffmann, E. Willing, C. Lanz, D. Weigel et al., 2009. b Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc. R. Soc. Ser. Lond. B 276 2195–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, Y., C. Nishida-Umehara, J. Ishijima, K. Yamada and Y. Matsuda, 2007. Comparison of the Z and W sex chromosomal architectures in elegant chrested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma 116 159–173. [DOI] [PubMed] [Google Scholar]
- Uno, Y., C. Nishida, Y. Oshima, S. Yokoyama, I. Miura et al., 2008. Comparative chromosome mapping of sex-linked genes and identification of sex chromosomal rearrangements in the Japanese wrinkled frog (Rana rugosa, Ranidae) with ZW and XY sex chromosome systems. Chromosome Res. 16 637–647. [DOI] [PubMed] [Google Scholar]
- Valenzano, D. R., J. Kirschner, R. A. Kamber, E. Zhang, D. Weber et al., 2009. Mapping loci associated with tail color and sex determination in the short-lived fish Nothobranchius furzeri. Genetics 183 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela, N., 2008. Evolution of the gene network underlying gonadogenesis in turtle with temperature-dependent and genotypic sex determination. Integr. Comp. Biol. 48 476–485. [DOI] [PubMed] [Google Scholar]
- Valenzuela, N., and T. Shikano, 2007. Embryological ontogeny of aromatase gene expression in Chrysemys picta and Apalone mutica turtles: comparative patterns within and across temperature-dependent and genotypic sex determining mechanisms. Dev. Genes Evol. 217 55–62. [DOI] [PubMed] [Google Scholar]
- Van Doorn, G. S., and M. Kirkpatrick, 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449 909–912. [DOI] [PubMed] [Google Scholar]
- Venkatesan, M., K. W. Broman, M. Sellers and J. L. Rasgon, 2009. An initial linkage map of the West Nile Virus vector Culex tarsalis. Insect Mol. Biol. 18 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst, E. C., L.W. Beukeboom and L. van de Zande, 2010. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 328 620–623. [DOI] [PubMed] [Google Scholar]
- Veyrunes, F., P. D. Waters, P. Miethke, W. Rens, D. McMillan et al., 2008. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso, B., and B. Charlesworth, 2006. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 7 645–653. [DOI] [PubMed] [Google Scholar]
- Volff, J. N., I. Nanda, M. Schmid and M. Schartl, 2007. Governing sex determination in fish: regulatory putsches and ephemeral dictators. Sex. Dev. 1 85–99. [DOI] [PubMed] [Google Scholar]
- Von Ubisch, G., 1936. Genetic studies on the nature of hermaphroditic plants in Antennaria dioica (L) Gaertn. Genetics 21 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier, S., R. Lande, J. J. M. Van Alphen and O. Seehausen, 2007. Invasion and fixation of sex-reversal genes. J. Evol. Biol. 20 913–920. [DOI] [PubMed] [Google Scholar]
- Waldbieser, G. C., B. G. Bosworth, D. J. Nonneman and W. R. Wolters, 2001. A microsatellite-based genetic linkage map for channel catfish, Ictalurus punctatus. Genetics 158 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, P. D., B. Duffy, C. J. Frost, M. L. Delbridge and J. A. M. Graves, 2001. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet. Cell Genet. 92 74–79. [DOI] [PubMed] [Google Scholar]
- West, S. A., 2009. Sex Allocation. Princeton University Press, Princeton, NJ.
- West, S. A., and B. C. Sheldon, 2002. Constraints on the evolution of sex ratio adjustment. Science 295 1685–1688. [DOI] [PubMed] [Google Scholar]
- Westergaard, M., 1958. The mechanism of sex determination in dioecious plants. Adv. Genet. 9 217–281. [DOI] [PubMed] [Google Scholar]
- White, M. J. D., 1973. Animal Cytology and Evolution. Cambridge University Press, Cambridge, UK.
- Wilkins, A. S., 1995. Moving up the hierarchy: a hypotheses on the evolution of a genetic sex determination pathway. BioEssays 17 71–77. [DOI] [PubMed] [Google Scholar]
- Williams, T. M., and S. B. Carroll, 2009. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 10 797–804. [DOI] [PubMed] [Google Scholar]
- Winge, O., 1927. The location of eighteen genes in Lebistes reticulatus. J. Genet. 18 1. [Google Scholar]
- Woram, R. A., K. Gharbi, T. Sakamoto, B. Hoyheim, L. E. Holm et al., 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 13 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woram, R. A., C. McGowan, J. A. Stout, K. Gharbi, M. M. Ferguson et al., 2004. A genetic linkage map for Arctic char (Salvelinus alpinus): evidence for higher recombination rates and segregation distortion in hybrid versus pure strain mapping parents. Genome 47 304–315. [DOI] [PubMed] [Google Scholar]
- Yamamoto, E., 1999. Studies on sex-manipulation and production of cloned populations in hirame, Paralichthys olivaceus (Temminch et Schlegel). Aquaculture 173 235–243. [Google Scholar]
- Yin, T., S. P. DiFazio, L. E. Gunter, X. Zhang, M. M. Sewell et al., 2008. Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res. 18 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, S., E. Okada, H. Umemoto, K. Tamura, Y. Uno et al., 2008. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 105 2469–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Q., R. Navajas-Pérez, E. Tong, J. Robertson, P. H. Moore et al., 2008. Recent origin of dioecious and gynodioecious Y chromosomes in Papaya. Trop. Plant Biol. 1 49–57. [Google Scholar]
- Zdobnov, E. M., C. von Mering, I. Letunic, D. Torrents, M. Suyama et al., 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298 149–159. [DOI] [PubMed] [Google Scholar]
- Zhang, L., M. P. Simmons, A. Kocyan and S. S. Renner, 2006. Phylogeny of the Cucurbitales based on DNA sequences of nine loci from three genomes: implications for morphological and sexual system evolution. Mol. Phylogenet. Evol. 39 305–322. [DOI] [PubMed] [Google Scholar]
- Zhang, L. S., C. J. Yang, Y. Zhang, L. Li, X. M. Zhang et al., 2007. A genetic linkage map of Pacific white shrimp (Litopenaeus vannamei): sex-linked microsatellite markers and high recombination rates. Genetica 131 37–49. [DOI] [PubMed] [Google Scholar]
- Zhang, R., and H. Wu, 1985. A study of sex chromosome in Carassius auratus by BrdU-Hoechst 33258-Giemsa technique. Acta Genet. Sin. 12 373–378. [Google Scholar]
- Zhao, D., D. McBride, S. Nandi, H.A. McQueen, M.J. McGrew et al., 2010. Somatic sex identity is cell autonomous in the chicken. Nature 464 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]