Abstract
The predicted structure and molecular trajectories from over 80 ns molecular dynamics simulation of the solvated double super helix (DSH) model of nascent-high density lipoprotein (HDL) was determined and compared with experimental data on reconstituted nascent HDL obtained from multiple biophysical platforms including small angle neutron scattering (SANS) with contrast variation, hydrogen-deuterium exchange tandem mass spectrometry (H/D-MS/MS), nuclear magnetic resonance spectroscopy (NMR), cross-linking tandem mass spectrometry (MS/MS), fluorescent resonance energy transfer (FRET), electron spin resonance spectroscopy (ESR), and electron microscopy. In general, biophysical constraints experimentally derived from the multiple platforms agree with the same quantities evaluated using the simulation trajectory. Notably, key structural features postulated for the recent DSH model of nascent HDL are retained during the simulation including: 1) the super helical conformation of the anti-parallel apolipoprotein A1 (apoA1) chains; 2) the lipid micellar-pseudolamellar organization; and 3) the solvent exposed Solar Flare loops, proposed sites of interaction with LCAT (lecithin cholesteryl acyltransferase). Analysis of salt bridge persistence during simulation provides insights into structural features of apoA1 that form the backbone of the lipoprotein. The combination of molecular dynamics simulation and experimental data from a broad range of biophysical platforms serves as a powerful approach to study large macromolecular assemblies such as lipoproteins. The present application to nascent HDL validates the DSH model proposed earlier, and suggests new structural details of nascent HDL.
Keywords: Apolipoprotein A1, high density lipoprotein (HDL), molecular dynamics simulation, structure
High density lipoproteins (HDL) are complex macromolecular assemblies that function to transport free and esterified cholesterol cargo from periphery cells to liver for ultimate elimination from the body through a complex physiological process called reverse cholesterol transport (1, 2). HDL continues to be the focus of intense clinical, biological and structural investigations due to substantial evidence indicating a crucial role for the lipoprotein in atherosclerosis (3–5). HDL historically are defined by their buoyant density characteristics, and constitute a complex array of apolipoprotein A1 (apoA1) containing particles that differ based upon their degree of maturation and composition, including the suite of proteins associated with the HDL particle. The heterogeneous nature of HDL, coupled with the plasticity and dynamics of the molten particle, are essential features for lipid carrying biological function. However, these same characteristics have impeded detailed structural interrogation of HDL and the ability to relate specific physiological functions of the particle to defined structural features. Theoretical models of nascent HDL, an early form of HDL formed upon lipidation of apolipoprotein A1 by the ABCA1 cell transporter, have evolved as new experimental data has accrued and been applied toward refinement of existing models (e.g. Picket Fence (6), Double Belt (7, 8), various Hairpin Loop types (9–11), Solar Flares (12), and most recently, the Double Super Helix model (DSH)(13)).
Traditional high resolution structural approaches such as X-ray crystallography and NMR have thus far proven elusive for structural characterization of intact HDL. Consequently, many global structure-function relationship of the lipoprotein have been inferred by molecular dynamics simulations, and most of the all atom structures of nascent HDL proposed thus far are computational models with limited experimental data (14–22). A growing movement within the structural biology field for the study of macromolecular complexes resistant to traditional high resolution approaches is the concept of combining synergistic data from multiple diverse experimental techniques through computationally intensive theoretical methodologies. In theory, it is thus possible to enhance lower resolution models to greater refinement, bridging gaps in results and conclusions from individual platforms (13, 23–25).
For example, mechanistic understanding of membrane protein structure and function can be achieved without crystallographic or alternative high resolution data by combining biophysical/biochemical constraints with computational analyses and cryo-EM low-resolution data (26). In this spirit, we have recently proposed a new structural model of nascent HDL, the Double Super Helix model (DSH), by combining for the first time small angle neutron scattering (SANS) with contrast variation and biophysical constraints from multiple platforms with computational modeling (13). Low-resolution structures of protein and lipid components of nascent HDL were directly visualized by SANS with contrast variation and selective isotopic deuteration of apoA1, and then enhanced by incorporating additional biophysical constraints (e.g. hydrogen/deuterium exchange tandem mass spectrometry (H/D-MS/MS), fluorescence resonance energy transfer (FRET), MS/MS cross-links, electron microscopy (EM) and electron spin resonance spectroscopy (ESR)).
Key structural features of the DSH model, namely the open helical shape of apoA1 and a non-bilayer organization of the lipid phase around the protein scaffold (13), are a dramatic departure from the widely held view of nascent HDL as a lipid bilayer disk enshrined by a circular belt of protein. While the DSH model proposed was derived from computational approaches that incorporated biophysical constraints from multiple diverse platforms, a detailed analysis of the model following prolonged molecular dynamics simulation has not yet been reported. In the present studies we more closely examine the relationship between available experimental data on reconstituted nascent HDL obtained by various biophysical approaches and an all-atom 80 ns molecular dynamics simulation analyses of the fully hydrated particle. The phospholipid : cholesterol : protein content of the reconstituted HDL particles examined in this study is comparable to that reported for ABCA1 generated nascent HDL (e.g. (27)), and structure/function studies of the reconstituted HDL particles by our group has previously shown they possess biological activities indistinguishable from HDL isolated from plasma, including promotion of cholesterol efflux activity, LCAT interaction and activation, anti-inflammatory and anti-apoptotic activity, and specific binding to the HDL receptor, scavenger receptor B1 (12, 13). Our main goal in this study was to investigate whether molecular dynamics simulations are congruent with SANS imaging data, i.e. we tried to answer the following question: is the helical shape of the protein component of nascent HDL thermodynamically feasible?
MATERIALS AND METHODS
Preparation and routine characterization of nascent HDL
All reconstituted nascent HDL (rHDL) preparations generated for SANS, NMR and EM analyses in the present studies were first characterized by an array of biophysical (e.g. dynamic light scattering, non-denaturing PAGE and far-UV circular dichroism), chemical (e.g. confirmation of chemical composition by LC/MS/MS analysis of select amino acids for protein quantification, cholesterol, phospholipid and cholate), and biochemical assays (i.e. total and ABCA1 dependent cholesterol efflux, interaction with the plasma enzyme lethicin cholesteryl acyltransferase (LCAT), binding to the HDL receptor (scavenger receptor B1, SRB1), and both anti-apoptotic and anti-inflammatory activities) to ensure generation of a relatively monodispersed, chemically and biologically defined, and functionally intact particle. A detailed presentation of the methods used to prepare rHDL and the above biophysical, chemical, biochemical and functional assays used for its characterization have recently been extensively described (13). In brief, human apoA1 was isolated from HDL recovered from plasma of healthy donors by ultracentrifugation, and lipid free human apoA1 was isolated by ion exchange chromatography of delipidated HDL (28). Recombinant human apoA1 was generated in E. coli (29) using the kanamycin resistant plasmid pET20b+ encoding 6xHis- tagged human apoA1 transformed into E. coli BL21(DE3) cells (13). Deuterated recombinant human apoA1 was similarly prepared from E. coli grown in >90% D2O and deuterated nutrients (13). Sodium cholate dialysis (30) was used to prepare rHDL using 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), cholesterol and apoA1 in a molar ratio of 100:10:1. Before use in SANS, EM or NMR studies, rHDL and isolated humans plasma derived HDL (pHDL) preparations were further purified by gel filtration chromatography using a large (approximately 3 in × 5 ft) Sephacryl S300 (GE healthcare) column.
Small angle neutron scattering analyses
SANS experiments were carried out on instrument KW2 (http://www.jcns.info/detailsKWS2), a classical pinhole camera that provides the highest neutron flux available at the Jülich Center for Neutron Science at the reactor FRM-II, (Garching, Germany). Data was collected from two positions of the detector (2 and 8 m from the sample) thus covering the momentum transfer (q) range from 0.008 to 0.35 Å-1. The wavelength λ of the neutron beam used was 4.5 Å. Similar to recently reported studies (13), samples were measured at multiple levels of contrast including those optimized to visualize the protein within rHDL (i.e. in 12% D2O, the match-point of lipid) and those optimized to visualize the lipid within rHDL (i.e. in 42% D2O, the match-point of protein). Analyses of the scattering intensities at each contrast level produced the radius of gyration, Rg (using Guinier approximation (31)), and the intensity extrapolated to 0 angle. First, the GNOM program (32) was used to deconvolute the scattering intensity curve into the distance distribution function, p(r), using the inverse Fourier transformation. The p(r) function was subsequently used for the program DAMMIN (32) to produce low resolution structures of both the protein and the lipid components of nascent HDL based on data from 12 and 42% D2O contrast levels, respectively. The signal to noise ratio of the sample in 12% D2O was enhanced using deuterated apoA1 for the reconstitution of the HDL particle.
31P NMR experiments on nascent HDL and small unilamellar vesicles
rHDL, pHDL and small unilamellar vesicles (SUV) comprised of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) generated by extrusion. POPC was selected for generating small unilamellar vesicles (SUV) because X-Ray diffraction studies have demonstrated this molecular species of glycerophospholipid preferentially adopts lamellar phase (33, 34). To prepare SUV, POPC in chloroform was initially dried under vacuum and then fully hydrated by addition of buffer (20 mM Hepes buffer, 100 mM NaCl in 10% D2O, pH 7.4) above the phase transition temperature (35). Following vortexing to disperse the hydrated lipids, SUV were generated by extrusion through polycarbonate filters with 0.4 mm, 0.1 mm, 0.05 mm and 0.03 mm pore size sequentially for multiple times using an Avanti Mini-Extruder Set (Avanti Polar Lipids, Alabaster, AL). Throughout all experiments, freshly prepared vesicles were maintained well above the phase transition temperature to avoid vesicle fusion. The hydrodynamic radius of SUV preparations was determined with a DynaPro-801 dynamic light scattering instrument as described (12), 31P NMR analyses of HDL and SUV preparations were performed on a Varian Inova 600 MHz spectrometer operating at 242.85 MHz equipped with a broadband probe. Spectra were measured with WALTZ-16 modulated proton decoupling (599.91 MHz) at 30 °C. The chemical shift positions of lipid dispersions and HDL were recorded relative to 85% H3PO4 (0.00 ppm).
Electron microscopy studies
Images of negatively stained HDL were performed as recently described (13) using a JEOL 1200 EX II microscope at 100 kV and selected negatives were digitized on a Zeiss scanner at a step size of 14 µm giving a pixel size of 3.5 Å at the specimen level. The resulting data were processed further with the Imagic package.
Solvation of the DSH model of nascent HDL
The Double Super Helix (DSH) model used as the starting structure of the simulation is deposited in the Protein Databank (PDB id: 2K2S). To shorten the simulation time, the all-atom model of nascent HDL (the DSH model) was converted to an united-atom model using well-established force fields within GROMACS molecular dynamics simulation program (36) to describe the interactions between atoms of molecules composing the lipoprotein and solvent. In this type of force field, (modified GROMOS87 force field in GROMACS (36)) hydrophobic hydrogen atoms, and the carbon atom they attach to (CH3, CH2 and CH), are replaced by united atoms that have a volume that accommodates all atoms in the group. To describe the interaction among atoms of phospholipids (POPC) a modified united-atom force field with special parameters for acyl chains was used (37, 38). Cholesterol was described by a united-atom force field developed by Höltje and Brandt (39). Next, the united-atom model of nascent HDL (referred herein as the DSH model) was solvated in water using a protocol as previously described (12). Briefly, the DSH model was immersed in a box of SPC water molecules (40). The distance from the particle to solvent box walls was set to 1.5 nm to minimize the interaction of the lipoprotein with its images in the replicas. Eight water molecules were replaced by sodium ions to neutralize the negative charge of the two chains of apoA1, and then 48 water molecules were substituted with 24 pairs of NaCl ions to produce a physiological salt concentration in the simulation box (300 milliosmollar). The solvated system was subjected to a series of energy minimizations, using GROMACS program, in which the system was allowed to relax gradually (first the solvent alone, then the lipid, and finally the protein). In the last step of energy minimization all atoms in the system were allowed to relax. The calculation of the total energy of the system has taken into account periodic boundary conditions for the solvated system, the cut-off for the van der Waals interactions was 1 nm, and for Coulomb interactions the Ewald summation was used as implemented in the PME method (41).
Molecular dynamics simulation on solvated nascent HDL
Prior to the 80 ns simulation of HDL, a preliminary 100 ps simulation of the particle was performed where movement of all non-hydrogen atoms of the lipoprotein were restrained in order to allow the solvent to rearrange and start equilibrating. In addition, the change in temperature, total energy, and the root mean square displacement (RMSD) of protein backbone was monitored during the entire simulation. By 20 ns all of these variables achieve a steady state level. The simulation was performed in a canonical ensemble, NVT (i.e. the variable of states number of particles, N, volume, V, and absolute temperature, T, were maintained constant). The simulation T was maintained at 300 K using the Berendsen thermostat (42). Random initial velocities were assigned for T=300K. The bonds between atoms in protein and lipid components were reset to their correct value by using the LINCS algorithm (43). Both the bond lengths and the bond angles in the water molecules were constrained by the SETTLE algorithm (44). The time integration step was 2 femtosecond and a coordinates frame was saved each 10 ps. All snapshots (over 8 thousands) constitute the trajectory of the particle during simulation, which was used to evaluate the quality of the simulation and the thermodynamic stability of the lipoprotein-solvent system. A trajectory made of 21 snapshots taken every ns between 0 and 10 ns, and every 5 ns between 15 and 60 ns was used for time consuming analyses. While the initial 20 ns of simulation can be seen as system equilibration, this period was included in the analysis in order to have a comprehensive understanding of where the model started, versus its steady state position following prolonged molecular dynamics simulation. This allowed us to more readily compare the impact of the simulation trajectory on global architecture of apoA1, as well as the experimentally derived DSH model versus the penultimate model derived following the prolonged molecular dynamics simulation.
Calculation of hydrogen-deuterium exchange incorporation factors from simulation trajectory
Hydrogen-deuterium exchange incorporation factors (Di0) were calculated for all residues of apoA1 using experimental data from overlapping apoA1 peptides of nascent HDL analyzed by H/D-MS/MS, as described in our previous publications (12, 13). To calculate per residue Di0 values for all snapshots extracted from the simulation trajectory of apoA1 we used the H/D exchange methodology implemented in the program DEXANAL (12, 13). This methodology was validated by previously applying it to a set of protein crystal structures with published NMR derived H/D exchange rate constants (kxci), and/or H/D-MS/MS derived D0 values for peptides (13). For completeness we briefly describe here the theoretical formalism behind this methodology. The program DEXANAL uses experimentally measured D0 values for overlapping peptides determined by H/D-MS/MS (in the case of apoA1 of rHDL there was >95% coverage), and a molecular model to produce a set of H/D exchange probabilities (XPi) that are used to predict per residue deuterium incorporation factors (D0i), residue unfolding equilibrium constants (Kui), and H/D exchange rate constants (kxci). Gauging the difference between experimental and calculated D0 values assesses the input model.
The H/D exchange rate constant, kxci, for each residue i relates to the experimentally-derived D0i and the exchange time, t, by the following equation:
| (1) |
The residue unfolding equilibrium constant, Kui, (the reciprocal of the NMR protection factor) is the ratio of kxci and the intrinsic H/D exchange rate constant (kchi) of individual residues in unstructured loops (45, 46):
| (2) |
For an ensemble of protein conformations, XPi defines the ratio of the number of protein molecules (NDi) where the backbone amide H of residue i exchanged to D, to the total number of molecules:
| (3) |
For a single conformation (and not an ensemble), XPi for each residue i can be calculated as the product of two quantities (RAIi, BAIi) encoding structural information about residue i's size, chemical composition, and interactions with the rest of the system:
| (4) |
This basic equation incorporates information about the degree of protection (both local solvent accessibility and dynamics) of the backbone amide H of residue i. BAIi and RAIi are the backbone and residue accessibility/dynamics indices, respectively, with values between 0 and 1. The probability correction factor (PCFi), is the ratio of the experimentally determined per residue deuterium incorporation factor D0i (obtained by partitioning the experimentally measured D0 for peptic peptides) and the calculated probability XPi:
| (5) |
PCF gauges the difference in protection to exchange of a backbone amide H atom in the molecular model with respect to the same H in the real structure, and is used to refine the conformation of individual residues in the model (i.e. adjust the protection of amide H’s). Indices BAI and RAI are calculated using an approach similar to COREX (47).
The BAI index is the ratio of the atomic (van der Waals) surface area of the backbone amide H atom in the model (AHi)model and the random coil conformation (AHi)rc, respectively.
| (6) |
If the amide H is involved in H-bonding BAI is modified by an exponential term that includes the length of the H-bond (dHB).
The RAIi index of residue i is the ratio of the solvent accessible surface area of the residue in the model (Ai)model and in the random coil (Ai)rc conformation, respectively:
| (7) |
RAI is modulated by a factor fiPD that takes into account the dynamics of the protein (12, 13), present in experimental data but not accounted for in a single frozen conformation.
Calculation of patch-hydrophobicity and solvent accessible surface area (SASA)
Patch hydrophobicity (PH) is a calculated per-residue index based on the hydrophobic indices of amino acid residues that takes into account the local environment of the residue within the predicted three-dimensional structure (48). PH was determined for each amino acid residue by averaging the hydrophobic indices of neighboring residues (within 15 Å) located on the same side of protein, i.e. either inside (facing lipid) or outside (facing solvent) (13). PH is thus a normalized residue index of solvent accessibility in range [0,1] that incorporates local structure and accounts for the fact that a hydrophobic residue exposed to solvent, when surrounded by mostly hydrophilic residues, destabilizes the local conformation of the protein to a lesser extent. Solvent accessible surface area (SASA) of the protein was calculated using the MSMS program (49) and was separated into hydrophobic and hydrophilic contributions based on the hydrophobic index of amino acid residues. The SASA exhibited by the acyl chains comes mainly from regions of acyl chains exposed to solvent (50) and to a lesser extent from the fact that acyl chains pack less dense than polar head groups and protein (51). Inspection of the simulation box shows that no water molecule was found inside the lipid core.
RESULTS
ApoA1 retains a super helical conformation during the entire simulation
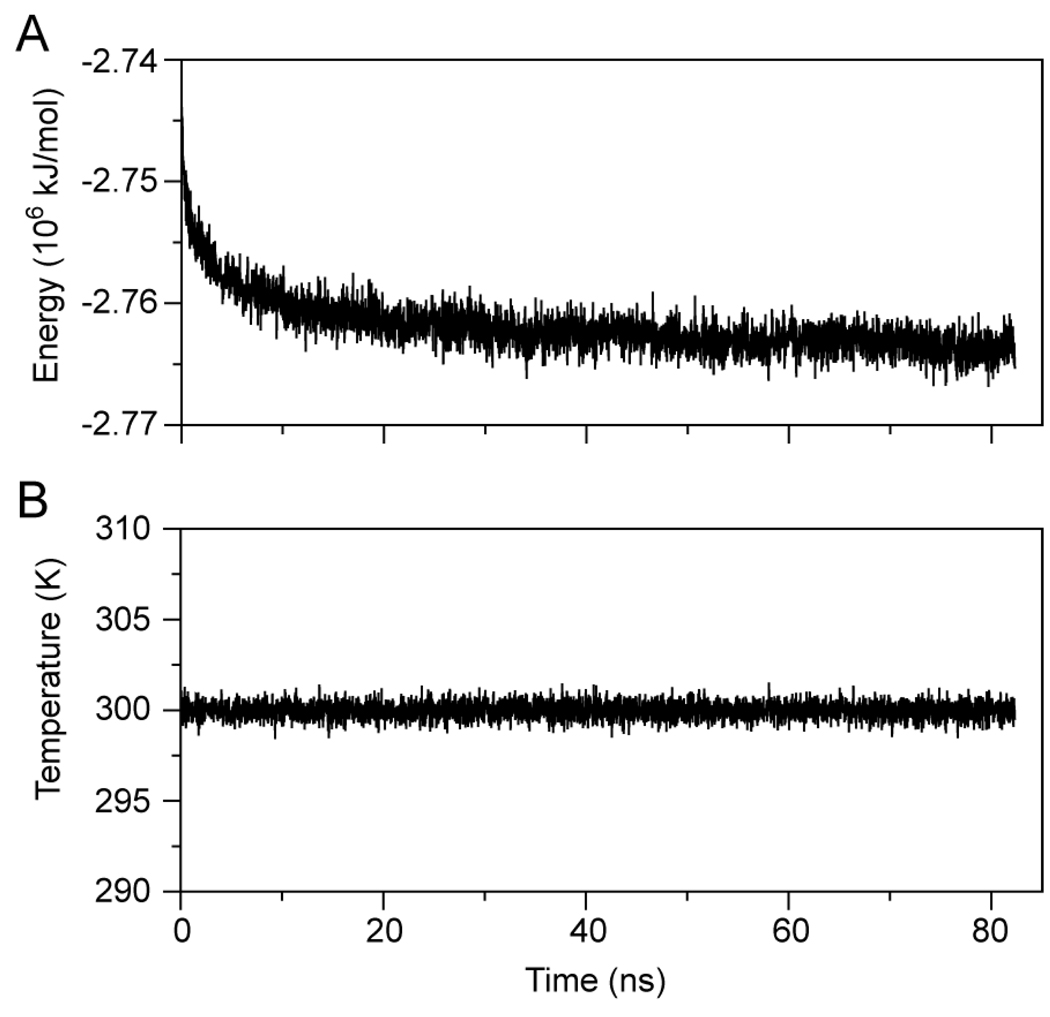
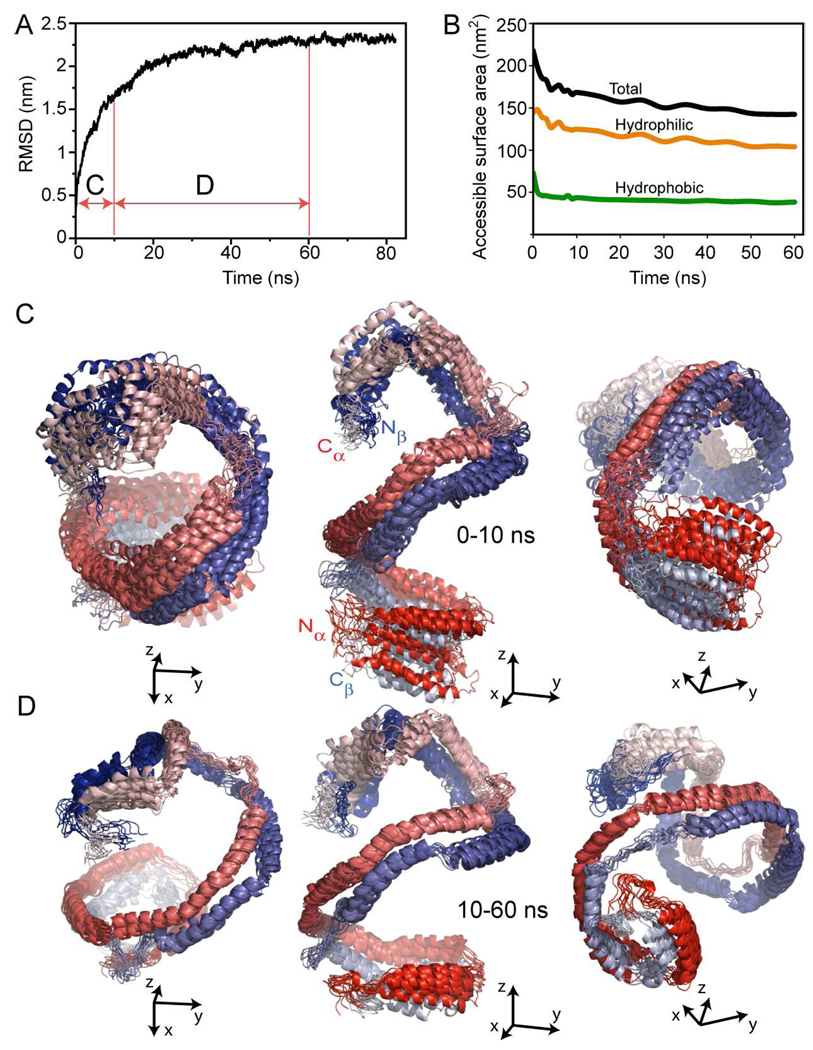
Nascent rHDL in solution reached thermodynamic equilibrium (i.e. constant average total energy (E), Figure 1A) after roughly 20–30 ns simulation. The average T of the system remained constant (300 K, Figure 1B) during the entire simulation. Analysis of the root mean square displacement (RMSD) of the apoA1 backbone during simulation indicates that the most significant change in the conformation of apoA1 (i.e. the decrease in the pitch of the overall helix) takes place during the first 10 ns (domain C of Figure 2A), after which the RMSD reaches a plateau (domain D of Figure 2A). Figure 2B shows the change in solvent accessible surface area (SASA) calculated throughout the simulation. Most of the SASA of apoA1 (~80%) corresponds to solvent exposed hydrophilic amino acid residues. It is evident that the proper orientation of apoA1 amphipathic chains during simulation contributes to the thermodynamic stability of the HDL particle as confirmed by the minimal variation in E (Fig. 1A).
FIGURE 1.
Energy and temperature fluctuations during the molecular dynamic simulation of nascent HDL in solution. A, Fluctuation in the total potential energy of the simulation cell. The graph shows that it takes about 30 ns simulation for the system to reach thermodynamic equilibrium. B, Fluctuation in the absolute temperature; the system maintains in average the temperature setup for simulation (300 K).
FIGURE 2.
Root mean square deviation (RMSD) of apoA1 backbone from the conformation of apoA1 in the Double Super Helix model of nascent HDL. A, Fluctuation in the root mean square displacement (RMSD) of apoA1 backbone conformation during simulation. The graph shows that most of the conformational change in apoA1 backbone occurs in the first 10 ns of simulation. B, Change in the total (black curve), hydrophilic (orange curve) and hydrophobic (green curve) solvent accessible surface area (SASA) of apoA1 during simulation. Most of solvent accessible surface of apoA1 (~80%) is made of hydrophilic amino acid residues. B, Superposition of 11 simulation snapshots taken 1 ns apart during the first 10 ns. ApoA1 chains are gradient colored red (chain A) and blue (chain B) with the N-termini colored dark red/blue and C-termini colored light red/blue. The left and right panels show the same superposition of frames from different vantage points with the N-terminal of chain A pointing outward in the left panel, and the N-terminal of chain B pointing outward in the right panel. C, Conformational changes of apoA1 backbone during the next 50 ns simulation: the three panes confirm the result in Fig 2A, that is, there is little change in the conformation of apoA1 backbone after the first 10 ns simulation. In the left and right panels the N/C-termini pairs of apoA1 point outward emphasizing the helical shape of apoA1, which is retained during the entire simulation.
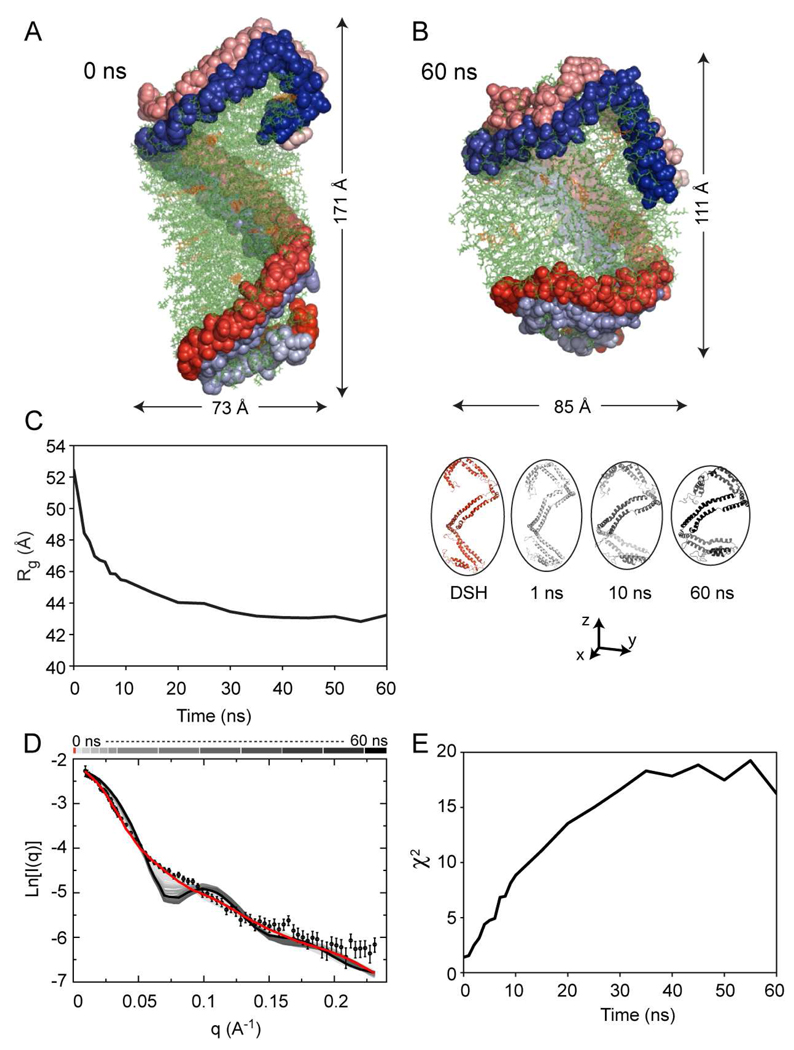
The overall conformation of apoA1 chains and the changes that occur during the molecular dynamics trajectory are illustrated in Figures 2C (first 10 ns) and 2D (10 ns though 60 ns). During the first 10 ns of the simulation, there is a noticeable decrease in the particle’s main axis (from 171Å to 111Å) and an increase of the shorter dimension (from 73Å to 85Å) leading to a more spheroidal shape. There is very little change in the conformation of apoA1 after 10 ns and the protein retains the overall helicity and conformational asymmetry during more prolonged simulation (Figure 2D). Importantly, despite the change in overall dimensions of the particle (from an elongated ellipsoid to a spheroid), the overall architecture of apoA1 remains unchanged. Namely the protein retains: 1) the overall helical conformation as proposed for the DSH model of nascent HDL; 2) the secondary structure (i.e. the same degree of α-helicity); and 3) a global conformational asymmetry (i.e. the two associated N/C-termini fold back and interact with the rest of the protein distinctly). We hypothesize that this asymmetry of apoA1 proposed for the first time in the DSH model is characteristic to nascent HDL, and is reflected in several structural features of the protein conformation, which will be discussed later (vide infra).
Simulation confirms that micellar/pseudo-lamellar organization of lipids in nascent HDL is thermodynamically stable
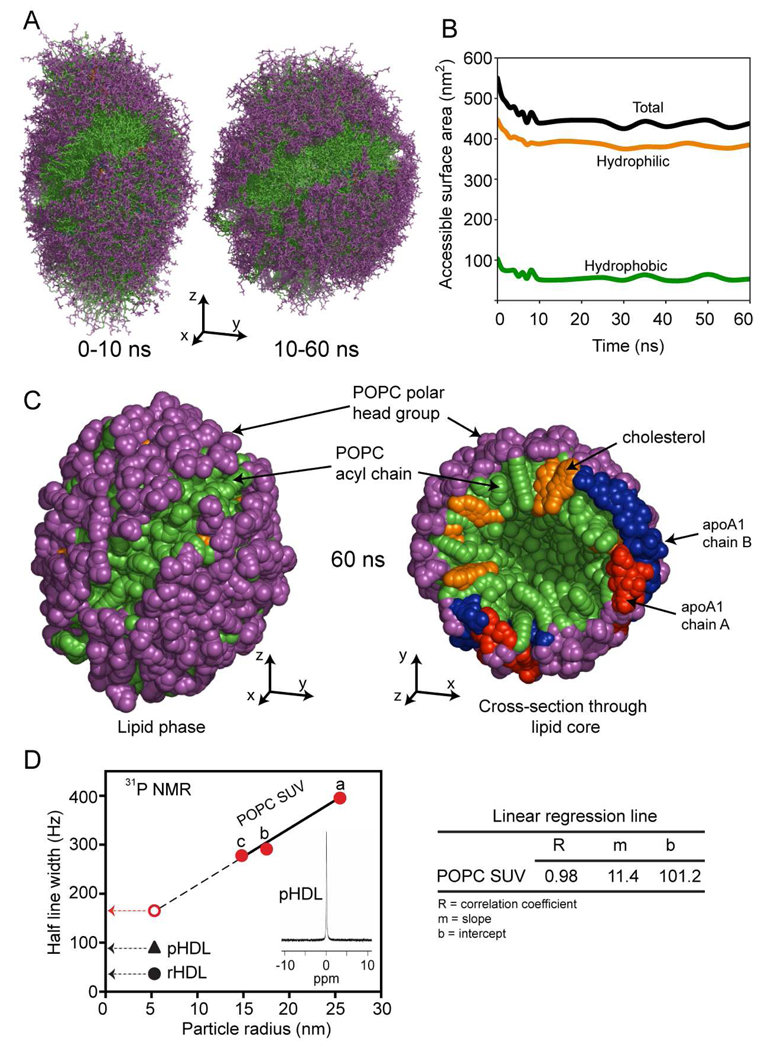
Similar to the analysis of apoA1 trajectory, Fig. 3A shows the superposition of various models of the lipid phase (POPC and cholesterol) corresponding to simulation snapshots. The left panel displays the superposition of 11 snapshots taken in the first 10 ns of simulation, while the right panel shows the superposition of 11 snapshots taken during the next 50 ns. While the change in shape from prolate ellipsoid to spheroid is visible during the first 10 ns, it is worth noting that the overall lipid organization (micellar/pseudo-lamellar packing) proposed for the DSH model of nascent HDL is retained during this part of the simulation and the lipid remains in the proper orientation with respect to solvent, i.e. polar head groups are oriented outward (toward solvent) while acyl chains are oriented inward (toward each other) and toward the hydrophobic surfaces of apoA1.
FIGURE 3.
The simulation trajectory of the lipid phase component of nascent HDL. A, Left panel: superposition of 11 snapshots taken during the first 10 ns simulation. The polar head groups of POPC are colored purple and the acyl chains are colored green. Cholesterol is colored orange. Right panel: superposition of 11 snapshots taken during simulation in the next 50 ns (from 10 to 60 ns). The superposition shows that the lipid core becomes more spheroidal, and that the most significant change in its shape takes place during the first 10 ns. B, The change in the hydrophobic (green), hydrophilic (orange) and total (black) solvent accessible surface area during simulation. C, Left panel: The packing of the lipid core after 60 ns simulation. The polar heads of POPC are shown as purple beads, while the acyl chains are shown as green beads. Cholesterol molecules are colored orange. Right panel: cross section through the lipid core and apoA1 double chain (red/blue). The panel shows the micellar-lamellar structure of the lipid core with the POPC polar heads groups oriented radially toward solvent and acyl chains pointing inside. D, Plot of half-line width of the 31P NMR spectrum recorded for multiple distinct small unilamellar vesicles (SUV) of differing size produced from POPC, reconstituted HDL (rHDL) and nascent HDL isolated from human plasma (pHDL). Preparation of SUV with diameters greater than 20 nm were generated by extrusion through polycarbonate filters with 0.4 mm, 0.1 mm, 0.05 mm and 0.03 mm pore sizes sequentially for 15 times. Preparation of POPC SUV with diameter 17.6 nm was generated similarly as POPC SUV with diameter of 25. 6 nm, but with 30 times extrusion. Preparation of POPC SUV with diameter of 14.9 nm was generated using cholate dialysis method.
Analyses of SASA during the trajectory of the simulation confirm that the lipid polar head groups and hydroxyl group of cholesterol contribute the most to SASA (Fig. 3B). The fact that the major contribution to solvent accessible surface comes from lipid polar head groups and is retained during simulation indicates that the micellar/pseudo-lamellar organization of the lipid in nascent HDL is thermodynamically stable when bound to protein. The left panel of Figure 3C shows the overall structure of the lipid core after 60 ns simulation (polar head groups in purple, fatty acyl chains in green). The visible diagonal green stripe on the lipid surface corresponds to the place where the anti parallel apoA1 chains are located in the model. The right panel displays a cross-section through the HDL particle, which reveals the micellar/pseudo-lamellar packing of the lipid with polar head groups pointing radially outward, and acyl chains and sterol core of cholesterol pointing inward. The protein chains are visible at the periphery of the particle, and cholesterol molecules are shown to intertwine with the acyl chains.
A noticeable feature of the simulated lipid phase is the presence of a central empty cavity. Inspection of the simulation box shows that no solvent molecule penetrated the lipid layer, and the cavity is empty (i.e. free of solvent). It is noteworthy that previous experimental scattering studies of micellar systems have shown the existence of a central void within micellar systems (52, 53), and molecular dynamics simulations of micellar systems have similarly predicted the presence of such voids (51, 54, 55), with ellipsoidal to spheroidal shape changes depending upon subtle changes in the molecular dynamics force fields used (56, 57). Prior molecular dynamic simulations of micellar systems (56, 57) suggest that the change in shape may be attributed to an imbalance between polar interactions among polar head groups of the lipid, on the one hand, and dispersion interactions among acyl chains, on the other hand (vide infra).
31P NMR chemical shift and half line width of POPC point to micellar packing of lipids in nascent HDL
The micellar organization of lipids posited in the DSH model of HDL and the present molecular dynamics simulation studies suggests a highly dynamic environment for the phospholipids within rHDL. We therefore sought to perform experimental studies to directly address this possibility. Prior studies have shown that 31P NMR of the polar head group phosphate moiety of phospholipids can distinguish between different smectic mesophases (molecular packing arrangements) since lamellar, micellar and hexagonal array arrangements of phospholipids demonstrate distinct 31P NMR spectral features including distinct chemical shifts, line shapes (symmetric vs. asymmetric with up-field or down-field tails) and line widths (33, 35, 58, 59). 31P NMR analyses of both nascent rHDL and pHDL demonstrate a sharp isotropic peak at 0 ppm (Fig. 3D, and inset). The chemical shift, line width and symmetrical shape are all consistent with a micellar smectic mesophase (33, 35, 58).
While the chemical shift of the 31P NMR spectra for HDL preparations indicates a micellar and non-lamellar organization, a small particle rapidly tumbling in all orientations could theoretically contribute to the isotropic spectral features observed for both pHDL and rHDL. We therefore compared the 31P NMR half-line width of the phospholipid within HDL with the spectral characteristics of phosphate within SUV of differing sizes. SUV of different radii (measured with dynamic light scattering) were made with the most abundant phosphatidylcholine molecular species present within rHDL and pHDL (POPC) by passage of the lipid multiple times through different extrusion pore sizes (Methods). POPC was chosen for generating SUV of varying radii because it is known to have strong lamellar phase packing preference as demonstrated by X-ray diffraction (34). The line width (in Hz) at half peak height observed during 31P NMR analyses of each vesicle preparation and both rHDL and pHDL are shown in Fig. 3D. As expected, a linear relationship (R=0.98) was observed between particle radius and 31P NMR line width for varying sized SUV (Fig. 3D). Extrapolating to the half-line width predicted for a vesicle (bilayer lipid packing) the same size of a nascent HDL particle (radius of gyration 51.3Å), the half-line width predicted for a lamellar phase particle is considerably larger than that observed with either rHDL or pHDL (Fig. 3D). Collectively, these results demonstrate that the environment experienced by the phosphate group of phospholipid within HDL is highly dynamic with greater isotropic motion than would be observed with a similar sized bilayer (SUV) organized particle.
Repeat small angle neutron scattering and electron microscopy studies visualize nascent HDL as a prolate ellipsoid
The shape changes of the HDL particle during simulation from elongated ellipsoid, as proposed in the DSH model (13), to spheroid. This is the only simulation result that differs from experimental evidence with all other features of SANS and other biophysical platforms (as outlined below) matching the simulation results. This was surprising since the prolate ellipsoid shape of the DSH model is based upon experimental observation, i.e. direct visualization of the particle by small angle neutron scattering (SANS) and electron microscopy (EM) (13).
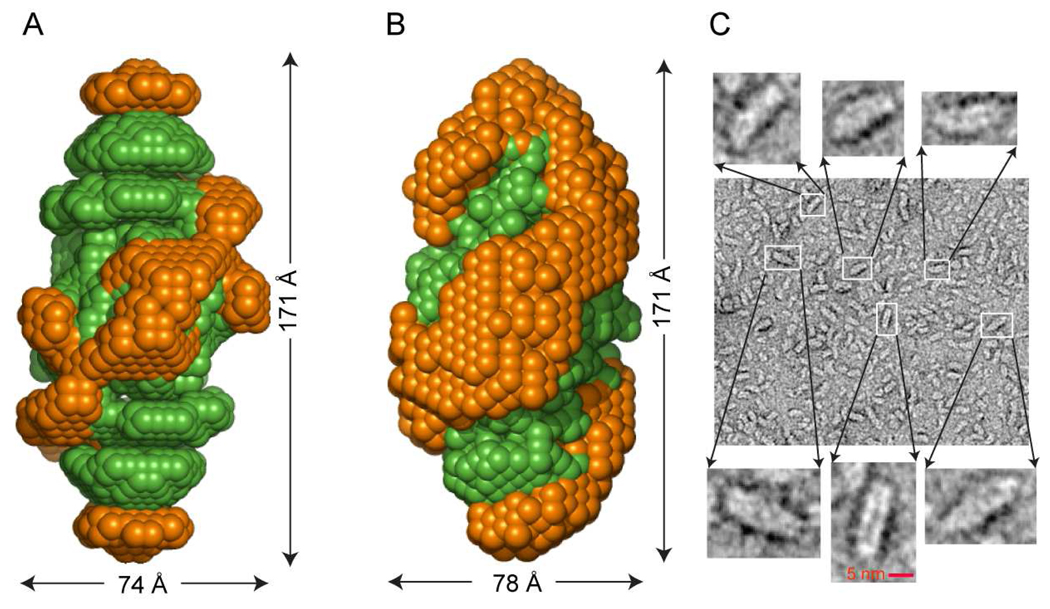
To experimentally reassess the global shape of rHDL new SANS experiments with contrast variation were performed. The experimental scattering data and predicted scattering curve for protein (at 12% contrast) and lipid (at 42% contrast) are shown in Supplemental Fig 1. Comparison of the low resolution structures of protein and lipid components of rHDL from the original SANS studies used for generation of the DSH model (Fig. 4A), and the new SANS shapes for protein and lipid obtained from experiments on new rHDL preparations (Fig. 4B) reveals striking similarity. The shape of the protein has the same spatial characteristics, i.e. a helix that wraps around a prolate ellipsoid lipid core. In addition, EM micrograph analyses of rHDL preparations (Fig. 4C) confirm that the rHDL particles are elongated ellipsoids of about 165–170 Å, in close agreement with the overall dimensions of the ellipsoid obtained from independent sets of SANS analyses. We conclude from the direct experimental observations using both SANS and EM that the overall shape of rHDL is a prolate ellipsoid.
FIGURE 4.
Low resolution structures of nascent HDL from small angle neutron scattering (SANS) and images from electron microscopy; the Double Super Helix (DSH) model and the model obtained after 60 ns simulation. A, Superposition of SANS low resolution structures of apoA1 (orange) and lipid core (green) of nascent HDL obtained from processing scattering intensities collected at Intitut Laue-Langevin in Grenoble, France as previously reported (13). B, Repeat low resolution structures for protein and lipid within rHDL preparations obtained from data collected at JCNS, Garshing, Germany, in the present studies. C, Electron microscopy micrograph of rHDL preparations. Detailed magnified insets show that the majority of the particles are prolate ellipsoids.
Further comparison of experimental SANS data, obtained from samples examined at 12 % D2O contrast, with theoretical results derived from simulation is illustrated in Fig. 5, along with the change of apoA1 shape and of the overall HDL particle during the course of simulation from an elongated ellipsoid (DSH model, Fig. 5A) to a spheroid (60 ns, Fig. 5B). During simulation, the radius of gyration, Rg, decreases from 52.3 Å to about 44 Å with the majority of change in Rg occurring during the first 10 ns of the simulation (Fig. 5C). A comparison of the experimental and calculated SANS scattering intensities during the molecular dynamics trajectory is shown in Figure 5D. The predicted scattering intensities of various simulation-derived models show that the scattering curves depart from the experimental data as simulation progresses, and the departure correlates with the change in the Rg of the model. In addition, the scattering curves for the simulation models develop humps, which accentuate as the simulation progresses. Figure 5E displays the goodness of fit (χ2) of experimental vs. calculated intensities for various simulation models during the course of the molecular simulation. It is worth noting that during the first 30 ns the goodness of fit deteriorates monotonically, and then starts oscillating, an indication that apoA1 starts changing conformation back to some extent. For example, the models obtained after 30 and 60 ns have the same χ2.
FIGURE 5.
Comparison of experimental and calculated small angle neutron scattering (SANS) intensities for the Double Super Helix (DSH) model of nascent HDL, and models obtained during 60 ns simulation. A, The DSH model of nascent HDL; apoA1 is represented with spheres with the two chains colored in red and blue fading from the N to the C terminal. The phospholipid is colored green and free cholesterol is colored orange. B, The model of nascent HDL obtained from simulation after 60 ns. C, Left: The radius of gyration (Rg) during the simulation trajectory is shown. Right: Conformations of apoA1 (cartoon representation) in the DSH model (red) and after 1, 10 and 60 ns simulation. D, Comparison of the experimental scattering intensity (open circles with error bars) with those obtained by calculation from the DSH model (red line) and all 22 snapshots during the simulation trajectory (various shades of gray). The gray shades bar at the top of the graph color codes the intensity curves: light gray shades are used for conformations of apoA1 at the beginning of the simulation and dark gray shades are used for conformation towards the end of the simulation. The intensity line corresponding to the last simulation snapshot (60 ns) is colored black. E, The χ2 statistics, that gauges goodness of fit between the experimental and theoretical intensities, is shown as a function of simulation time. χ2 = 1.37 for the DSH model.
H/D exchange predicted from simulation agrees with experimental data
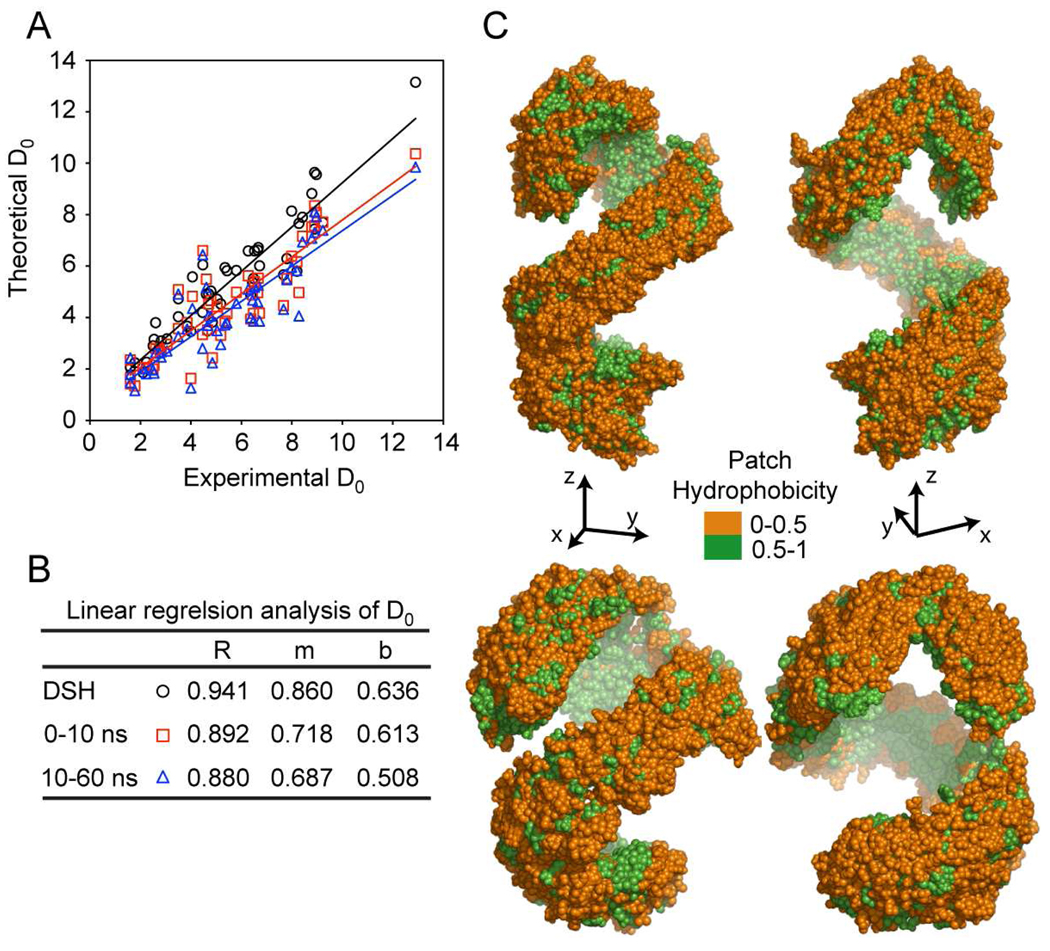
The analysis of the fluctuations in apoA1 backbone amide H/D exchange rate constants (kex), deuterium incorporation factors (D0) and per residue unfolding constants (Ku) during the course of the simulation indicates that the average D0 predicted from simulation are consistent with experimental data (Fig. 6, Supplemental Table 1). Comparisons of experimental and predicted D0, correlation coefficients (R) and patch hydrophobicity (PH) for trajectory 0–10 ns and 10–60 ns are also shown (Fig. 6). Notably, the trajectory analyses show D0 and R in close agreement with the same experimental quantities, and that the partition of the amino acid residues (based on PH made in the DSH model) is retained during the entire simulation. The overall goodness of fit of the experimental H/D exchange data derived from analysis of rHDL and the calculated H/D exchange data for the starting DSH model and molecular dynamics simulation models of nascent HDL are shown in Table 1. Both the correlation between the experimental and theoretical H/D exchange incorporation factors of peptic peptides (D0) and the RMSD of the difference between the experimental H/D exchange data and the calculated ones show excellent agreement throughout the course of the simulation.
FIGURE 6.
Comparison between experimental and calculated deuterium incorporation factors (D0) using molecular dynamics trajectory snapshots. A, Experimental vs. theoretical D0 values and regression lines corresponding to the Double Super Helix model (black, open circle), the average over 11 snapshots in the first 10 ns (red, open square), and average over 11 snapshots between 10 to 60 ns (blue, open triangle). B, The correlation coefficients (R) between experimental vs. theoretical D0 values, and the slope (m) and the intercept (b) of the linear regression lines for the D0 values. C, Top: Patch hydrophobicity of apoA1 in nascent HDL from simulation frames in the range 0–10 ns. The hydrophilic surface is colored orange and the hydrophobic surface is colored green. Bottom: Patch hydrophobicity of apoA1 in nascent HDL from simulation frames in the range 10–60 ns.
Table 1.
Goodness of fit between experimental hydrogen / deuterium exchange data and the calculated hydrogen / deuterium exchange data for the Double Super Helix and molecular dynamics simulation models of nascent HDL
| Model | Correlation coefficient a | RMSD b |
|---|---|---|
| Double Super Helix | 0.946 | 0.82 |
| 0–10 ns c | 0.892 | 1.47 |
| 10–60 ns d | 0.880 | 1.70 |
| 0–60 ns e | 0.888 | 1.56 |
Correlation between the theoretical hydrogen / deuterium exchange incorporation factors of peptic peptides (D0) in the DSH or simulation model and the actual experimentally measured deuterium incorporation within apoA1 peptic peptides.
RMSD, root-mean square deviation, quantifies the difference between the experimental hydrogen / deuterium exchange data and the calculated ones. Smaller RMSD indicates a better agreement between experiment and theory.
Eleven snapshots in the range 0–10 ns separated by 1 ns were used to perform H/D-MS/MS calculations and average over all snapshots.
Eleven snapshots in the range 10–60 ns separated by 5 ns were used to perform H/D-MS/MS calculations and average over all snapshots.
Twenty two snapshots in the range 0–60 ns have been used to perform H/D-MS/MS calculations and average over all snapshots.
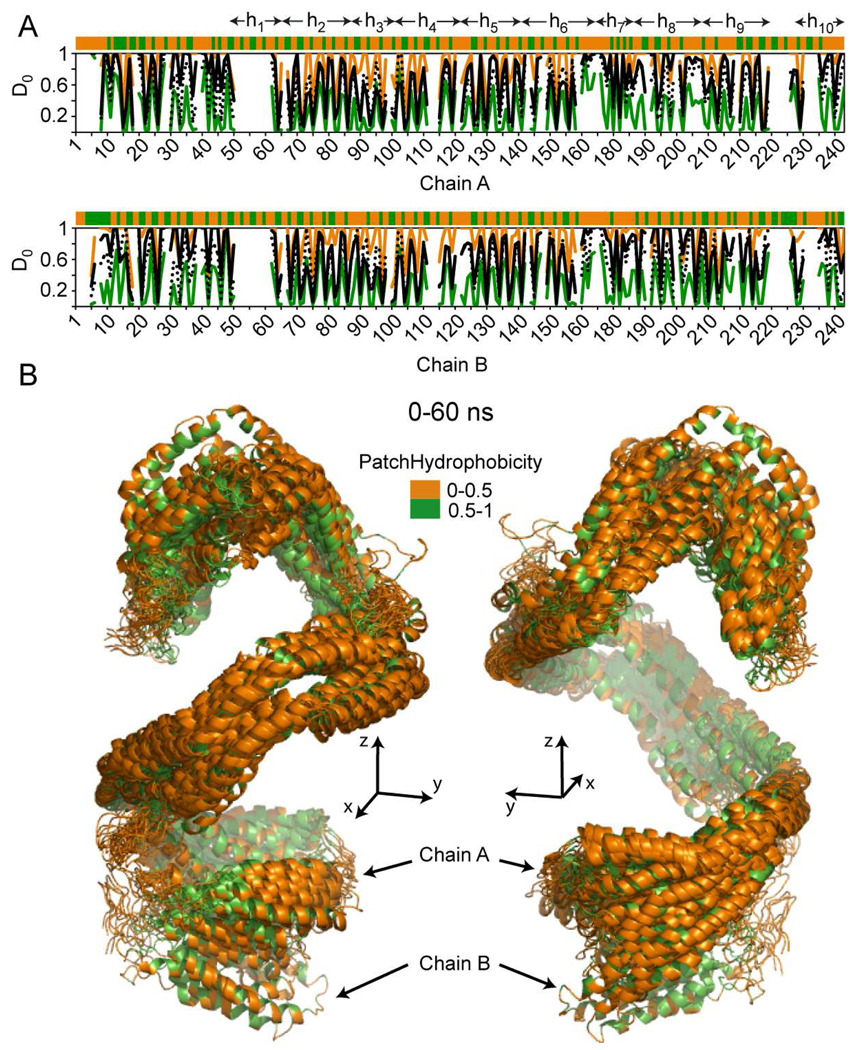
A residue level analysis of the H/D exchange data and the correlation with PH of individual residues is shown for the entire particle in Figure 7, and for two distinct regions of the lipoprotein in Figures 8 and 9. Figure 7A displays a combination of average per residue PH (top stripe) and both simulated and experimentally derived per-residue D0i (graphs below) for apoA1. Data for the 0–60 ns simulation shown are the maximum (in orange), minimum (in green) and mean (in black dotted) calculated D0i values plotted versus residue number, with superimposed experimentally determined D0i (solid line) from H/D-MS/MS analyses of rHDL. All values show remarkable periodicity that correlates with the secondary structure of the protein (vide infra). Figure 7B displays the 60 ns trajectory of apoA1 made of all 21 snapshots (see Methods), and the overlaid snapshots show that most hydrophilic residues (in terms of PH, shown in orange) are exposed to solvent, and the majority of hydrophobic residues (in terms of PH, shown in green) face inward where they are in contact with the lipid. This also points to the fact that the hydrophilic/hydrophobic partition of apoA1 residues proposed in the DSH model produces a thermodynamically stable structure.
FIGURE 7.
Hydrogen deuterium exchange incorporation factors (D0) and patch hydrophobicity of full length apoA1 in nascent HDL. A, Top: patch hydrophobicity (top orange/green stripe), and comparison between experimental (black line) and calculated D0 values for residues in of apoA1 chain A. The orange and green lines identify the maximum and minimum of D0 range of fluctuations calculated from simulation snapshots. The black dotted line represents the average of calculated D0 values obtained from all 22 snapshots taken during the first 60 ns simulation and used for analysis. On top of the patch hydrophobicity stripe the traditional α-helix domains on apoA1 (h1 to h10) are identified. Bottom: similar information is displayed for residues in chain B of apoA1. These two plots show that the average D0 obtained from simulation follows the same pattern as the experimentally-derived D0. B, Superposition of all 22 snapshots of apoA1 (chain A and B) taken during first 60 ns simulation; the apoA1 chains are shown in cartoon representation. The residues are colored according to their patch hydrophobicity (orange = solvent accessible residues and green = buried residues). Left and Right panels show the trajectory of apoA1 in two different orientations. In the left panel helix 5 is facing upfront, while in the right panel helix 5 is facing away from the viewer.
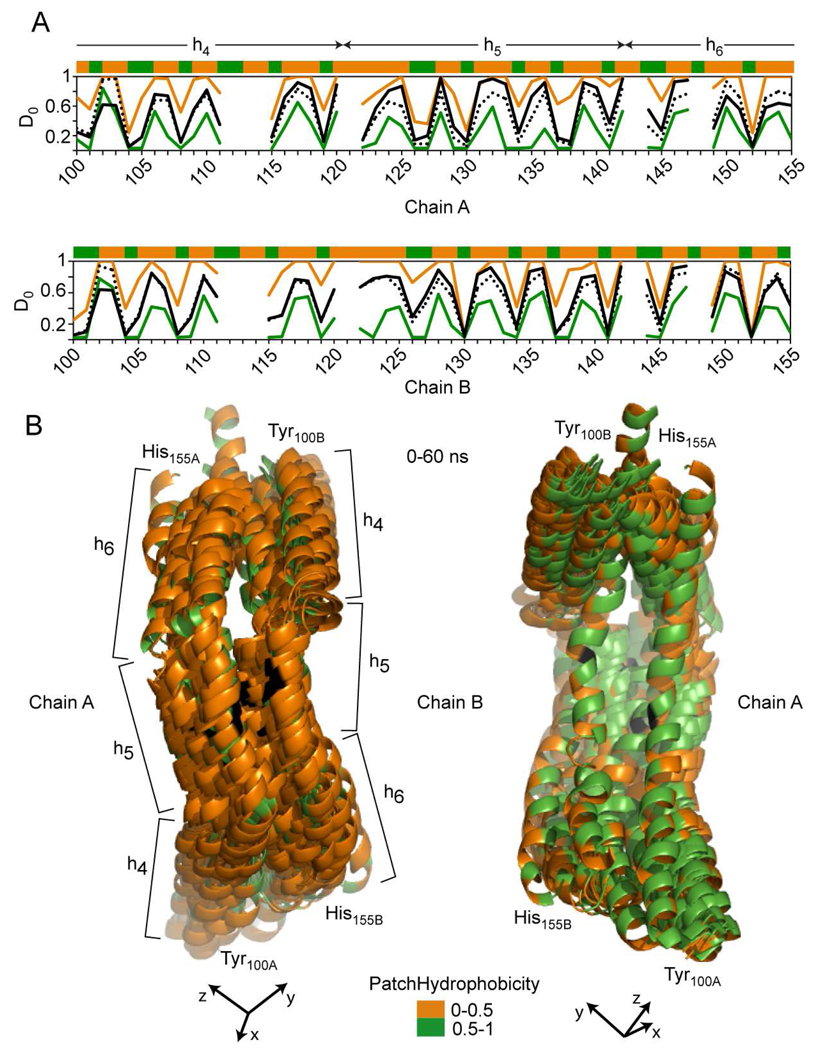
FIGURE 8.
Hydrogen deuterium exchange incorporation factors (D0) and patch hydrophobicity in an α-helix region of apoA1 (Y100-H155) that hosts helix h4 (P99-E120), helix h5 (P121-S142) and part of helix h6 (P143-A164). A, Top: patch hydrophobicity (top stripe), and comparison between experimental (black line) and calculated D0 values for residues in Tyr100-His155 region of apoA1 chain A. The orange and green lines identify the maximum and minimum for range of fluctuations of D0 values calculated from simulation snapshots. The black dotted line shows the average value of calculated D0 obtained from all 22 snapshots taken during the 60 ns simulation and used for analysis. Bottom: similar information is displayed for Tyr100-His155 region of chain B of apoA1. These two plots show that the average values of D0 obtained from simulation follow the same pattern as the experimentally-derived D0 values, and agree well with the latter. B, Superposition of all 22 snapshots of the Tyr100-His155 region (chain A and B) taken during 60 ns simulation. The apoA1 chains are shown in cartoon representation indicating that this region of the protein maintains α-helix secondary structure during the entire simulation. The residues are colored according to their patch hydrophobicity (orange = solvent accessible residues and green = berried residues). h5 of apoA1 is facing upward (out of the page) due to the curvature of the apoA1 chains. As a reference point, Gly129, the middle point of h5, is shown in black. The left, and right panels show the trajectory of Tyr100-His155 region in different orientations. The left panel shows the trajectory having the solvent accessible surface (orange) of residues facing upfront, while the right panel shows the trajectory with the buried residues surface (green) facing upfront. h5 is facing away (into the page).
FIGURE 9.
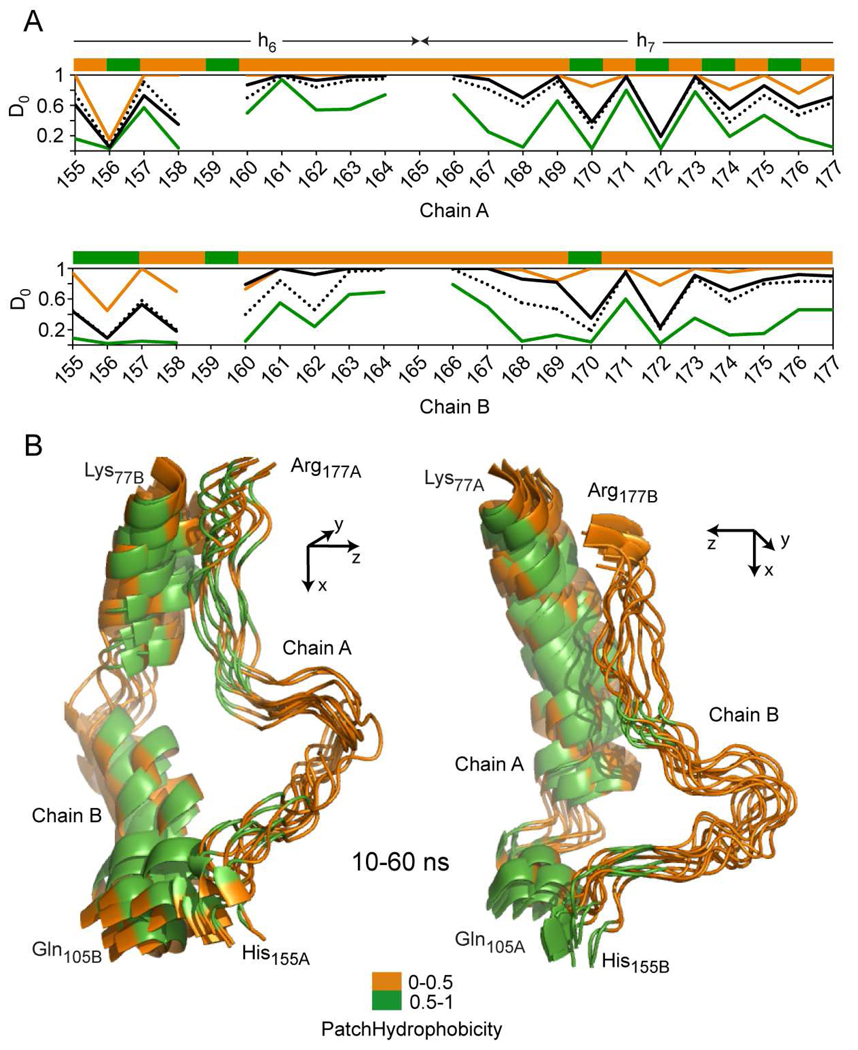
Hydrogen deuterium exchange incorporation factors (D0) and patch hydrophobicity in the Solar Flare region of apoA1 (H155-R177) that hosts helix part of helix h6 (P143-A164), and part of helix h7 (P165-G186). A, Top: patch hydrophobicity (top stripe), and comparison between experimental (black line) and calculated D0 values for residues in His155-Arg177 region of apoA1 chain A. The orange and green lines identify the maximum and minimum for range of fluctuations of D0 values calculated from simulation snapshots. The black dotted line shows the average value of calculated D0 obtained from all 22 snapshots taken during the 60 ns simulation and used for analysis. Bottom: similar information is displayed for His155-Arg177 region of chain B of apoA1. These two plots show that the average values of D0 obtained from simulation follow the same pattern as the experimentally-derived D0 values, and agree well with the latter. B, For clarity, this picture shows the superposition only of 11 snapshots of the His155-Arg177 region (chain A and B) taken between 10 and 60 ns. The apoA1 chains are shown in cartoon representation and Tyr166 (one of the sites for selective oxidation) is shown in stick-representation and colored black. The residues are colored according to their patch hydrophobicity (orange = solvent accessible residues and green = berried residues). Left panel shows the trajectory of the Solar Flare region of chain A (His155A-Arg177A) and the Right panel shows the trajectory of the Solar Flare region of chain B (His155B-Arg177B).
In further analyses we investigated the relation between the secondary structure of apoA1 and the observed periodicity of H/D exchange data by performing separate analyses of H/D exchange and PH on domains of apoA1 with distinct secondary structure (i.e. either α-helical or random coil loop). For example, Figure 8 displays the results of the analysis performed on a predominantly α-helical domain of apoA1 (Tyr100-His155) that hosts what has historically been called helix h4 (Pro99-Glu120), h5 (Pro121-Ser142) and part of h6 (Pro143-Ala164). Simulation results shown in Fig. 8B reveal that this domain of apoA1 remains α-helical during the entire trajectory. The top graph of Fig. 8A displays oscillations in the experimental (solid line) and simulation derived D0i (maximum, minimum and average simulation data shown in orange, green and black dotted lines). The PH for individual residues is shown in the top stripe. For example, in HDL models extracted from simulation snapshots residues Ala152 (both chains of apoA1) are predicted to face the lipid core and are predicted to be involved in H-bonding. Amide H of Ala152A persists in H-bonding 100% of the simulation; H of Ala152B persists 90%. The experimentally derived D0i for these amino acid residues are 0.05 (chain A) and 0.04 (chain B), and the average, minimum and maximum values of the predicted D0i agree well with the former (chain A: 0.04, 0.03, 0.05, chain B: 0.05, 0.03, 0.24, respectively). Thus, both experimental and simulation results indicate that Ala152 of both apoA1 chains experience minimal hydrogen/deuterium exchange, and minimal dynamics. In contrast, Ser142 of both apoA1 chains face the solvent. Amide H of Ser142A is involved in H bonding only 24% of the simulation while H of Ser142B persists 67%. The experimentally derived D0i for these amino acid residues are 0.97 (chain A) and 0.93 (chain B). The average, minimum and maximum values of predicted D0i are for chain A: 0.86, 0.53, 1.00 and for chain B: 0.83, 0.55, 1.00, respectively. The average values for both chains (0.86 and 0.83) are in reasonable agreement with the experimental D0i (0.97 and 0.93). The experimental and simulation predicted D0i values for Ser142 are congruent and show that the amide proton on this residue rapidly exchanges with solvent and experiences much more dynamics than Ala152. It is also interesting to note that even though some of the residues in apoA1 are extremely dynamic with a large range of predicted D0i values, there is remarkable concordance between the predicted average D0i across the simulation trajectory (dotted line, Fig. 8A) and the experimental D0i derived from H/D-MS/MS analyses (solid line, Fig. 8A).
The marked oscillatory behavior of the experimental and simulation derived per residue deuterium incorporation factors, D0i, are consistent with the predominantly α-helical secondary structure and lipid interfacial environment of apoA1. Thus, while in globular proteins hydrogen bonding can play a dominant role in overall rates of H/D exchange of amide protons (60), in HDL, the lipid environment has a dramatic impact on overall H/D exchange in apoA1 amide hydrogens. Figure 8B displays a superposition of the models of apoA1 corresponding to the 0–60 ns trajectory. The superposition of these apoA1 domain models shows that “patch” hydrophilic residues (orange) remain solvent exposed (left panel) while “patch” hydrophobic residues (green) remain buried during the simulation, facing the lipid core.
A similar analysis was performed for the loop domain His155-Arg177 (Solar Flare, SF) of apoA1 chains A and B, and is shown in Fig. 9. These loops host part of helices h6 (Pro143-Ala164), and h7 (Pro165-Gly186). Figure 9A displays graphs with changes in experimental and simulation-predicted D0i values, and the PH on the top stripes. A notable feature of this region of apoA1 within rHDL is that the variation in D0i values does not follow the same oscillatory pattern as in the case of predominantly α-helical domains (e.g. Fig. 8). Most residues in the SF loops show markedly rapid H/D exchange rates, and have corresponding enhanced predicted dynamics exemplified by the range of predicted D0i, kxci, and Kui values (Supplemental Table 1). Some residues within the SF loops show markedly dynamic character. For clarity, Fig. 9B shows the superposition of 11 snapshots only (every 5 ns between 10 ns and 60 ns simulation trajectory) of these loop domains of apoA1 in rHDL. The PH is displayed in the stripes on top of the graphs and color coded in the models. Visual inspection show that the majority of the residues in the SF region remain as protruding solvent exposed loops during the entire simulation, consistent with the experimental H/D data. These findings support the idea that these loops are thermodynamically stable structures that remain highly exposed to solution.
Simulation models preserve experimental distance constraints and overall asymmetry of apoA1 shape proposed for the DSH model
A wealth of experimental investigations (FRET, MS/MS cross-linking, ESR) report various distance constraints in apoA1 of rHDL (9, 11, 61–63). We therefore analyzed the simulation models for goodness of fit with available reported distance constraints from diverse biophysical approaches. Remarkably, we found that during the trajectory of the simulation, virtually all distance constraints can be accommodated. Table 2 lists the distance constraints obtained from the different experimental platforms, together with those found in the DSH model. Also shown are predicted minimum, average and maximum values of the constraints obtained from simulation by the analysis of the simulation trajectory. The distance constraints listed for the DSH model incorporate specific modifications of amino acid residues according to the biophysical method used for their determination. For example, to compare distance constraints from the DSH model with those obtained from FRET experiments (11), the distance was measured between a Trp residue and a residue modified in silico to cysteine and attaching the AEDANS acceptor (covalent adduct with 5-[2-[(1-oxoethyl)amino]ethylamino]-1-naphthalenesulfonic acid). When ESR data were used (9), both residues were modified to cysteine and the methane thiosulfonate nitroxide spin label (MTSSL) probe was attached. To compare with distance constraints obtained from MS/MS cross-linking (9, 61–63), in the DSH model we freely rotated the lysine residue to obtain the minimum distance between them. None of these transformations have been applied to models produced from simulation because it was impracticable to do this for such a large number of structures. Thus, the distances obtained from simulation models are between the extremities of non-modified amino acid residues involved in the distance constraints, and underestimate the distances that would be observed with the probes covalently attached for the FRET and ESR studies. The additional transformations on residues that we performed for the DSH model in principle lead to a decrease in the distance between residues because the AEDANS and MTSSL attachments are rather large (15 Å and 6.5 Å, respectively). Thus, if the distance between residue extremities in simulation models is close (e.g. < 40 Å) to the experimental constraint, then the attachment of the AEDANS or MTSSL probes will bring the experimental and theoretical constraints in agreement.
Table 2.
Distance constraints (MS/MS cross-links, FRET, ESR) in nascent HDL and their fluctuations during molecular dynamics simulation
| Geometrical constraint |
Experiment | DSH model a | Simulation b | ||
|---|---|---|---|---|---|
| Distance (Å) | Distance (Å) | Minimum distance (Å) |
Average distance (Å) |
Maximum distance (Å) |
|
| W50A-L230B | FRET (22.7)c | 22.8a | 28.2 | 36.2 | 46.1 |
| W50B-L230A | 22.2 | 28.1 | 34.2 | ||
| W72A-A210B | FRET (23.5)c | 30.2 | 32.9 | 38.7 | 49.2 |
| W72B-A210A | 42.1 | 48.4 | 55.1 | ||
| L90A-A190B | FRET (24.0)c | 26.2 | 30.8 | 34.3 | 39.8 |
| L90B-A190A | 43.1 | 48.3 | 54.3 | ||
| W108A-A170B | FRET (28.8)c | 26.6 | 13.8 | 20.9 | 28.4 |
| W108B-A170A | 20.2 | 25.3 | 35.3 | ||
| Q132A-Q132B | FRET (30-35)c | 17.3d | 1.9 | 8.3 | 16.6 |
| K12A-K182B | MS/MS (12)e,g | 16.1 | 15.9 | 21.0 | 26.5 |
| K12B-K182A | 50.1 | 59.6 | 68.4 | ||
| K40A-K239B | MS/MS (12)e,f,g | 30.1 | 35.7 | 41.8 | 51.0 |
| K40B-K239A | 26.3 | 35.7 | 52.1 | ||
| K59A-K208B | MS/MS (12)f | 15.6 | 14.7 | 23.5 | 32.6 |
| K59B-K208A | 21.9 | 28.5 | 39.1 | ||
| K77A-K195B | MS/MS (12)f | 18.1 | 13.7 | 19.8 | 35.0 |
| K77B-K195A | 35.0 | 41.5 | 49.8 | ||
| K88A-K118B | MS/MS (12)f | 69.1 | 27.9 | 64.3 | 93.8 |
| K88B-K118A h | 62.3 | 71.2 | 78.4 | ||
| K96A-K118B h | MS/MS (12)f | 64.2 | 37.9 | 63.2 | 80.1 |
| K96B-K118A | 63.7 | 69.5 | 74.3 | ||
| K118A-K140B | MS/MS (12)e,f,g | 10.7 | 4.5 | 13.4 | 19.7 |
| K118B-K140A | 6.4 | 16.6 | 26.3 | ||
| K133A-K140B | MS/MS (7.7)g | 6.4 | 7.8 | 12.9 | 25.2 |
| K133B-K140A | 12.0 | 17.6 | 24.6 | ||
| K208A-K208B h | MS/MS (12)f | 160.1 | 59.2 | 89.5 | 111.5 |
| K133A-K133B i | ESR (<15)e | 6.2 | 3.0 | 6.6 | 12.7 |
| L134A-L134B i | ESR (<15)e | 10.9 | 6.0 | 9.2 | 21.9 |
| E146A-E146B i | ESR (17–19)e | 43.8 | 41.7 | 47.2 | 53.2 |
Wu et al, 2009 JBC;
The distances reported from simulation do not include any conformational adjustments, like residue free rotation, Cys mutation/attachment of methane thiosulfonate nitroxide spin label (MTSSL) for ESR, or attachment of 5-[2-[(2-Iodo-1-oxoethyl)amino]ethylamino]-1-naphthalenesulfonic acid (AEDANS) acceptor for FRET experiments; rather they are distances between the extremities of the residues in that particular snapshot from the simulation.
Martin et al, 2006 JBC;
Lietal.,2001,JBC;
Bhat et al, 2005, JBC;
Silva et al, 2005, Biochemistry;
Bhat et al., 2007, Biochemistry;
This constraint is not compatible with 5/5registry;
Distances between two residues in the DSH model are from the oxygen atom of MTSSL on one residue to the oxygen atom of MTSSL of the other residue.
As shown in Table 2, all distance constraints satisfied in the DSH model are also satisfied in the simulation models. In addition, there are a couple of intra-chain MS/MS cross-linking constraints not satisfied in the DSH model (K88-K118, K96-K118) that have minimum distances much shorter in simulation models (K88-K118: 69.1 → 27.9 Å; K96-K118: 64.2 → 37.9 Å). Another interesting aspect is that the analysis of distance constraints points to a lack of symmetry in constraints between protein chains. For example, the constraint between K12A-K182B (min: 15.9 Å) is not detected for K12B-K182A (min: 50.1 Å). These situations highlight the prediction that the conformations of the two chains of apoA1 within rHDL are dissimilar.
The Solar Flare loops remain solvent accessible and do not collapse during simulation
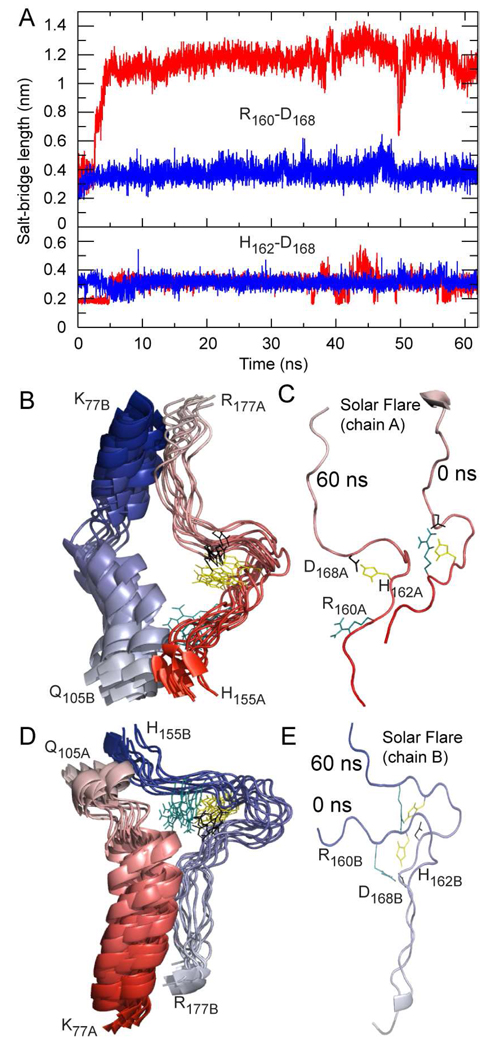
The original application of H/D exchange to the study of rHDL resulted in identification of an unusual region of apoA1 demonstrating a particularly rapid rate of H/D exchange, consistent with highly solvent exposed protruding loops named the Solar Flares (12). Experimental studies involving apoA1 mutagenesis and peptide competition binding studies confirmed this region corresponded to an LCAT interaction site, facilitating LCAT activation (12). In the DSH model of nascent rHDL the Solar Flare loops are proposed to be retained, with charged residues (Arg160, His162, and Asp168) of apoA1 forming a pair of salt bridges. Despite the experimental data demonstrating marked H/D exchange and the subsequent functional studies, the presence of stable Solar Flare loops has been debated. Using only a computational approach Shih et al. (64) reported a brief 1 ns simulation and suggested the SFs "collapse" because a salt bridge within the SF was not preserved throughout the simulation (64). In the present studies we therefore carried out more in depth analyses of the SF regions, including analysis of formation and breaking of predicted salt bridges (e.g. Arg160–Asp168– His162) in the Solar Flare loops during simulation. The graph in Fig. 10A displays the change in predicted distance (nm) between the charged residues involved in the ternary salt bridges during the course of the simulation (i.e. Arg160–Asp168 and His162–Asp168 in both chains of apoA1; chain A: red, chain B: blue). This analysis shows that salt bridge His162–Asp168 persists during the entire simulation in both chains, while salt bridge Arg160–Asp168 in chain A breaks after approximately 3 ns. Of note, the latter salt bridge is predicted to again be reformed briefly around 50 ns of simulation. On the other hand, the salt bridge made of same residues located on chain B (Arg160B–Asp168B) is predicted to persist over the entire simulation. The analysis of salt bridge dynamics in the SF loops confirms that salt bridges are not irreversible covalent bonds, but rather, are ephemeral, and may be broken and remade during simulation. Importantly, these domains retain their overall solvent exposed conformation (Fig 10) regardless the fate of the salt bridges.
FIGURE 10.
Molecular dynamics simulation trajectory of the Solar Flare loops (His155-Arg177) and the predominantly α-helix sequences (Lys77-Gln105), from the adjacent chain of apoA1 in the Double Super Helix model of Nascent HDL. The trajectory is composed of 11 snapshots in the interval 10–60 ns. Chain A of apoA1 is colored red and chain B is colored blue with the color fading from the N-terminal toward the C-terminal. A, Fluctuations in the length of salt-bridges R160-D168 (upper panel) and H162-D168 (lower panel, red for chain A and blue for chain B) during simulation. The upper panel graph shows that the R160-D168 salt-bridge in chain A breaks after approximately 2 ns and remains mainly broken during the entire simulation, but is reformed briefly when the simulation reaches 50 ns. On the contrary, the R160-D168 salt-bridge in chain B is conserved during the simulation. The lower panel shows that the H162-D168 salt-bridges in both chains are conserved during the entire simulation. B, Superposition of the eleven snapshots of the Solar Flare of chain A and the adjacent α-helix of chain B taken during 10–60 ns of simulation. The residues involved in salt-bridges are colored and shown as sticks (cyan = R160, yellow = H162, black = D168). The superposition shows that the Solar Flare conformation (chain A) changes little during simulation and remains unfolded and solvent exposed as proposed in the Double Super Helix model of HDL. C, The conformation of the Solar Flare loop (chain A) at the beginning of the simulation (0 ns) and after 60 ns. A comparison of the two snapshots indicates that the shape of the Solar Flare loop remains approximately the same regardless of one salt-bridge (R160-D168) being broken. D, Superposition of the eleven snapshots of the Solar Flare (chain B) and the adjacent α-helix of chain A taken during 10–60 ns of simulation. This trajectory shows that the two salt-bridges are conserved in all frames displayed. E, The conformation of the Solar Flare loop (chain B) at the beginning of the simulation (0 ns) and after 60 ns. The two snapshots indicate that the shape of the Solar Flare loop remains approximately the same.
Salt bridges provide stability to the apoA1 chains that serve as a backbone for the HDL particle
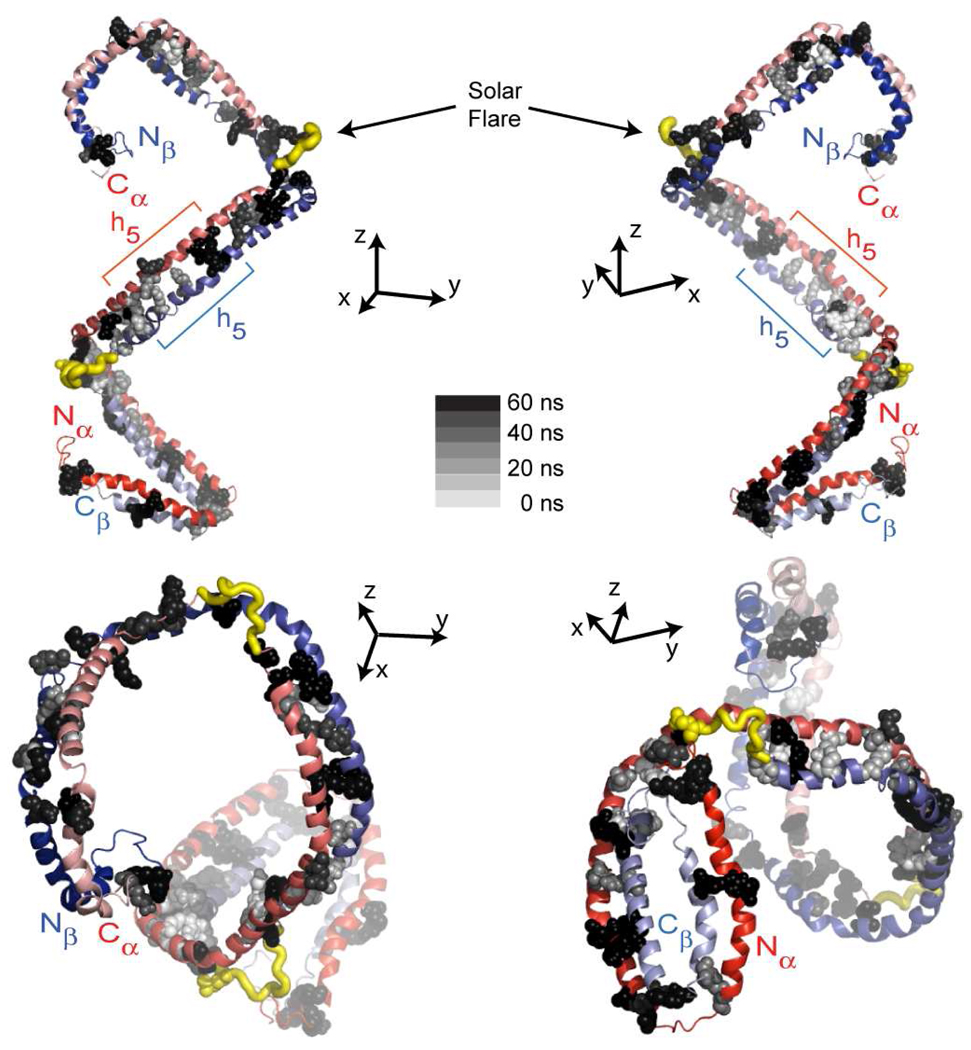
The persistence of salt bridges over the course of molecular dynamics simulation can give insights into structures of potential importance in HDL function. We therefore monitored salt bridge formation and breaking throughout the particle during the simulation and analyzed the stability and dynamics of inter-chain salt bridges in terms of persistence over the trajectory. Figure 11 displays the location and persistence (in gray scale) of the inter-chain salt bridges on apoA1 dimer identified during simulation. The salt bridges are displayed on the conformation of apoA1 present at the beginning of the simulation. Table 3 provides the minimum, maximum and average distance between charged residues for each salt bridge identified during simulation and its persistence. There are 25 inter-chain salt bridges predicted to be located in the Nα/Cβ–half-domain of apoA1 dimer, and 22 predicted to reside in the other half-domain (Nβ/Cα) during the course of the entire simulation. Of note, only 7 salt bridges in the Nα/Cβ–half-domain and 2 in the Nβ/Cα–half-domain were observed to persist more than 80% of the simulation (Table 3). Table 3 also lists predicted distances between the charged residues of the salt bridges suggested in the original DSH model. Interestingly, some of the salt bridges identified in the original DSH model are very short lived in the simulation (e.g. Arg27A–Glu223B, Lys59A–Glu198B, Glu62A–Lys195B), while other salt bridges not observed in the DSH model are predicted to form during the simulation (e.g. Glu85A–Arg177B, Glu125A–Lys133B, His199A–Asp48B). Among the 28 inter-chain salt bridges reported in the DSH model (13), 12 are found to persist more than 50% of the simulation.
FIGURE 11.
Predicted salt bridge distribution in apoA1 during 60 ns of molecular dynamics simulation. The conformation of apoA1 shown from a different vantage point with the N/C-termini pointing toward/away from the reader. The figure shows that the salt bridges located toward N/C-termini are more persistent (colored black or dark grey) during simulation while those located at the middle region of apoA1 are less persistent (colored light grey).
Table 3.
Predicted inter-chain salt bridges in nascent HDL during molecular dynamics simulation
| Salt bridge | DSH model | Simulation trajectory | |||
|---|---|---|---|---|---|
| Distance (Å) |
Minimum a distance (Å) |
Average b distance (Å) |
Maximum c distance (Å) |
Persistence d (%) |
|
| Arg10A–Glu234B | 1.53 | 2.17 | 2.72 | 3.21 | 14.3 |
| Lys12A–Glu234B | 1.67 | 1.97 | 2.83 | 3.96 | 14.3 |
| Arg27A–Glu223B | 1.68 | 2.47 | 2.47 | 2.47 | 4.8 |
| Lys40A–Asp213B | 1.65 | 2.23 | 2.50 | 2.77 | 57.1 |
| Asp48A–Lys206B | 1.69 | 2.19 | 2.70 | 3.94 | 23.8 |
| Asp48A–Lys208B | 7.25 | 2.02 | 3.16 | 3.85 | 57.1 |
| Asp51A–Lys206B | 5.88 | 3.95 | 3.95 | 3.95 | 4.8 |
| Lys59A–Glu198B | 1.52 | 2.30 | 2.30 | 2.3 | 4.8 |
| Glu62A–Lys195B | 1.61 | 2.08 | 2.08 | 2.08 | 4.8 |
| Glu70A–Arg188B | 1.53 | 1.92 | 3.07 | 3.75 | 57.1 |
| Lys77A–Glu183B | 1.55 | 2.03 | 2.03 | 2.03 | 4.8 |
| Glu78A–Arg177B | 1.68 | 1.95 | 2.69 | 4.00 | 85.7 |
| Glu85A–Arg177B | 10.55 | 1.95 | 2.71 | 3.87 | 66.7 |
| Asp89A–Arg173B | 1.65 | 1.81 | 2.14 | 3.34 | 100 |
| Glu92A–Arg171B | 1.53 | 1.72 | 2.66 | 3.99 | 33.3 |
| Glu92A-Arg173B | 1.53 | 1.94 | 2.31 | 3.14 | 95.2 |
| Lys96A-Glu169B | 1.56 | 2.73 | 2.73 | 2.73 | 4.8 |
| Lys107A-Asp157B | 1.5 | 2.10 | 2.63 | 3.97 | 80.9 |
| Glu110A-Arg151B | 9.11 | 3.66 | 3.66 | 3.66 | 4.8 |
| Glu111A-Arg151B | 1.54 | 2.82 | 2.82 | 2.82 | 4.8 |
| Glu111A-His155B | 1.42 | 1.61 | 1.79 | 2.01 | 100 |
| Lys118A-Glu147B | 1.51 | 2.02 | 2.64 | 3.81 | 85.7 |
| Glu125A-Lys133B | 10.33 | 2.03 | 2.41 | 3.16 | 66.7 |
| Glu125A-Lys140B | 1.59 | 2.06 | 2.42 | 3.30 | 80.9 |
| Glu128A-Lys133B | 10.25 | 2.26 | 2.98 | 3.92 | 28.6 |
| Glu136A-Arg123B | 1.6 | 2.21 | 2.21 | 2.21 | 4.8 |
| Lys140A-Glu125B | 1.76 | 2.23 | 2.50 | 2.77 | 9.5 |
| Glu147A-Lys118B | 1.62 | 2.04 | 2.59 | 3.09 | 33.3 |
| Arg151A-Glu111B | 1.65 | 1.82 | 2.83 | 3.80 | 61.9 |
| His155A-Glu110B | 4.2 | 3.84 | 3.84 | 3.84 | 4.8 |
| His155A-Glu111B | 1.49 | 1.61 | 1.83 | 2.40 | 95.2 |
| Asp157A-Lys107B | 9.2 | 3.90 | 3.90 | 3.90 | 4.8 |
| Arg171A-Glu85B | 7.29 | 3.14 | 3.55 | 3.99 | 19.0 |
| Arg171A-Glu92B | 13.81 | 3.97 | 3.97 | 3.97 | 4.8 |
| Arg173A-Glu78B | 1.51 | 2.27 | 2.58 | 3.45 | 28.6 |
| Arg173A-Glu85B | 5.55 | 2.02 | 2.62 | 3.83 | 38.1 |
| Arg177A-Glu70B | 19.97 | 2.28 | 3.31 | 3.96 | 38.1 |
| Arg177A-Asp73B | 13.68 | 2.95 | 3.47 | 3.95 | 14.3 |
| Glu191A-Lys59B | 1.47 | 2.10 | 2.78 | 3.45 | 66.7 |
| Lys195A-Asp48B | 13.46 | 2.19 | 2.96 | 3.52 | 19.0 |
| Lys195A-Asp51B | 11.74 | 2.10 | 3.26 | 3.96 | 38.1 |
| Glu198A-Lys45B | 8.82 | 3.63 | 3.84 | 3.97 | 14.3 |
| His199A-Asp48B | 6.37 | 1.63 | 1.91 | 2.71 | 95.2 |
| Glu212A-Lys40B | 7.34 | 1.99 | 2.69 | 3.96 | 19.0 |
| Asp213A-Lys40B | 7.64 | 3.08 | 3.08 | 3.08 | 4.8 |
| Lys238A-Glu2B | 2.69 | 2.43 | 3.35 | 3.94 | 42.8 |
| Lys239A-Glu2B | 1.66 | 2.27 | 2.79 | 3.30 | 9.5 |
Minimum distance between charged residues detected during the entire trajectory.
Average distance between charged residues calculated from all snapshots.
Maximum distance between charged residues detected during the entire trajectory.
Salt bridge persistence calculated as the fraction of the snapshots in which the salt bridge was detected (i.e. that distance between charged residue was less than 4 Å. Salt bridges with light shaded background are located in the half domain of the apoA1 dimer that contains the Nβ/Cα termini pair. Salt bridges with a darker background persist more than 80% of the simulation.
DISCUSSION
Prolonged molecular dynamics simulation of the DSH model of rHDL supports the major conclusions reached about the structure of nascent HDL based on a multitude of experimental data (SANS, EM, H/D-MS/MS, NMR, distance constraints from FRET, ESR and MS/MS cross-linking). Notably, the overall super-helical architecture of the protein directly visualized by SANS with contrast variation is retained during the entire simulation despite the change in the overall dimensions of the particle. This scaffolding dictates the organization of the lipid phase of the particle. Importantly, the proposed micellar lipid organization posited in the DSH model is preserved throughout the simulation, though the particle shape changes from prolate ellipsoid to spheroid. The micellar lipid packing proposed is consistent with new experimental 31P NMR data (chemical shift, line shape and half line width) on both plasma isolated HDL and rHDL preparations. The H/D exchange deuterium incorporation factors and H/D exchange rate constants predicted for apoA1 amide hydrogens from the simulation trajectory are also in excellent agreement with the experimental H/D-MS/MS data. Finally, molecular models produced by simulation accommodate a larger number of distance constraints derived from various biophysical experiments than any other model. Thus, lengthy all-atom molecular dynamics simulation suggests the structure posited for nascent HDL in the DSH model is a thermodynamically stable particle with properties congruent with experimental data obtained from multiple biophysical platforms.
One of the prominent features of apoA1 architecture in the DSH model of nascent HDL is its super-helical shape. SANS analyses with contrast variation have allowed direct visualization of the protein shape within rHDL. The open helical shape for the anti-parallel apoA1 chains appears to form a scaffold upon which lipids assemble in the lipoprotein particle. It should be noted that while the depicted illustration of apoA1 helical conformation has a specific "handedness", programs used for deconvolution of scattering intensities into low-resolution structures cannot distinguish between mirror images of the helical protein (Supplemental Figure 2) (65). This raises an intriguing possibility that an enzymatic reaction like ABCA1 catalyzed nascent HDL formation may actually produce particles containing only one protein helical "handedness". Unfortunately, being able to distinguish between HDL particles containing mirror images of the helical apoA1 is beyond the state of the art in the field at present.
One major question raised from the present molecular dynamics simulation is the origins for the change in the overall dimensions of the DSH model during simulation, since this feature contradicts direct experimental data (SANS and EM). Previous molecular dynamics simulations of micellar systems have shown similar artifacts (i.e. preference for spherical micelles when molecular mechanics force fields used to describe the intermolecular interactions among polar head groups are strong relative to weak dispersion interactions among acyl chains (56, 57)). The force field used in our simulation has been used in multiple prior HDL simulations, and was extensively tested on lipids packed as bilayers (37, 38). Based upon the present studies, these molecular mechanics parameters may need subtle further refinements to be able to reproduce the overall time-averaged ellipsoidal shape of nascent HDL.
One structural element that no doubt plays an important role in defining apoA1 architecture in nascent HDL is the location and persistence of salt bridges. It is generally believed (8) that inter-chain salt bridges between the amphipathic apoA1 α-helices provide mechanical strength to the particle through dynamic interactions between the two chains, reducing their flexibility in a fashion similar to that of two strands of DNA reversibly tethered together through H-bonds. The results of the molecular dynamics simulation suggest candidate residues that may play an important role in providing structural support to the particle (Fig. 12). They also reaffirm the idea of asymmetry in the local structure of the two half-domains of apoA1, which underscores the dissimilarity in the conformations of the two Nt/Ct domains of apoA1, as shown in Fig. 11 (bottom). The predicted ends of the two half domains are distinct, and the Nα/Cβ–termini are predicted to fold back more tightly than the Nβ/Cα–termini.
FIGURE 12.
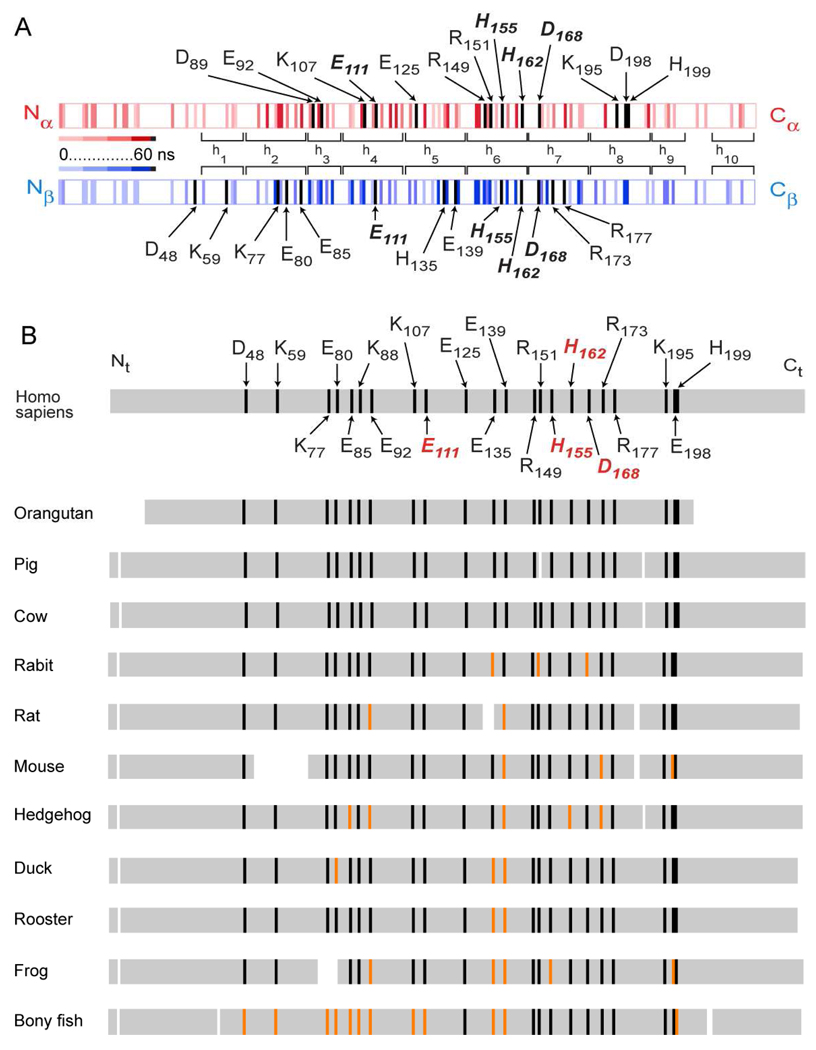
Map of charged amino acid residues involved in salt bridges during the simulation and their conservation between species. A, Distribution of residues predicted to form inter- and intra-chain salt bridges in apoA1 during simulation. ApoA1 dimer is shown as a double stripe colored red for chain A and blue for chain B. Residues predicted to be involved in a salt bridge during the simulation trajectory are shown as vertical stripes. The predicted persistence of residues within a salt bridge during the simulation trajectory is indicated by the intensity of color, with lighter color used for less persistence and darker color for longer persistence. Residues that persist more than 95% of the simulation are colored black. B, Analysis of species conservation of apoA1 residues predicted to persist (> 95%) within inter- and intra-chain salt bridges throughout the simulation trajectory. Residues (n=22) that persist more than 95% of the simulation are indicated on Homo sapiens (black) and those that are common to both chains of apoA1 are labeled in red. Multiple alignment was performed using the COBALT multiple alignment tool (http://blast.ncbi.nim.nih.gov) of the BLASTP program (76) relative to Homo sapiens. Of the 22 residues, those showing either identity or conservation within the acid / base side chain group are indicated by black stripe, whereas those not conserved are shown as an orange stripe. Additional species aligned included: Pongo pygmaeus (orangutan), Sus scrofa (pig), Bos taurus (cow), Oryctolagus cuniculus (rabit), Rattus norvegicus (rat), Mus musculus (mouse), Erinaceus europaeus (hedgehog), Anas platyrhynchos (duck), Gallus gallus (rooster), Xenopus (frog), Anguilla japonica (bony fish).
Analyses of the simulation trajectory for residues in either intra- or inter-molecular salt bridges suggest a large number (over 80 in each chain) can participate in reversible charge-charge associations within the apoA1 dimer (Fig. 12A). Interestingly, 13 residues are observed in each chain of apoA1 to persist >95% of the simulation within close enough proximity (4 Å) to a charged counterpart. Of these, four apoA1 residues are predicted to exist within salt bridges throughout the entire simulation in both chains. Remarkably, two of the four are predicted to make a salt bridge within the SF loop and are highly conserved across species (i.e. His162–Asp168; Fig. 12B). It is also noteworthy that many additional residues that are predicted to form stable salt bridges based upon the simulation are also highly conserved across multiple species (Fig. 12B). It is tempting to speculate that these residues may serve essential roles in promoting HDL function, whether it involves generation of a structural element essential for activity of an HDL associated protein-like LCAT (e.g. as proposed in the case of His162–Asp168 within the SF loop) or perhaps by acting as a structural "zipper" that helps to tether the apoA1 chains together. While such salt bridges may provide mechanical strength, their reversible nature also allows for a highly dynamic particle that is capable of adapting and changing shape to meet its function of serving as a carrier of lipid cargo during particle maturation and remodeling.
Another intriguing finding of the molecular dynamics simulation is that while the lipid phase retains micellar packing as in the original DSH model and appears to be thermodynamically stable over the entire simulation, unexpectedly, it develops a noticeable empty cavity at the center of the particle. Frankly, this finding was at first met with skepticism by the authors. However, we were surprised to find that several decades ago, when micellar systems were being intensively structurally studied, that direct experimental data on micelles (51–56, 66), including small angle neutron scattering (67–69) and small angle X-ray scattering (53) have confirmed the presence of a cavity at the center of some spherical micelles. Moreover, computer simulations (51, 54–57) have previously posited the presence of a central void within micelles, and theoretical arguments (50) have postulated that it in part arises from the unfavorable packing of acyl chain ends into a small volume at the center of the micelle. Further, simulation models have postulated that the size of this cavity may be minimized in larger micelles by adopting alternative shapes like ellipsoidal and cylindrical micelles (56, 57, 66).
The shape of the DSH model of nascent HDL is consistent with experimental observation (ie - SANS and EM). Still, its lipid packing exhibits a small void along the main axis of the ellipsoid. We speculate that the existence of a void at the center of the micellar lipid phase in nascent HDL supports the particle carrier function because the storage of hydrophobic neutral lipids such as cholesterol esters and triglycerides at the center of the particle will require a lower free energy for lipid reorganization. The work required to create a cavity in a condensed system is known in statistical thermodynamics as the free energy of cavity formation (70). The central void noted within the micellar structure suggests it may not be necessary to push away as many lipids from the center of the nascent HDL during particle maturation. It is also worth noting that the molecular dynamics simulation studies of the DSH model of nascent HDL is more easily reconciled with one that can be predicted for spherical HDL. It also suggests an overall structure that helps envision how a nascent HDL particle that undergoes maturation starts to accrue neutral lipids in its void core and gradually changes shape from a prolate ellipsoid to a spheroid without altering the overall packing of the lipid phase at the surface, which retains its micellar / pseudolamellar (where protein overlies) organization.
Early seminal studies by Brewer and colleagues over 3 decades ago first reported that the 31P NMR spectrum of HDL demonstrates an unusually unique isotropic character compared to that observed in other lipoproteins, indicating that the phospholipids within HDL are far more dynamic than observed with other lipoproteins (71). Despite these early studies, further 31P NMR studies into the lipid organization (e.g. lamellar vs. micellar) of HDL have not been reported. 31P NMR can be used to distinguish between micellar and lamellar mesophases (33, 35, 58). This is because the 31P NMR chemical shift, line shape and line width arising from the phosphate head group of phospholipids in aqueous dispersions are sensitive to the dynamics and organization of model and biological membranes. For example, micellar phase phospholipids show a single sharp isotropic peak at 0 ppm, whereas lamellar, hexagonal array and multilamellar phases show different chemical shifts and significant line broadening (both up-field and down-field, depending upon the mesophase), consistent with limitation in rotation of lipid in these alternative organizations (33, 35, 58). The sharp isotropic 31P NMR peak at 0 ppm observed in phospholipid within rHDL and pHDL is consistent with lipid that is highly dynamic and tumbling in all orientations - more so than predicted for a small unilamellar vesicle of similar size.
From a biological function stand point a micellar lipid organization can be more adaptable and flexible than a bilayer. For example, HDL associated proteins that catalyze direct lipid transfer reactions with the particle will not require any type of mechanism to tackle an asymmetry that becomes inherent in a disk model of HDL (i.e. with two lipid leaflets of a bilayer, there is substantial barrier to "flip flop", whereas the lipids in a micellar particle can diffuse freely throughout the particle). This limitation in freedom of motion likely underlies the origins in the differences in isotropic motion of phospholipid within lamellar (bilayer) phase vs. HDL noted in the 31P NMR studies. Finally, a micellar packing of lipid may actually represent a conserved structural motif amongst many lipoproteins. For example, Weisgraber and colleagues used SAXS to explore the structure of apolipoprotein E - DPPC particles in solution and observed a spheroidal particle with contorted apolipoprotein E and an ellipsoidal lipid core with phospholipids posited to be packed in a micellar organization (72).
Prior models of HDL are characterized by protein conformation that is highly symmetrical (6–11). An inference that follows from these models is that HDL associated proteins can interact on two locations of apoA1 within HDL. In contrast, with the improved resolution afforded by SANS for directly visualizing the protein shape within nascent HDL, the overall conformation of apoA1 in nascent HDL proposed in the DSH model and the present simulation studies indicate that the protein helical shape is not symmetric. Interestingly, as experimental data on HDL has accrued, we see that some regions of apoA1 must have imprinted in them some structural specificity for a high degree of recognition and selectivity for HDL associated proteins. For example, myeloperoxidase (MPO), a leukocyte derived enzyme that binds to HDL in human atherosclerotic plaque and plasma, has been shown to interact with a small region of apoA1 corresponding to residues Ala190-Leu203 (12). Another example is the site of LCAT interaction on apoA1 of nascent HDL, the SF region (approximately residues His155-Arg177) (12). While the SF loops are highly dynamic, these regions are predicted by molecular dynamics simulation to be thermodynamically stable when exposed to solvent, and the overall “random coil” loop conformation is conserved during simulation (Fig. 10). It is thus remarkable that the two SF loops are predicted to possess distinct structural environments. While there is clearly a high degree of specificity in both MPO and LCAT binding to apoA1 observed experimentally by H/D-MS/MS and peptide competition studies (12, 73), the precise stoichiometry of these and other HDL - HDL associated protein interactions have not been quantitatively investigated. The present simulation studies show intriguing differences in the two SF regions, suggesting distinct steric environments exist for each loop. This raises the possibility that LCAT may interact preferentially with one of the SFs. Experiments to actually discriminate whether LCAT, MPO or other HDL associated proteins differentially bind nascent HDL at more than one site per particle with different affinities is an intriguing hypothesis that awaits future testing. It is tempting to speculate that the SF loop may serve as a generalized structural motif that facilitates lipid exchange with more than one HDL associated protein. It remains to be seen if LCAT interacts with just one of the SF loops, and an alternative lipid exchange protein docks at the other SF loop. Further studies are necessary to investigate these hypotheses.
Limitations of the study are worth noting. While tremendous increases in computational power have permitted computer simulations techniques to evolve significantly over the last decades and their use in structural/computational biology has become more ubiquitous, it is important to keep in mind that simulation techniques, while increasingly sophisticated, have significant limitations. They can test if a proposed molecular model is thermodynamically plausible. However, they cannot easily discriminate amongst alternative structural models if these are relatively stable alternatives. Molecular dynamics simulations have been performed on alternative structural models of nascent HDL and suggested they are feasible, including the picket fence model (6), and multiple double belt structural models (8, 14, 15, 17–20, 22, 64). Thus, molecular dynamics simulations alone cannot determine what the “true” structure(s) of HDL are. Rather, it is direct experimental data that allows for determination of the overall architecture of the particle. In this regard, none of the alternative models examined by molecular dynamics simulations show structures that match the results of the broad array of biophysical platforms examined in the present investigation. The recent application of contrast variation SANS (ref (13) and the present study) enables direct visualization of the overall structure of apoA1 within nascent HDL and reveals that the overall architecture of the protein is open and helical. Similarly, the use of 31P NMR in the study of HDL in the present investigation reveals that the dynamic motion of the phospholipids within HDL are incompatible with lamellar phase organization in a particle the size of HDL, and indeed is consistent with a micellar organization. It is also worth noting that while 80 ns simulation performed in the present study is the longest yet reported for an all-atom simulation of HDL, it is still small in duration to account for the full dynamics of the particle. Further, based upon reported diffusion coefficients for a phospholipid within a small unilamellar vesicle or lipoprotein (74, 75), one can predict that the average diffusion path length of a lipid during 80 ns for a particle the size of HDL is approximately the particle dimension (~ 100 Å). Thus, one may argue that the system is not fully equilibrated after 80 ns of simulation, in the sense that not all individual phospholipids had a chance to explore the entire volume of the particle on this time scale. We note, however, that the total energy and most importantly, the RMSD, reach steady state after 10–20 ns of the simulation, suggesting that the system reached thermodynamic equilibrium. To produce longer simulations for such large macromolecular assemblies practically requires use of coarse-grained force fields that describe molecules as a collection of beads (and make prior assumptions about secondary structure). While allowing for longer simulations, the simplifications in the force fields required produce additional artifacts. Perhaps the major strength of molecular dynamics simulations is they can serve as hypothesis generating exercises, and provide insights into what interactions (e.g. salt bridges, H-bonds, hydrophobic contacts, etc.) may account for specific and global structural features.
Structural studies through traditional high-resolution approaches of large macromolecular complexes remain a challenge. The recent introduction of SANS with contrast variation to the HDL field provides much greater resolution for molecular shape than prior data used for modeling, such as crude estimates of particle size derived from native gels, light scattering, electron microscopy and gel exclusion chromatography. The present study also explored the congruency of experimental data from multiple additional and synergistic biophysical methods (e.g. constraints from H/D –MS/MS, mass spectrometry cross-linking, FRET and ESR) with molecular dynamics simulations. Remarkably, a high degree of concordance was observed between experimental data from all platforms and those predicted by the simulation trajectory. Finally, the present studies are hypotheses generating in as much that they have identified candidate residues that may play important structural and functional role(s), and are also notable for being remarkably conserved throughout evolution.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by National Institutes of Health grants P01 HL098055, P01 HL076491-055328, P01 HL087018-02001 and R01 DK080732-01. Computational resources were provided by the Ohio Supercomputer Center and the National Center for Supercomputer Applications (NCSA, University of Illinois).
ABBREVIATIONS
- apoA1
apolipoprotein A1
- DSH
Double Super Helix
- EM
electron microscopy
- ESR
electron spin resonance spectroscopy
- FRET
fluorescent resonance energy transfer
- H/D-MS/MS
hydrogen-deuterium exchange tandem mass spectrometry
- HDL
High density lipoproteins
- LCAT
lecithin cholesteryl acyltransferase
- MPO
myeloperoxidase
- MS/MS
cross-linking tandem mass spectrometry
- NMR
nuclear magnetic resonance spectroscopy
- PH
Patch hydrophobicity
- pHDL
plasma derived HDL
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- rHDL
reconstituted nascent-high density lipoprotein
- RMSD
root mean square displacement
- SANS
small angle neutron scattering
- SASA
Solvent accessible surface area
- SUV
small unilamellar vesicles
Footnotes
SUPPORTING INFORMATION AVAILABLE. The supporting information contains Suplemmental Figures 1 and 2 that depict small angle neutron scattering intensities (I(q)) versus the scattering vector (q), and distance distribution functions (p(r)) versus the distance between scattering centers (r), and low-resolution structures of nascent rHDL in 12% and 42% D2O. The Supplemental Table 1 lists experimental and calculated per residue deuterium incorporation factors, H/D exchange rate constants, and residue unfolding constants. This material is available free of charge via the Internet at http://pubs.acs.org. Additionally, the following information is available for download from http://www.lerner.ccf.org/cellbio/hazen/data/ - neutron scattering intensities for HDL samples analyzed in 12% and 42% D2O (text files), low resolution structures of nascent HDL (pdb files), and calculated H/D exchange deuterium incorporation factors for all residues in apoA1 dimer (Excel file).
REFERENCES
- 1.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. Cell Metab. 2008;7:365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Trigatti B, Rigotti A, Krieger M. Curr. Opin. Lipidol. 2000;11:123–131. doi: 10.1097/00041433-200004000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Assmann G, Gotto AM., Jr Circulation. 2004;109:III8–III14. doi: 10.1161/01.CIR.0000131512.50667.46. [DOI] [PubMed] [Google Scholar]
- 4.Barter PJ, Rye KA. Curr. Opin. Lipidol. 2006;17:399–403. doi: 10.1097/01.mol.0000236365.40969.af. [DOI] [PubMed] [Google Scholar]
- 5.Rader DJ. Nat. Clin. Pract. Cardiovasc. Med. 2007;4:102–109. doi: 10.1038/ncpcardio0768. [DOI] [PubMed] [Google Scholar]
- 6.Phillips JC, Wriggers W, Li Z, Jonas A, Schulten K. Predicting the structure of apolipoprotein A-I in reconstituted high-density lipoprotein disks. Biophys. J. 1997;73:2337–2346. doi: 10.1016/S0006-3495(97)78264-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koppaka V, Silvestro L, Engler JA, Brouillette CG, Axelsen PH. J. Biol. Chem. 1999;274:14541–14544. doi: 10.1074/jbc.274.21.14541. [DOI] [PubMed] [Google Scholar]
- 8.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, De Loof H, Harvey SC. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 9.Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular Contact between Globular N-terminal Fold and C terminal Domain of ApoA-I Stabilizes Its Lipid-bound Conformation. J. Biol. Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- 10.Maiorano JN, Jandacek RJ, Horace EM, Davidson WS. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal figh-density lipoproteins of different size. Biochemistry. 2004;43:11717–11726. doi: 10.1021/bi0496642. [DOI] [PubMed] [Google Scholar]
- 11.Martin DD, Budamagunta MS, Ryan RO, Voss JC, Oda MN. J. Biol. Chem. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- 12.Wu Z, Wagner MA, Zheng L, Parks JS, Shy JM, III, Smith JD, Gogonea V, Hazen SL. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z, Gogonea V, Lee X, Wagner MA, Li X-M, Huang Y, Arundhati U, May RP, Haertlein M, Moulin M, Gutsche I, Zaccai G, DiDonato J, Hazen SL. The double super helix model of high density lipoprotein. J. Biol. Chem. 2009;284:36605–36619. doi: 10.1074/jbc.M109.039537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catte A, Patterson JC, Bashtovyy D, Jones MK, Gu F, Li L, Rampioni A, Sengupta D, Vuorela T, Niemela P, Karttunen M, Marrink SJ, Vattulainen I, Segrest JP. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys. J. 2008;94:2306–2319. doi: 10.1529/biophysj.107.115857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catte A, Patterson JC, Jones MK, Jerome WG, Bashtovyy D, Su Z, Gu F, Chen J, Aliste MP, Harvey SC, Li L, Weinstein G, Segrest JP. Novel Changes in Discoidal High Density Lipoprotein Morphology: A Molecular Dynamics Study. Biophys. J. 2006;90:4345–4360. doi: 10.1529/biophysj.105.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones MK, Catte A, Patterson JC, Gu F, Chen J, Li L, Segrest JP. Thermal stability of apolipoprotein A-I in high-density lipoproteins by molecular dynamics. Biophys. J. 2009;96:354–371. doi: 10.1016/j.bpj.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klon AE, Segrest JP, Harvey SC. Molecular dynamics simulations on discoidal HDL particles suggest a mechanism for rotation in the apo A-I belt model. J. Mol. Biol. 2002;324:703–721. doi: 10.1016/s0022-2836(02)01143-9. [DOI] [PubMed] [Google Scholar]
- 18.Shih AY, Arkhipov A, Freddolino PL, Schulten K. Coarse grained protein-lipid model with application to lipoprotein particles. J. Phys. Chem. B. 2006;110:3674–3684. doi: 10.1021/jp0550816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih AY, Arkhipov A, Freddolino PL, Sligar SG, Schulten K. Assembly of lipids and proteins into lipoprotein particles. J. Phys. Chem. B. 2007;111:11095–11104. doi: 10.1021/jp072320b. [DOI] [PubMed] [Google Scholar]
- 20.Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophys. J. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu F, Jones MK, Chen J, Patterson JC, Catte A, Jerome WG, Li L, Segrest JP. Structures of discoidal high density lipoproteins: A combined computational-experimental approach. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.069914. Epub ahead of print, PMID: 19948731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocco AG, Gianazza E, Calabresi L, Sensi C, Franceschini G, Sirtori CR, Eberini I. Structural features and dynamics properties of human apolipoprotein A-I in a model of synthetic HDL. J. Mol. Graph. Model. 2009;28:305–312. doi: 10.1016/j.jmgm.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Cowieson NP, Kobe B, Martin JL. United we stand: combining structural methods. Curr. Opin. Struct. Biol. 2008;18:617–622. doi: 10.1016/j.sbi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Gingras AR, Bate N, Goult BT, Hazelwood L, Canestrelli I, Grossmann JG, Liu H, Putz NS, Roberts GC, Volkmann N, Hanein D, Barsukov IL, Critchley DR. Embo J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radford SE, Dobson CM, Evans PA. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature. 1992;358:302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- 26.Fleishman SJ, Unger VM, Ben-Tal N. Trends Biochem. Sci. 2006;31:106–113. doi: 10.1016/j.tibs.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Zannis VI, Chroni A, Krieger M. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J. Mol. Med. 2006;84:276–294. doi: 10.1007/s00109-005-0030-4. [DOI] [PubMed] [Google Scholar]
- 28.Matz CE, Jonas A. J. Biol. Chem. 1982;257:4535–4540. [PubMed] [Google Scholar]
- 29.Peng DQ, Wu Z, Brubaker G, Zheng L, Settle M, Gross E, Kinter M, Hazen SL, Smith JD. J. Biol. Chem. 2005;280:33775–33784. doi: 10.1074/jbc.M504092200. [DOI] [PubMed] [Google Scholar]
- 30.Baker PW, Rye KA, Gamble JR, Vadas MA, Barter PJ. J. Lipid Res. 2000;41:1261–1267. [PubMed] [Google Scholar]
- 31.Guinier A. Ann. Phys. 1939;12:161–237. [Google Scholar]
- 32.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cullis PR, de Kruijff B. Biochim. Biophys. Acta. 1979;559:399–420. doi: 10.1016/0304-4157(79)90012-1. [DOI] [PubMed] [Google Scholar]
- 34.Luzzati V, Gulik-Krzywicki T, Tardieu A. Nature. 1968;218:1031–1034. doi: 10.1038/2181031a0. [DOI] [PubMed] [Google Scholar]
- 35.Li XM, Salomon RG, Qin J, Hazen SL. Biochemistry. 2007;46:5009–5017. doi: 10.1021/bi700163y. [DOI] [PubMed] [Google Scholar]
- 36.Lindahl E, Hess B, van der Spoel D. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 37.Sapay N, Tieleman DP. Computer simulation of lipid-protein interactions. Curr. Top. Memb. 2008;60:111–130. [Google Scholar]
- 38.Kandt C, Ash WL, Tieleman DP. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 2007;41:475–488. doi: 10.1016/j.ymeth.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Höltje M, Förster T, Brandt B, Engels T, von Rybinski W, Höltje HD. Molecular dynamics simulations of stratum corneum lipid models: fatty acids and cholesterol. Biochim Biophys Acta. 2001;1511:156–167. doi: 10.1016/s0005-2736(01)00270-x. [DOI] [PubMed] [Google Scholar]
- 40.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular Forces. Dordrecht: D. Reidel Publishing Company; 1981. pp. 331–342. [Google Scholar]
- 41.Darden T, York D, Pedersen L. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 42.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 43.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J. Comp. Chem. 1997;18:1463–1472. [Google Scholar]
- 44.Miyamoto S, Kollman PA. Settle: an analytical version of the shake and rattle algorithms for rigid water models. J. Comp. Chem. 1992;13:952–962. [Google Scholar]
- 45.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins: Struc. Funct. Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chettya PS, Mayneb L, Lund-Katza S, Stranzc D, Englander SW, Phillips MC. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2009;106:19005–19010. doi: 10.1073/pnas.0909708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilser VJ, Freire E. Structure-based Calculation of the Equilibrium Folding Pathway of Proteins. Correlation with Hydrogen Exchange Protection Factors. J. Mol. Biol. 1996;262:756–772. doi: 10.1006/jmbi.1996.0550. [DOI] [PubMed] [Google Scholar]
- 48.Black SD, Mould DR. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal. Biochem. 1991;193:72–82. doi: 10.1016/0003-2697(91)90045-u. [DOI] [PubMed] [Google Scholar]
- 49.Sanner MF, Spehner J-C, Olson AJ. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 50.Dill KA, Flory PJ. Molecular organization in micelles and vesicles. Proc. Natl. Acad. Sci. USA. 1981;78:676–680. doi: 10.1073/pnas.78.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruce CD, Berkowitz ML, Perera L, Forbes MDE. Molecular dynamics simulation of sodium dodecyl sulfate micelle in water: micellar structural characteristics and counterion distribution. J. Phys. Chem. B. 2002;106:3788–3793. [Google Scholar]
- 52.Itri R, Amaral LQ. Distance distribution function of sodium dodecyl sulfate micelles by X-ray scattering. J. Phys. Chem. 1991;95:423–427. [Google Scholar]
- 53.Lipfert J, Columbus L, Chu VB, Lesley SA, Doniach S. Size and shape of detergent micelles determined by small-angle X-ray scattering. J. Phys. Chem. B. 2007;111:12427–12438. doi: 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- 54.Tieleman DP, van der Spoel D, Berendsen HJC. Molecular dynamics simulations of dodecylphosphocholine micelles at three different aggregate sizes: micellar structure and chain relaxation. J. Phys. Chem. B. 2000;104:6380–6388. [Google Scholar]
- 55.Wendoloski JJ, Kimatian SJ, Schutt CE, Salemme FR. Molecular dynamics simulation of a phospholipid micelle. Science. 1989;243:636–638. doi: 10.1126/science.2916118. [DOI] [PubMed] [Google Scholar]
- 56.Yoshii N, Okazaki S. A molecular dynamics study of structure and dynamics of surfactant molecules in SDS spherical micelle. Cond. Mat. Phys. 2007;10:573–578. [Google Scholar]
- 57.Fujiwara S, Itoh T, Hashimoto M, Tamura Y. Molecular dynamics simulation of micelle formation in amphiphilic solution. Mol. Simul. 2007;33:115–119. [Google Scholar]
- 58.Tilcock CP, Cullis PR. Ann. N.Y. Acad. Sci. 1987;492:88–102. doi: 10.1111/j.1749-6632.1987.tb48657.x. [DOI] [PubMed] [Google Scholar]
- 59.Traikia M, Warschawski DE, Recouvreur M, Cartaud J, Devaux PF. Formation of unilamellar vesicles by repetitive freeze-thaw cycles: characterization by electron microscopy and 31P-nuclear magnetic resonace. Eur. Biophys. J. 2000;29:184–195. doi: 10.1007/s002490000077. [DOI] [PubMed] [Google Scholar]
- 60.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J. Am. Mass. Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaption of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silva RAGD, Hillard GM, Li L, Segrest JP, Davidson WS. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 2005;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- 63.Li H-H, Lyles DS, Pan W, Alexander E, Thomas MJ, Sorci-Thomas MG. ApoA-I Structure on discs and spheres. Variable helix registry and conformational states. J. Biol. Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 64.Shih AY, Sligar SG, Schulten K. Biophys. J. 2008;94:L87–L89. doi: 10.1529/biophysj.108.131581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep. Progr. Phys. 2003;66:1735–1782. [Google Scholar]
- 66.Chevalier Y, Zemb T. The structure of micelles and microemulsions. Rep. Progr. Phys. 1990;53:279–371. [Google Scholar]
- 67.Dill KA, Koppel DE, Cantor RS, Dill JD, Bendedouch D, Chen S-H. Molecular conformations in surfactant micelles. Nature. 1984;309:42–45. [Google Scholar]
- 68.Cabane B, Zemb T. Water in the hydrocarbon core of micelles. Nature. 1985;314:385–386. [Google Scholar]
- 69.Goyal PS, Aswal VK. Micellar structure and inter-micelle interactions in micellar solutions: Results of small angle neutron scattering studies. Curr. Sci. 2001;80:972–979. [Google Scholar]
- 70.Hill TL. Statistical Mechanics. Principles and Selected Applications. New York: McGraw-Hill; 1956. [Google Scholar]
- 71.Assmann G, Highet RJ, Sokoloski EA, Brewer HB., Jr Proc. Natl. Acad. Sci. USA. 1974;71:3701–3705. doi: 10.1073/pnas.71.9.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peters-Libeu CA, Newhouse Y, Hall SC, Witkowska HE, Weisgraber KH. J. Lipid Res. 2007;48:1035–1044. doi: 10.1194/jlr.M600545-JLR200. [DOI] [PubMed] [Google Scholar]
- 73.Zheng L, Settle M, Brubaker G, Schmitt D, Hazen SL, Smith JD, Kinter M. Localization of nitration and chlorination sites on apolipoprotein A-I catalyzed by myeloperoxidase in human atheroma and associated oxidative impairment in ABCA1-dependent cholesterol efflux from macrophages. J. Biol. Chem. 2005;280:38–47. doi: 10.1074/jbc.M407019200. [DOI] [PubMed] [Google Scholar]
- 74.Ellena JF, Lepore LS, Cafiso DS. Estimating lipid lateral diffusion in phospholipid vesicles fro 13C spin-spin relaxation. J. Phys. Chem. 1993;97:2952–2957. [Google Scholar]
- 75.Vauhkonen M, Sassaroli M, Somerharju P, Eisinger J. Lateral diffusion of phospholipids in the lipid surface of human low-density lipoprotein measured with a pyrenyl phospholipid probe. Eur. J. Biochem. 1989;186:465–471. doi: 10.1111/j.1432-1033.1989.tb15230.x. [DOI] [PubMed] [Google Scholar]
- 76.Altschul SF, Madden TL, Schaefer AAJZ, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a generation of protein database search programs. Nucleic Acids Res. 1997;25:5101–5109. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.