Abstract
Defining bacterial species and understanding the relative cohesiveness of different components of their genomes remains a fundamental problem in microbiology. Bacterial species tend to be comprised of both a set of core and dispensable genes, with the sum of these two components forming the species pan-genome. The role of the core and dispensable genes in defining bacterial species and the question of whether pan-genomes are finite or infinite remain unclear. Here we demonstrate, through the analysis of 96 genome sequences derived from two closely related sympatric sister species of pathogenic bacteria (Campylobacter coli and C. jejuni), that their pan-genome is indeed finite and that there are unique and cohesive features to each of their genomes defining their genomic identity. The two species have a similar pan-genome size; however, C. coli has acquired a larger core genome and each species has evolved a number of species-specific core genes, possibly reflecting different adaptive strategies. Genome-wide assessment of the level of lateral gene transfer within and between the two sister species, as well as within the core and non-core genes, demonstrates a resistance to interspecies recombination in the core genome of the two species and therefore provides persuasive support for the core genome hypothesis for bacterial species.
Keywords: pan- and core genome, lateral gene transfer, speciation, prokaryote
Introduction
One of the more significant recent discoveries in bacterial genomics is that bacterial species appear to be comprised of both a set of core and dispensable genes, with only the former present in all isolates of that species and with the sum of the two components forming the species pan-genome (or supra-genome) (Tettelin et al. 2005). Much speculation has centered around the origin, composition, and size of bacteria pan-genomes and whether they are finite or infinite (Tettelin et al. 2008; Lapierre and Gogarten 2009). At the same time, actually defining bacterial species has remained somewhat of a conundrum, and there is no clear understanding of the relative roles of the different genomic components in a possible biological definition of bacterial species. The difficulty lies principally in the fact that bacteria exchange genetic material in unique and unusual ways, distinguishing them from eukaryotic genomics and species concepts.
Campylobacter species are a leading bacterial cause of gastroenteritis within the United States and throughout much of the rest of the developed world (Ketley 1997). Campylobacter jejuni and C. coli are closely related sister species (Lefébure and Stanhope 2009) that infect humans as well as a wide range of agricultural organisms, including bovine, swine, and poultry. Both species are commonly isolated from the gastrointestinal tract of the same host organism and thus can be regarded as sympatric (Moore et al. 2005). It has been argued that they may have a very recent divergence of the order of thousands of years (Wilson et al. 2009), whereas another study suggests that the two taxa may actually be converging (Sheppard et al. 2008). Paralleling our interest in understanding the history of genetic exchange in the pan-genome components of these two taxa was our intention to use these data to assess the core genome hypothesis for defining bacterial species. Dykhuizen and Green (1991) first introduced the idea of a biological species concept for bacteria, which was subsequently refined by Lan and Reeves (2000, 2001) who proposed a model suggesting the core genome is the principal genomic unit defining bacterial species. The logic suggests that there is little selective advantage in acquiring core genes from another species and that the gradual divergence of core genes, combined with mechanisms such as mismatch repair and restriction modification systems, will act to inhibit interspecific homologous recombination in the core genome (Denamur et al. 2000; Lawrence and Retchless 2009). Because C. coli and C. jejuni are so closely related, increasing the probability of reciprocal homologous recombination (Roberts and Cohan 1993; Zawadzki et al. 1995; Vulić et al. 1997; Majewski et al. 2000), and are overlapping in their distributions, they provide an ideal species pair in which to test the core genome hypothesis.
Materials and Methods
Strains and Genome Sequencing
A total of 42 strains of C. coli and 43 strains of C. jejuni were selected to represent different hosts, countries, and sequence types (supplementary table S1, Supplementary Material online). Genomic DNA was sequenced using the Illumina GA II instrument at the Cornell Biotechnology Resource Center. One lane per strain yielded between 4 and 8 million 36 bp reads. Illumina traces have been deposited with the National Center for Biotechnology Information (NCBI) sequence read archive database under the accession numbers SRP001790 and SRA010929.
Read Mapping and De Novo Assembly
Sequence reads of each species were first aligned to the C. coli RM2228 and C. jejuni NCTC11168 genomes, respectively, using MAQ (Li et al. 2008) (fig. 1). De novo assemblies were performed using Velvet (Zerbino and Birney 2008). Several hash lengths (from 21 to 31) and coverage cutoffs (0–80) were used, and the best assembly per strain was selected based on the N50, which refers to a weighted median, with 50% of the entire assembly contained in contigs equal to or greater than this value. Open reading frames were called using Glimmer (Delcher et al. 2007). A preliminary analysis revealed that several regions of both genomes were too divergent to map the reads. To resolve these regions, using Blast, each MAQ consensus coding sequence (CDS) containing unresolved positions was searched against the de novo assemblies. When a single hit was found, the corresponding open reading frame was then aligned to the consensus sequence and whenever possible the undetermined positions resolved.
FIG. 1.—
Pipeline combining de novo assemblies and read mapping, yielding a gene content table and core gene alignments.
Construction of the three Species Alignments
For the one-to-one orthologs shared among C. jejuni, C. coli, and C. upsaliensis, the MAQ consensus data, as well as the coding genes from C. jejuni and C. upsaliensis genomes available on NCBI (supplementary table S2, Supplementary Material online), were merged and aligned to produce 967 multiple alignments each containing 98 sequences. Intragenic recombination was assessed for each of these gene alignments using GARD (Kosakovsky Pond et al. 2006). Genes with significant breakpoints (12%) were split into gene fragments.
Orthology Reconstruction
An orthology search was performed using the de novo assembled genomes and 17 Campylobacter genomes available on NCBI (supplementary table S2, Supplementary Material online). An all-versus-all Blast search was performed and orthologs delimited using MCL (van Dongen 2000). A common problem with bacteria draft genomes is that many genes are split into fragments because they span over two contigs. Because the resulting partial genes show no homology, clustering methods will exclude one fragment from the correct cluster and assign it to a new one (supplementary fig. S1, Supplementary Material online). The clusters made of split proteins were searched by looking for 1) clusters made of draft genome proteins only and 2) proteins that showed strong homology with proteins of another cluster, with the exception of proteins coming from the same genome. To correct for missed genes in the de novo assembly, the MAQ consensus sequences were used to predict gene presence/absence by counting the percentage of sites absent in each CDS using a 50% absent sites threshold to delimit the present and absent genes. The C. jejuni GenBank draft genome CG8486 appeared to be missing large sections of the genome and was excluded from the core genome size estimation.
Simulations
To assess the pipeline sensitivity and specificity, we simulated Illumina reads using MAQ. One hundred independent simulations of 5 million reads were produced using the C. jejuni RM1221 genome and run through the pipeline. False positives were counted as the number of genes found in the simulations but absent in the RM1221 genome and false negatives the number of missed genes in the simulations.
Annotation and Enrichment Tests
Gene ontology (Ashburner et al. 2000) (GO) annotations were obtained for each orthologous cluster using HMMER searches (Eddy 2008) on the Pfam A and B databases (Finn et al. 2010) and using Blast against the Uniref90 database (Suzek et al. 2007). Best hits were then mapped to GO terms using the pfam2go mapping (Hunter et al. 2009) and the GOA database (Barrell et al. 2009), respectively. GO enrichment tests were conducted using GO::TermFinder (Boyle et al. 2004) with false discovery rate (FDR) correction and a 5% significance level.
Principal Component Analysis
To characterize genes of different occurrence frequencies, we calculated the guanine and cytosine (GC) content, standardized per genome, the codon usage distance relative to the average genomic codon usage using B(g|C) (Karlin et al. 1998), and the U/C choice in degenerate codon position index with P2 (Gouy and Gautier 1982). These variables can be used to differentiate genes under high translational selection as well as laterally transferred genes. We also used the gene lengths, Glimmer scores, Blast best E value and number of hits on non-redundant (NR), as well as HMMER best score on Pfam. We used a centered and scaled between-group principal component analysis (Dolédec and Chessel 1987) to discriminate the genes belonging to five different occurrence frequency groups, defined using the k-means method, and implemented in ade4 (Dray and Dufour 2007).
Gene Trees, Interspecies, and Intraspecies Lateral Gene Transfer Detection
Gene trees were reconstructed with PhyML (Guindon and Gascuel 2003) using a GTR + G model and the subtree pruning and regrafting branch swapping. Phylogenetic signal was assessed using nonparametric bootstrap with 500 pseudoreplicates. Because species monophyly was supported by most gene trees, a quantitative assessment of the number of interspecies lateral gene transfer (LGT) per gene could be performed using the species tree as a control (supplementary fig. S2, Supplementary Material online). Gene trees were filtered that did not contain a node giving rise to a C. coli and C. jejuni clade, each supported with at least 50% bootstrap, with a branch length at least 1% of the tree length, and containing at least 60% of the species strains. For the remaining gene trees (85%), the number of interspecies LGTs were determined by counting the number of species sequences found within the other species' clade. The absence of congruent phylogenetic signal within the species genealogies prevented us from using the same method for assessing intraspecific recombination. Thus, for each species independently, we used three substitution pattern–based recombination detection methods: the pairwise homoplasy index (Bruen et al. 2006) (PHI), neighbor similarity score (NSS) (Jakobsen and Easteal 1996), and MaxChi (Maynard Smith 1992), as implemented in Phipack (Bruen et al. 2006).
Interspecies LGTs in the Core versus Dispensable Genes
Because of the lack of a control tree for the dispensable genes, we used a distance approach to obtain a comparative estimate of the influence of interspecies LGT in the core versus dispensable genes. The logic lies in the fact that with increasing levels of interspecies LGT, the overlap between the intra- and interspecies pairwise divergences increases. By comparing the overlap across the core and dispensable genes, one can obtain a comparative estimate of the level of interspecies LGT. Pairwise divergences were calculated using the Hasegawa–Kishino–Yano substitution model (Hasegawa et al. 1985) using PAUP* (Swofford 2002).
Results and Discussion
Sampling Gene Repertoire with Illumina
The development of high throughput sequencing technologies that produce millions of short reads are becoming widely used as an efficient strategy to determine genome-wide polymorphism by mapping the reads against a reference genome (Bentley et al. 2008). Sequencing the bacterial pan-genome requires a de novo assembly strategy, and genome de novo assemblies are typically accomplished with more expensive technologies such as 454 Roche (Margulies et al. 2005), involving longer reads. Recently, however, it has been demonstrated that the majority of genes in prokaryotic genomes can be reconstructed with short reads, even if the complete closed genomes cannot (Kingsford et al. 2010). In parallel to this work, we developed a pipeline that combines mapping and de novo assembly of short read sequence data to produce an exhaustive assessment of polymorphism in the core genome and a thorough representation of the gene content of each studied genome. Using simulations of illumina reads from a complete genome, we first estimated a rate of false negatives (i.e., missed genes) of 2.4% and a percentage of false positives (false genes) of 4.1% in the simulated genomes. The errors were concentrated on a few genes that were either false positive or false negative in all the simulations. A closer look revealed that the regions encoding these genes were either present in the de novo assemblies but not considered genes or not considered genes in the genome used as template for the simulations (RM1221). A comparison between the recently reannotated C. jejuni NCTC11168 genome (Gundogdu et al. 2007) and the older RM1221 annotation supported most of the simulation gene calls. If one considers the NCTC11168 annotation as the correct one, we now obtain a false-negative rate of 0.06% and a percentage of false-positive calls of 0.6%, which attest that Illumina technology can be used on small genomes to get an accurate view of a species pan-genome and that most mistakes are made at the gene call step, not during assembly.
Estimating Core and Pan-Genomes
The question of whether sufficient genomes have been sequenced to describe the core and pan-genome content of each species was assessed by observing the change in size of these components with increasing numbers of sampled genomes. The decrease in core genome size quickly leveled off, for both C. coli and C. jejuni, although continuing to decrease by one or a few genes even after more than 40 samples (fig. 2A). However, with a more liberal core gene definition that allows a single strain to miss a core gene out of the 43 and 51 genomes per species, both core genomes quickly reached a plateau. The C. coli core genome was larger than that of C. jejuni, by 121 orthologs, but the average C. coli genome size was also larger (1,732 vs. 1,609 orthologs), yielding about the same proportion of core genes per genome (83.5% vs. 82.4%) for each species.
FIG. 2.—
Core genome (A) and pan-genome (B) size estimates, as well as number of newly discovered genes (C), as a function of the number of sequenced genomes. The genome input order was randomly permuted 1,000 times. The lines describe the average number of genes (using median statistics), whereas the vertical bars delimit the second and third quartiles, with the exception of panel (C), where quartiles are represented by short dashed lines. On panel (A), the long dashed lines correspond to the average core genome size when one taxon is allowed a missing core gene, whereas on the (B and C) panels, they describe the pan-genome size or number of new genes for the combined species data set when the putative pseudogenes are excluded.
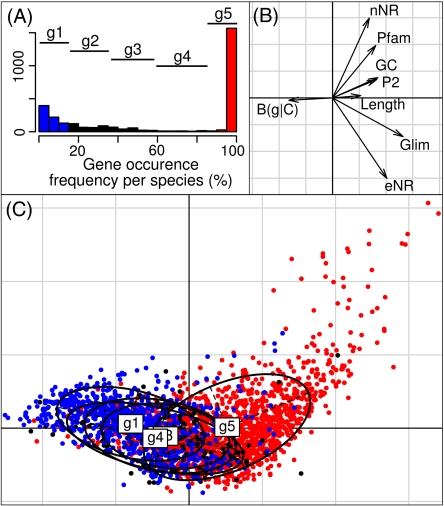
The same logic was applied to the pan-genomes of both species, with the result that each taxon has a pan-genome size of around 2,600 genes, whereas the pan-genome of both species combined reaches approximately 3,000 genes. For both species, the increase in pan-genome size clearly reaches a plateau, particularly in comparison to the pan-genome of eight different Campylobacter species (fig. 2B). With 40 genomes sequenced, on the average, only two new C. coli and three C. jejuni genes are discovered (fig. 2C), for each newly sequenced genome, and with both species combined, after more than 80 genomes, one would expect to discover only a single new gene per newly sequenced genome. Furthermore, if one examines in detail the characteristics of these rare genes, a significant number has the hallmark of putative pseudogenes. To characterize genes of different occurrence frequencies, from core genes to unique genes, we assembled a data set that included information pertaining to gene composition, structure, and homology to known proteins (fig. 3). All the variables were correlated and provided good discrimination of rare genes from core genes (g1–g5, respectively). The rare genes exhibited a divergent codon usage, lower GC content, shorter proteins, as well as lower Glimmer, HMMER, and Blast scores, whereas the core genes displayed opposite patterns. Within the rare genes, we could distinguish a set of genes with no hits at all on the NR and Pfam databases, as well as being short (median of 195 bp), having peculiar base composition and low Glimmer scores (supplementary fig. S4, Supplementary Material online). We consider this subset of the rare genes (425 proteins) to be putative pseudogenes generated during the open reading frame prediction. When using a different reading frame, about 20% of these putative pseudogenes turn out to have Blast hits against the NR database and other Campylobacter clusters. This indicates that these genes are likely to be actual CDS translated in the wrong reading frame, but their re-inclusion in the correct reading frame would not change our pan-genome estimate. It is also the case that 43 putative pseudogene clusters (10%) were exclusively found at contig ends and were therefore incomplete, increasing their chances of being artificial genes generated during the gene call step. Thus, for about a third of the putative pseudogenes, we have additional arguments for their exclusion; for the remaining genes, additional analysis such as genome closure or transcriptome screening would be necessary to definitively exclude them from the pan-genome. Removing these putative pseudogenes from the analysis drops the pan-genome size for both species (fig. 2B), the estimate now approaches an even more complete plateau, and with both species combined, the average number of newly discovered genes quickly reaches zero (fig. 2C). Thus, our analysis indicates that the pan-genome of C. coli and C. jejuni is indeed finite and that sequencing any further genomes would likely recover only a few putative pseudogenes.
FIG. 3.—
Principal component analysis (PCA) of Campylobacter coli and C. jejuni gene characteristics aimed at discriminating different gene frequency groups (between-group PCA). The first and second components of the PCA summarized 45% and 21% of the total inertia, respectively, whereas the gene frequency group factor accounted for 17% of the total inertia. (A) Histogram of gene occurrence frequency and the frequency groups g1–g5 that were used in the analysis. (B) Canonical graph, with B(g|C) representing the codon usage distance to the average genomic codon usage; nNR, the number of hits with the NR database; Pfam, the best Pfam score; GC, the GC content; P2, the U/C choice in degenerate codon position index; length, protein length; Glim, Glimmer score; and eNR, the log transformed best NR E value. (C) Projection of gene orthologs on the first and second components, with red dots representing genes belonging to the g5 group, black dots the g2–g4 groups, and blue dots g1. Ellipses and gravity centers are used to represent the frequency group distribution.
Species Pan-Genome Overlap
A comparison of the two species pan-genomes indicates that the greatest overlap involved the shared core genomes, followed by the shared dispensable genes, followed distantly by the specific components of each species, and, in particular, the sets of species-specific dispensable genes (fig. 4). Overall, the two species have a gene repertoire that is almost completely overlapping. This pattern is not due to a low level of variability in the gene repertoire of the genus Campylobacter because despite completely sequencing the pan-genomes of C. coli and C. jejuni, we missed more than 1,600 other Campylobacter genes (fig. 4). Regarding core genome overlap, C. coli showed more unique core genes (86 vs. 35) as well as core genes in one species that are dispensable in the other (82 vs. 17). The 86 unique core genes in C. coli included many (75%) that were not found in any other Campylobacter species and were therefore probably gained during C. coli evolution. Many of these genes formed clusters around particular genomic locations (supplementary table S7, Supplementary Material online) and were likely acquired through LGT events involving large blocks of sequence. This set of genes was significantly enriched in unannotated genes (supplementary table S3, Supplementary Material online) but nonetheless are likely important in defining the unique features of the biology of this species and therefore warrant further characterization. The unique dispensable set of genes from each species was composed of many putative pseudogenes but also genes with a viral signature as well as genes involved in DNA modification and regulation (supplementary table S3, Supplementary Material online), suggesting that each species may have evolved its own set of mobile genetic elements (Hwang et al. 2009). Another set of C. coli core genes was dispensable in C. jejuni and generally present as genomic clusters in C. coli (supplementary table S8, Supplementary Material online). Although these genes are clearly found in the ecosystem of both species, only C. coli integrated them in its core genome. Thus, contrary to the former set of unique core genes that could have been gained during an opportunistic transfer event in the C. coli ancestor, these genes were specifically selected by C. coli but rejected by C. jejuni, increasing their relevance in demarcating the biology of the two species. A similar pattern is apparent for the C. jejuni core, though involving many fewer genes. The unique C. jejuni core genes were enriched in transporter activity and membrane components (e.g., kdp and pglG genes), whereas the core set that was dispensable in C. coli was enriched in citrate transport and response to chemical stimulus (fig. 5; supplementary table S3, Supplementary Material online). Taken collectively, these observations support the view that C. coli evolved a larger genome by incorporating additional genes in its core genome since its split from common ancestry with C. jejuni. A commonly accepted model suggests that bacterial genomes constantly lose genes to compensate for gene gain and duplication (Mira et al. 2001). Additionally, we suggest that selection against the loss of the most important genes ultimately shapes and delimits the species core genome, whereas dispensable genes are comprised of nonessential genes that go through an intense turnover. In this context, the inclusion of a new gene in the C. coli core genome probably coincides with new environmental requirements.
FIG. 4.—
Overlap between the core and dispensable (disp.) genomic components of Campylobacter coli and C. jejuni; core genes were allowed to be missing in one strain per species. The absent/absent section represents genes that were found in other Campylobacter species but absent in C. coli and C. jejuni. Cirle radii are proportional to the number of genes. The black surface represents the proportion of putative pseudogenes.
FIG. 5.—
Campylobacter jejuni NCTC11168 genome map with genes (CDS) displayed on either strand as tracks 1 and 2 (tracks numbered from outside in). Gene frequencies and core genes are displayed for C. jejuni (tracks 3 and 4) and C. coli (tracks 5 and 6), as well as recombinant genes that showed evidence of interspecies (tracks 7 and 9, the height being proportional to the number of LGT events) and intraspecies (tracks 8 and 10) LGTs. Coding genes that are core genes in C. jejuni but absent (in black) or dispensable (in gray) in C. coli are labeled with gene names or locus names. Color code for each track is given at the top of the figure.
Gene Content and Host Adaptation
Campylobacter coli and C. jejuni tend to differ in their relative prevalence in animal host species and various environmental sources (Rosef et al. 1983; Waldenström et al. 2002; Bull et al. 2006), and there is some evidence that both taxa may include groups of host-specific putative ecotype strains (Meinersmann et al. 2005; Miller et al. 2006; McCarthy et al. 2007; Colles et al. 2008; Sopwith et al. 2010). This in turn suggests the possibility that some genes may be linked to particular host species. Whereas there was no link between gene content and host in C. jejuni (Fisher exact test P value = 0.9), in C. coli, there was an overall link (P value < 0.01) but no clear association between any specific gene and host (post hoc Fisher exact test P values with FDR correction >0.05). Though we cannot rule out the possibility that some genes have a role in differential host adaptation, gene content does not seem to be a major factor, suggesting that other processes, such as gene regulation, should be investigated.
Testing the Core Genome Hypothesis
Lan and Reeves (2001) core genome hypothesis suggests that the core genome is the primary cohesive unit defining a bacterial species. More specifically, they suggest that if the core genome is relatively free of interspecific recombination, while still harboring examples of intraspecific recombination, then this is analogous to a biological species concept for bacteria. Using three different substitution pattern–based methods (PHI, MaxChi, and NSS), we found significant levels of intraspecies recombination in C. jejuni and C. coli genes (61% and 33%, respectively, using PHI; supplementary table S4, Supplementary Material online; fig. 5). In contrast, using a phylogenetic signal approach based on the identification of one species sequence in the other species lineage, we found very few instances of recombination between species (fig. 5). A total of 80% of the core genes were free of any between-species recombination (fig. 5), and the frequency of transfer per strain and per gene was lower than 1.15% in C. coli and 0.29% in C. jejuni. If two or more monophyletic sequences were found in the other species, they were counted as a single transfer event, and this slightly reduces the rates of transfer to 1.12% and 0.25%, respectively. Using the index of dispersion (D = σ2/μ) with a gene window size of 10 or 20 genes, the distribution of the intraspecies LGTs was underdispersed in C. jejuni (0.5 < D < 0.6) and slightly overdispersed in C. coli (1.8 < D < 2.4), whereas interspecies LGT genes were strongly aggregated for both species (7.8 < D < 19.2; fig. 5). At least for C. jejuni, this might indicate that the inter- and intraspecies LGT events are controlled by different mechanisms. There was no clear accumulation of recombination in particular genes or strains, with the exception of one C. coli strain (cco71), which had 9.6% of its core genes harboring C. jejuni alleles. Interestingly, most of the between-species transfers were between the two sister species, with only 5% of them having a donor other than these two species. A different approach, based on gene tree comparisons, showed that with the exception of species monophyly, there was virtually no consensus history between genes (supplementary fig. S7, Supplementary Material online). Within species, lack of gene tree consensus can be due to either weak overall phylogenetic signal or extensive recombination. We tested both options by screening gene tree bipartition support and the best conflicting bipartition (fig. 6), as done in quartet analysis (Zhaxybayeva et al. 2006). The species monophyly bipartitions were commonly supported by gene trees, although they were rarely rejected. There were also a few supported bipartitions, as well as a set of more than 4,000 bipartitions, most of which were supported by a single gene, while being rejected by many others. This latter set of bipartitions was not in conflict with species monophyly and indicates the presence of many intraspecies LGT events involving unique loci in both species. Finally, there was a set of bipartitions (extreme right of fig. 6) conflicting with species monophyly, supported by very few genes, and rejected by a wide majority of genes. Thus, although it is true that the methods used to assess the level of LGT have very different assumptions, sensitivity and specificity, they all nonetheless support the idea that in Campylobacter, recombination within the core genome is frequent within species, rare between sister species, and extremely rare with other species.
FIG. 6.—
Gene tree bipartition support. This graph displays the percentage of gene tree supporting or rejecting a set of 5,264 bipartitions that are supported by at least one gene tree. Support was assessed using nonparametric bootstrap, with support higher than 70% and 90% in red and blue, respectively. Bipartitions are sorted from the least rejected to the most rejected, resulting in bipartitions supporting species paraphyly at the extreme right. The five most commonly supported bipartitions are labeled as “(C.jejuni),” Campylobacter jejuni monophyly; “(C.coli),” C. coli monophyly; “a,” cje12 and cjj81116 monophyly; “b,” cje23 and cjjhb9313 monophyly; and “c,” monophyly of the C. coli species with the exception of cco71.
The level of interspecific recombination in the dispensable components was assessed by comparing the amount of overlap between the intra- and interspecies divergences (fig. 7). In agreement with the overall picture of a cohesive core genome and the absence of cohesiveness in the dispensable genome, the core genes displayed little divergence overlap, whereas there was considerable intra- and interspecies overlap involving the dispensable genes.
FIG. 7.—
Overlap between the intra- (intra) and interspecies (inter) divergences in the core (white) and dispensable (gray) genes.
Conclusion
Given the reduced number of LGTs between the two Campylobacter species and the maintenance of species-specific features like species-specific core genes, this analysis does not support the idea that these two species are converging or “despeciating” (Sheppard et al. 2008). Although some strains and genes show a higher level of interspecies transfer, they likely are rare exceptions. As mentioned by Caro-Quintero et al. (2009), the Sheppard et al. (2008) conclusions may have been biased by the use of very few loci (seven partial genes) and the use of haplotype sequences instead of all the original individual sequences, thus overweighting certain rare recombinant alleles. That said, a more formal test would involve the comparison of a model of constant or decreasing gene flow between the two species against a model where the gene flow is increasing.
Our combined analyses of gene repertoire and recombination in Campylobacter indicate that despite the fact these two species share the same habitat, they have both evolved a small number of unique core genes and developed a species recombination barrier. Given our relative lack of specific knowledge of the function of many of these unique core genes, it is difficult to assess if adaptation of their respective core genomes was an instrumental factor associated with an ecological shift that triggered speciation of these two organisms or if additional mechanisms, potentially nonadaptive, were involved. Additional characterization of these genes as well as the differences between the species biology and ecology will be necessary to definitively settle this issue.
Thus, it seems that a tractable species concept for at least some groups of bacteria may be close at hand. However, the degree to which the core genome concept can be widely applied across all bacteria may well depend on factors such as the relative ease with which the species is naturally transformable, as well as the existence and specificity of restriction modification systems. Campylobacter is known to be highly competent, but at the same time, there is experimental evidence that it may be quite selective for Campylobacter DNA (Wilson et al. 2003), perhaps reflecting a high efficiency in its existing restriction modification systems (Miller et al. 2005). Pan-genome dynamic studies of bacteria differing in recombination-related features such as competence, mismatch repair, and restriction modification systems will be necessary before a thorough picture of bacteria genome species concepts can be attained.
Supplementary Material
Supplementary tables and figures are available at Genome Biology and Evolution Online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This project was supported by the Cornell University Zoonotic Research Unit of the Food and Waterborne Diseases Integrated Research Network, which is funded by the National Institute of Allergy and Infectious Diseases, US National Institutes of Health, under contract number N01-AI-30054 (ZC003-05 and ZC010-09) awarded to M.J.S.
References
- Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell D, et al. The GOA database in 2009—an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, et al. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull SA, et al. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl Environ Microbiol. 2006;72:645–652. doi: 10.1128/AEM.72.1.645-652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Quintero A, Rodriguez-Castaño GP, Konstantinidis KT. Genomic insights into the convergence and pathogenicity factors of Campylobacter jejuni and Campylobacter coli species. J Bacteriol. 2009;191:5824–5831. doi: 10.1128/JB.00519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles FM, Dingle KE, Cody AJ, Maiden MC. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl Environ Microbiol. 2008;74:3583–3590. doi: 10.1128/AEM.02491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E, et al. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell. 2000;103:711–721. doi: 10.1016/s0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- Dolédec S, Chessel D. Rythmes saisonniers et composantes stationnelles en milieu aquatique I—Description d'un plan d'observations complet par projection de variables. Acta Oecol Oec Gen. 1987;8:403–426. [Google Scholar]
- Dray S, Dufour AB. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. 2007;22:1–20. [Google Scholar]
- Dykhuizen DE, Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput Biol. 2008;4:e1000172. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gundogdu O, et al. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics. 2007;8:162. doi: 10.1186/1471-2164-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hunter S, et al. Interpro: the integrative protein signature database. Nucleic Acids Res. 2009;37:211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, et al. Isolation and characterization of bacteriophages specific for Campylobacter jejuni. Microbiol Immunol. 2009;53:559–566. doi: 10.1111/j.1348-0421.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- Jakobsen IB, Easteal S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci. 1996;12:291–295. doi: 10.1093/bioinformatics/12.4.291. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrázek J, Campbell AM. Codon usages in different gene classes of the Escherichia coli genome. Mol Microbiol. 1998;29:1341–1355. doi: 10.1046/j.1365-2958.1998.01008.x. [DOI] [PubMed] [Google Scholar]
- Ketley JM. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143:5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- Kingsford C, Schatz MC, Pop M. Assembly complexity of prokaryotic genomes using short reads. BMC Bioinformatics. 2010;11:21. doi: 10.1186/1471-2105-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Lan R, Reeves PR. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 2000;8:396–401. doi: 10.1016/s0966-842x(00)01791-1. [DOI] [PubMed] [Google Scholar]
- Lan R, Reeves PR. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 2001;9:419–424. doi: 10.1016/s0966-842x(01)02133-3. [DOI] [PubMed] [Google Scholar]
- Lapierre P, Gogarten JP. Estimating the size of the bacterial pan-genome. Trends Genet. 2009;25:107–110. doi: 10.1016/j.tig.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Retchless AC. The interplay of homologous recombination and horizontal gene transfer in bacterial speciation. Methods Mol Biol. 2009;532:29–53. doi: 10.1007/978-1-60327-853-9_3. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Stanhope MJ. Pervasive, genome-wide positive selection leading to functional divergence in the bacterial genus Campylobacter. Genome Res. 2009;19:1224–1232. doi: 10.1101/gr.089250.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Zawadzki P, Pickerill P, Cohan FM, Dowson CG. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J Bacteriol. 2000;182:1016–1023. doi: 10.1128/jb.182.4.1016-1023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- McCarthy ND, et al. Host-associated genetic import in Campylobacter jejuni. Emerging Infect Dis. 2007;13:267–272. doi: 10.3201/eid1302.060620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann RJ, Phillips RW, Hiett KL, Fedorka-Cray P. Differentiation of Campylobacter populations as demonstrated by flagellin short variable region sequences. Appl Environ Microbiol. 2005;71:6368–6374. doi: 10.1128/AEM.71.10.6368-6374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, et al. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology. 2006;152:245–255. doi: 10.1099/mic.0.28348-0. [DOI] [PubMed] [Google Scholar]
- Miller WG, et al. Diversity within the Campylobacter jejuni type I restriction-modification loci. Microbiology. 2005;151:337–351. doi: 10.1099/mic.0.27327-0. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Moore JE, et al. Campylobacter. Vet Res. 2005;36:351–382. doi: 10.1051/vetres:2005012. [DOI] [PubMed] [Google Scholar]
- Roberts MS, Cohan FM. The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics. 1993;134:401–408. doi: 10.1093/genetics/134.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosef O, Gondrosen B, Kapperud G, Underdal B. Isolation and characterization of Campylobacter jejuni and Campylobacter coli from domestic and wild mammals in norway. Appl Environ Microbiol. 1983;46:855–859. doi: 10.1128/aem.46.4.855-859.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Falush D, Maiden MC. Convergence of Campylobacter species: implications for bacterial evolution. Science. 2008;320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- Sopwith W, et al. Investigation of food and environmental exposures relating to the epidemiology of Campylobacter coli in humans in Northwest England. Appl Environ Microbiol. 2010;76:129–135. doi: 10.1128/AEM.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. Uniref: comprehensive and non-redundant uniprot reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- Swofford DL PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- van Dongen S. Graph clustering by flow simulation [Ph.D. thesis] [Utrecht(Netherlands)]: University of Utrecht; 2000. [Google Scholar]
- Vulić M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci U S A. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenström J, et al. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl Environ Microbiol. 2002;68:5911–5917. doi: 10.1128/AEM.68.12.5911-5917.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DL, et al. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology. 2003;149:3603–3615. doi: 10.1099/mic.0.26531-0. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, et al. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol Biol Evol. 2009;26:385–397. doi: 10.1093/molbev/msn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki P, Roberts MS, Cohan FM. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics. 1995;140:917–932. doi: 10.1093/genetics/140.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]