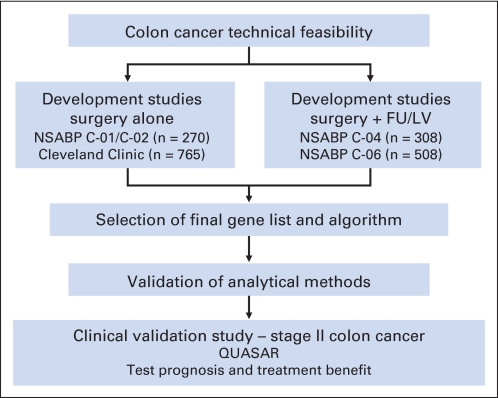
Abstract
Purpose
These studies were conducted to determine the relationship between quantitative tumor gene expression and risk of cancer recurrence in patients with stage II or III colon cancer treated with surgery alone or surgery plus fluorouracil (FU) and leucovorin (LV) to develop multigene algorithms to quantify the risk of recurrence as well as the likelihood of differential treatment benefit of FU/LV adjuvant chemotherapy for individual patients.
Patients and Methods
We performed quantitative reverse transcription polymerase chain reaction (RT-qPCR) on RNA extracted from fixed, paraffin-embedded (FPE) tumor blocks from patients with stage II or III colon cancer who were treated with surgery alone (n = 270 from National Surgical Adjuvant Breast and Bowel Project [NSABP] C-01/C-02 and n = 765 from Cleveland Clinic [CC]) or surgery plus FU/LV (n = 308 from NSABP C-04 and n = 508 from NSABP C-06). Overall, 761 candidate genes were studied in C-01/C-02 and C-04, and a subset of 375 genes was studied in CC/C-06.
Results
A combined analysis of the four studies identified 48 genes significantly associated with risk of recurrence and 66 genes significantly associated with FU/LV benefit (with four genes in common). Seven recurrence-risk genes, six FU/LV-benefit genes, and five reference genes were selected, and algorithms were developed to identify groups of patients with low, intermediate, and high likelihood of recurrence and benefit from FU/LV.
Conclusion
RT-qPCR of FPE colon cancer tissue applied to four large independent populations has been used to develop multigene algorithms for estimating recurrence risk and benefit from FU/LV. These algorithms are being independently validated, and their clinical utility is being evaluated in the Quick and Simple and Reliable (QUASAR) study.
INTRODUCTION
Although adjuvant chemotherapy is the standard of care in stage III colon cancer, its routine use in patients with stage II colon cancer is controversial.1–10 The Quick and Simple and Reliable (QUASAR) study11 showed that adjuvant chemotherapy with fluorouracil (FU) plus leucovorin (LV) produces a small (approximately 3%) survival benefit in stage II colon cancer, which must be balanced with its toxicity, including toxic deaths (approximately 0.5%). This narrow therapeutic index underscores the importance of selecting the appropriate patients for adjuvant treatment.
In current practice, clinical and pathologic markers (ie, intestinal perforation/obstruction, pathologic stage T4, presence of lymphatic/vascular invasion, high tumor grade, < 12 nodes examined) can identify a minority of patients with stage II disease who have higher recurrence risk, but they do not adequately assess recurrence risk for individual patients. To address this issue, the use of molecular markers, such as microsatellite instability (MSI)/mismatch repair (MMR), LOH 18q, and levels of expression of individual genes or groups of genes12–21 has been investigated. Some recent studies suggest MMR deficiency (ie, MSI high) may identify a small percentage (approximately 15%) of patients with stage II disease who receive little benefit from FU/LV.22 However, the clinical utility of these markers remains under study.23
Here, we report the application of the quantitative reverse transcription polymerase chain reaction (RT-qPCR) platform developed for the Oncotype DX Breast Cancer Assay (Genomic Health, Inc, Redwood City, CA)24–26 in four independent colon cancer studies to generate the 12-gene recurrence score and 11-gene treatment score algorithms that, if validated, will quantify the risk of recurrence as well as the likelihood of differential treatment benefit of FU/LV adjuvant chemotherapy for individual patients with stage II colon cancer.
PATIENTS AND METHODS
Patients and Samples
Samples from four independent cohorts of patients with stage II or stage III colon cancer treated with surgery alone (National Surgical Adjuvant Breast and Bowel Project [NSABP] C-01/C-02 or Cleveland Clinic [CC] study) or surgery plus FU/LV (NSABP C-04, NSABP C-06) were studied (Appendix Table A1, online only; Appendix Fig A1, online only).27–30 Prespecified criteria for being evaluable were as follows: eligibility for the parent clinical studies; availability of the fixed, paraffin-embedded (FPE) tumor block from initial diagnosis; presence of sufficient tumor (ie, ≥ 5% of tissue area occupied by invasive cancer cells in the guide hematoxylin and eosin slide); pathology diagnosis of colon adenocarcinoma (excluding signet ring carcinoma); adequate RNA to perform quantitative RT-qPCR analysis (≥ 1,069 ng for C-01/C-02 and C-04 and ≥ 587 ng for CC and C-06); and sufficient RNA quality by predefined metrics.
Sample Preparation
For each patient, RNA was extracted from three pooled 10-μm sections obtained from archived FPE colon tumor tissue. Nontumor elements were commonly identified on the guide hematoxylin and eosin slide reviewed for each patient and were removed by manual microdissection before transfer to the extraction tube.
Pathology, Assay Methods, Gene Selection, Reference Gene Normalization
Assessment of tumor grade was performed according to WHO criteria31 by an academic surgical pathologist with sub-specialty expertise in gastrointestinal pathology. The extracted RNA was quantified and then analyzed by RT-qPCR.32 For the C-01/C-02 and C-04 cohorts, two 384 well plates, which contained a total of 761 unique assays (ie, 761-gene panel), were used for each sample. With the exception of four assays (three K-ras mutations and one BRAF mutation), all assays were designed to detect the expression levels of wild-type genes. The panel of 761 candidate genes (Appendix Table A2, online only) was constructed from published gene expression profiling data and from biologic pathways identified as functionally important in colon cancer.17–21 For the CC and C-06 cohorts, one 384-well plate, which contained 375 unique assays (ie, 375-gene panel), was used for each sample. The genes for the 375-gene panel were chosen from the 761-gene panel on the basis of the strength of the association of their level of expression with recurrence risk and chemotherapy benefit in the C-01/C-02 and C-04 studies. Gene expression measurements were normalized relative to five reference genes. Among the available samples across the four cohorts, only eight were excluded because of inadequate RT-qPCR expression.
MMR status was assessed by immunohistochemistry for MLH1 and MSH2 (which identify > 90% of the MMR-deficient tumors) on fixed, primary colon tumor tissue in the CC study.33
Blinding and Data Preparation
FPE tissue sections were prepared by either NSABP or CC personnel and were shipped to Genomic Health, Inc (Redwood City, CA), where the expression profiling was performed, blinded to the clinical data. The expression data and the clinical/pathology data were independently locked and then merged to construct the analysis data set for each study.
Study Design, Objectives, and End Points
The primary objective of all four studies was to identify genes associated with recurrence-free interval (RFI), defined as the time from surgery to first colon cancer recurrence. Deaths before recurrence were considered censoring events. Second primary cancers were considered neither events nor censoring events. Secondary end points were disease-free survival and overall survival.
Analysis Methods
Prespecified univariate (primary analysis) and multivariate relationships between clinical outcomes and categorical or continuous variables (eg, gene expression) were modeled using Cox proportional hazards regression.34 All baseline patient characteristics related to RFI (P < .20) were included in the multivariate analysis for a given study. Hazard ratios (HRs) were tested for significance using the likelihood ratio test.35 For univariate models of gene expression and RFI, an unadjusted P value less than .05 was considered significant. A test of interaction was used to identify genes that predict treatment benefit; because such tests have lower power compared with the main effect tests, an unadjusted P value of less than .10 was considered significant. No adjustment for multiplicity was applied. To estimate the false discovery rate (FDR), the Benjamini-Hochberg method was used within each study,36 and a permutation-based method37 was used across studies.
For each of the 375 genes assessed in the four studies, univariate t tests were performed to identify mean differences in gene expression between patients with stage II and stage III disease in each study. Additionally, Cox proportional hazards regression models of gene expression, stage, and the interaction of gene expression and stage, stratified by study, were examined, and a P value of less than .10 for interaction was considered significant. In the absence of strong evidence of stage differences, data across stages were combined for gene discovery and algorithm development.
To identify clusters of coexpressed genes and to facilitate the understanding of important biologic pathways, unsupervised hierarchical clustering of genes was performed using Pearson r as the distance measure for gene expression and the unweighted pair-group average as the amalgamation method.35 Similar results were obtained using other methods, such as principal component analysis.
A smaller subset of genes significantly and consistently related to risk of recurrence was identified by examining the results across studies. Multiple factors were considered for gene inclusion in algorithm development, including, but not limited to, the known role of the genes in important biologic pathways, analytic performance, and range of expression. The final gene panels and algorithms for prediction of recurrence risk (ie, recurrence score; Table 1) and chemotherapy benefit (ie, treatment score; Table 2) were derived as described in the text of the Appendix (online only).
Table 1.
Prediction of Recurrence Risk: Kaplan-Meier Estimates of Recurrence Risk at 3 Years and Associated 95% CIs from Bootstrap Analysis for Patients With Stage II Disease in Surgery-Alone Studies
| Recurrence Risk Group | Patients(median %) | Risk of Recurrence at3 Years (%) | 95% CI |
|---|---|---|---|
| Low (RS < 30) | 25 | 8 | 5 to 12 |
| Intermediate (RS 31-40) | 39 | 11 | 7 to 15 |
| High (RS ≥ 41) | 37 | 25 | 18 to 32 |
Abbreviation: RS, recurrence score.
Table 2.
Prediction of Chemotherapy Benefit: Kaplan-Meier Estimates of Recurrence Risk at 3 Years by Treatment and FU/LV Benefit and Associated 95% CIs From Bootstrap Analysis for Patients With Stage II Disease
| Chemotherapy Benefit Group* | Recurrence at 3 Years |
Result at 3 Years |
||||
|---|---|---|---|---|---|---|
| Surgery Alone |
Surgery + FU/LV |
|||||
| Risk (%) | 95% CI | Risk (%) | 95% CI | Benefit (%) | 95% CI | |
| Low | 16 | 11 to 23 | 19 | 9 to 31 | −3 | −16 to 9 |
| Intermediate | 10 | 6 to 14 | 7 | 2 to 14 | 3 | −5 to 10 |
| High | 15 | 10 to 20 | 7 | 2 to 13 | 8 | 0 to 15 |
Bootstrap methods38,39 were used to evaluate the extent to which recurrence risk differed among the recurrence risk groups defined by recurrence score for patients with stage II disease. A total of 1,000 bootstrap samples were drawn randomly with replacement from the pooled data set, taking variability between studies into account. Kaplan-Meier estimates of recurrence risk at 3 years were obtained for each recurrence risk group. Median recurrence risk estimates across all bootstrap samples and percentile CIs were reported. A similar approach was used to assess the results of the final multigene algorithm to predict FU/LV benefit, in which patients were divided in chemotherapy benefit groups on the basis of both their recurrence scores and treatment scores (Appendix Table A3, online only). Data were analyzed independently by the NSABP Biostatistical Center, Cleveland Clinic, and Genomic Health, Inc for individual studies. Analyses across four studies were conducted by Genomic Health.
RESULTS
The final numbers of evaluable patients were 270 in the C-01/C-02, 765 in the CC, 308 in the C-04, and 508 in the C-06 cohort. The outcomes and clinical/demographic characteristics of evaluable patients with tumor blocks were similar to those observed in the parent NSABP studies.
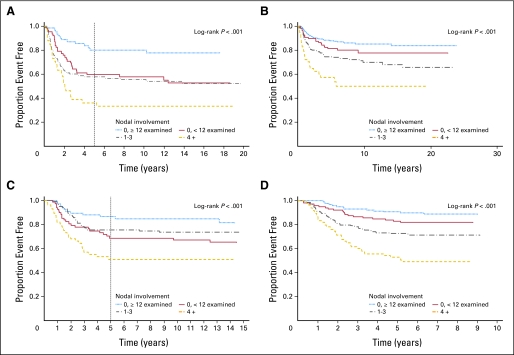
The baseline characteristics of the three NSABP cohorts were generally similar; patients from CC differed in age, percentage of right-sided tumors, number of lymph nodes examined, and percentage of stage II versus stage III disease (Appendix Table A1). Univariate Cox proportional hazards regression identified nodal status (0 positive nodes and ≥ 12 nodes examined, 0 positive nodes and < 12 nodes examined, 1 to 3 or ≥ 4 positive nodes) as the most significant clinical/pathologic predictor of RFI (P < .001) in all studies (Appendix Tables A4, A5, A6, and A7, online only; Appendix Figs A2A, A2B, A2C, and A2D, online only). T stage, available in adequate numbers of patients in CC, was associated with RFI (T4 v other; P = .003; Appendix Table A5). MMR was not associated with RFI in CC (P = .27; Appendix Table A5).
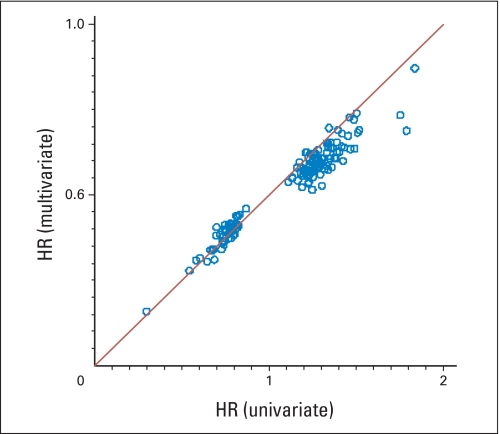
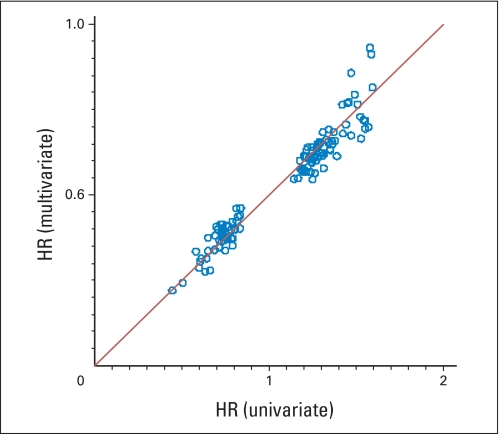
Univariate analysis identified 143 genes as significantly related to RFI in the C-01/C-02 cohort, 119 in the CC cohort, 143 in the C-04 cohort, and 169 in the C-06 cohort; 27%, 16%, 27%, and 11% of these genes, respectively, were expected to be false discoveries. When studies were pooled, the FDR was markedly lower. In the multivariate analysis, 43%, 74%, 50%, and 84% of the genes identified in the univariate analysis retained significance in C-01/C-02, CC, C-04, and C-06, respectively, and had similar HRs in both analyses (Appendix Figs A3 and A4, online only, for surgery-alone studies; similar results for studies of surgery + FU/LV not shown), which suggests that gene expression contributes information about recurrence beyond standard clinical and pathologic covariates. In these multivariate analyses, the contribution of nodal status was consistently statistically significant.
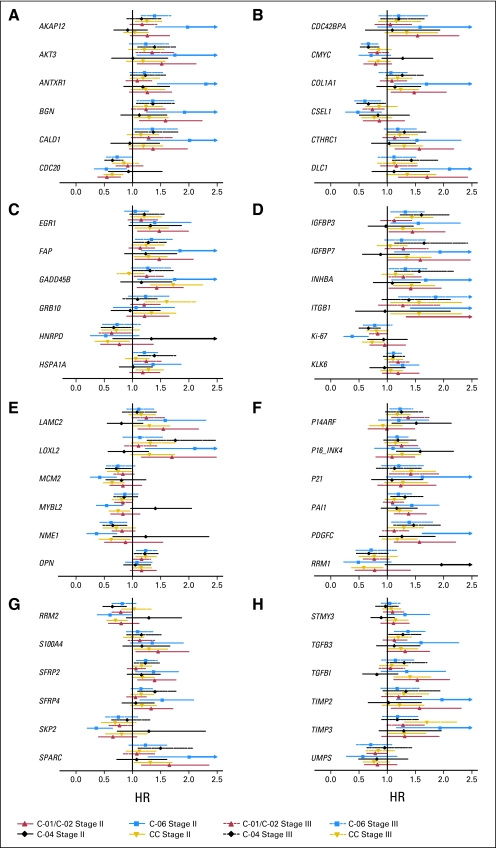
The relationship between gene expression, tumor stage, and RFI was investigated across studies. In univariate analyses, six of the 375 genes had significant (P < .05) mean differences in expression between patients with stage II and stage III disease in all four studies. Thirty-three genes had a significant interaction of gene expression and stage (P < .1) in Cox proportional hazards models stratified by study; 32 of these 33 genes were potential false positives (FDR = 97%), which suggests that interaction between gene expression and stage was weak. The coexpression of genes examined using cluster analysis was virtually identical in patients with stage II and stage III disease. Agreement between univariate HRs for patients with stage II and stage III disease is illustrated in Figure 1 for genes significantly associated with RFI in both surgery-alone studies and at least one study of surgery plus FU/LV. HRs were generally similar with overlapping CIs, with a few exceptions that could be chance findings due to multiplicity of testing across multiple genes and studies. These results support pooling data across stages for gene discovery and algorithm development.
Fig 1.
Hazard ratio estimates and 95% CIs for gene expression from univariate Cox PH regression models of recurrence-free interval in patients on studies C-01/C-02, CC, C-04, and C-06 by tumor stage for the 48 genes that were significantly related to recurrence-free interval (A) in both of the surgery-only studies as well as (B) in at least one study of surgery + fluorouracil and leucovorin.
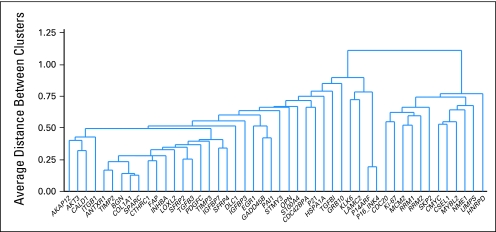
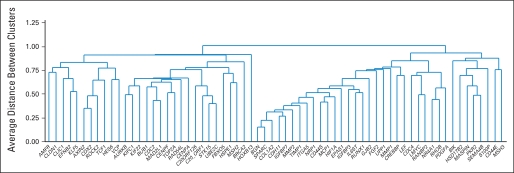
Recurrence risk genes were expected to have a similar relationship with RFI when measured in patients treated with surgery alone or surgery followed by FU/LV. A total of 48 (13%) of the 375 genes studied in all four development studies were significantly (P < .05) associated with RFI in both surgery alone studies and at least one study of surgery plus FU/LV. Fewer than one of these 48 genes is expected to be a false discovery. Cluster analysis identified two relatively distinct gene groups: a stromal gene group (containing several subgroups such as early response) and a cell cycle gene group (Fig 2). Higher expression of stromal genes (eg, BGN, FAP, GADD45B, and PAI) was associated with higher risk of recurrence, whereas higher expression of cell cycle genes (eg, Ki–67, MYBL2, and MCM2) was associated with lower risk of recurrence.
Fig 2.
Unsupervised hierarchical clustering of the 48 genes significantly related to recurrence-free interval in both surgery-only studies and at least one study of surgery + fluorouracil and leucovorin using data from all four studies.
In contrast to recurrence risk genes, the genes predictive of differential FU/LV benefit are required to exhibit a different relationship with outcome (ie, different HRs) in patients treated with surgery alone compared with patients treated with surgery plus FU/LV. A total of 66 (18%) of 375 genes studied in all four development studies had interactions of gene expression and treatment that were significant at the less than .10 level if the data across the four studies were pooled (13 genes with P < .01; 45 genes with P < .05); four of these genes were also associated with risk of recurrence at the P < .05 level. Approximately 37 of these 66 genes are expected to be false discoveries; this was expected, given the lower statistical power associated with the analysis of interaction. Among 66 potentially predictive genes, there were a large number of genes involved in multiple stages of the cell cycle and apoptosis (ie, MAD2L1, AURKB, BIK, BUB1, CDC2), and higher expression was associated with greater differential benefit from FU/LV (Appendix Fig A5, online only). There were also a prominent stress response/hypoxia signature (ie, HSPE1, NR4A1, RhoB, HIF1A); multifunctional transcription factors (RUNX1, CREBBP, KLF5); and genes associated with wnt signaling (AXIN2 and LEF), MMR (MSH2 and MSH3), and angiogenesis (EFNB2). Higher expression of some of these genes (eg, RUNX1, CREBBP, KLF5, and EFNB2) was associated with lower benefit from FU/LV.
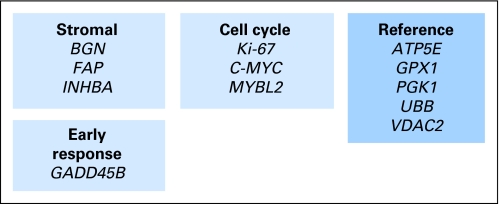
Seven of the 48 recurrence risk genes and six of the 66 chemotherapy benefit genes were selected to create the final recurrence score and treatment score algorithms (Appendix Figs A1 and A5; described in the Appendix). The results of bootstrap analyses to assess the predictive ability of the recurrence score are listed in Table 1 for patients with stage II disease treated with surgery alone (C-01/C-02 and CC cohorts). Patients were divided into three recurrence risk groups on the basis of the calculated recurrence score (ie, < 30, 30-40, and ≥ 41). The recurrence score separated the 632 patients with stage II disease into groups that had a sizable difference in estimates of risk of recurrence between the high- and the low-risk groups.
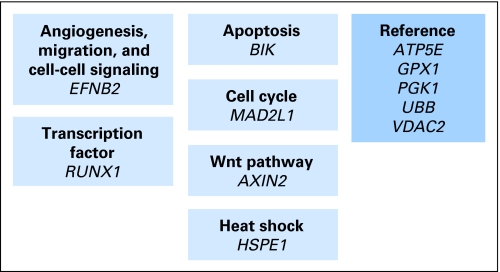
The results of bootstrap analyses to assess the predictive ability of the treatment score are listed in Table 2 for the 870 patients with stage II disease treated with surgery alone or surgery plus FU/LV. Patients were divided into three benefit groups (Appendix). For comparison, the overall 3-year risk of recurrence of patients with stage II disease was 14% for the surgery-alone group and was 10% for patients treated with surgery plus FU/LV. The correlation between the recurrence score and treatment score was relatively low (r = −0.4) in these studies, which suggests that the determinants of recurrence risk and differential FU/LV benefit may be distinct.
The performance of these algorithms was evaluated on the data set used for algorithm development; hence, the results in Tables 1 and 2 are likely to be optimistic. These algorithms will be validated on an independent data set of patients with stage II colon cancer from the QUASAR study.11
DISCUSSION
Our strategy for discovering genes related to recurrence risk and differential benefit with adjuvant FU/LV chemotherapy has been to perform multiple, large, independent studies to identify those genes most consistently and strongly related to clinical outcome (Appendix Fig A1).40,41 The results reported here are based on data from 1,851 patients, using standardized assay technology in a single laboratory. This approach is in contrast to algorithms developed from much larger numbers of genes and significantly smaller sample sizes.17–21
We have identified 48 genes that have significant and similar relationships with RFI and 66 genes that have different relationships with RFI in patients treated with surgery alone compared with patients treated with surgery plus FU/LV: the former genes are likely to predict recurrence, whereas the latter genes are likely to predict differential benefit with adjuvant FU/LV therapy. A large proportion of these genes remain significantly associated with RFI after analysis is controlled for the effects of numerous clinical/pathologic covariates, including nodal status, which also contributed significantly to prediction of recurrence risk.
This report highlights several challenges to biomarker development in colon cancer. First, our ability to identify genes predictive of differential treatment benefit was limited by the lack of large, randomized clinical trials with tumor specimens (beyond the QUASAR validation trial). Second, the lower power of the test of interaction leads to a high FDR among the candidate predictive genes, of greater than 50% in our studies. Finally, the question of whether stage II and III disease are biologically similar or dissimilar is unresolved. However, for the vast majority of genes, we saw no strong difference between stages in the relationship of gene expression and RFI, and an additional analysis that compared patients who had stage II disease and ≥ 12 nodes examined with patients who had stage III disease confirmed these findings (data not shown).
Our strategy has led to the discovery of recurrence risk genes that can be confidently associated with clinical outcome and are generally different from the genes identified in our breast cancer studies (with the exceptions of Ki-67 and MYBL2)42 or the genes previously reported in colon cancer.17–21 Higher expression of cell cycle genes was associated with an increased RFI; this observation is similar to that reported for Ki-67 in colon cancer43,44 but is opposite to the relationship observed in breast cancer studies.42 The association of stromal gene expression with colon cancer recurrence provides an elegant molecular explanation for Dukes' original observation that invasion is the critical characteristic that should be used in staging colorectal cancer.45,46
Some of the genes that were identified as predictive of chemotherapy benefit are not unexpected. Sensitivity to FU should be affected by factors related to the level of proliferation (ie, cell-cycle related genes), the induction of apoptosis and hypoxia,47 FU metabolism, and MMR.48 However, it is not clear why the expression of other genes, such as GJB2 or HES6, which are associated with gap junction communication and notch signaling respectively, would predict FU benefit. The quantitative expression of the genes related to FU activation/metabolism (TS, DPD, TP)15,49–54 or to the markers (hMLH1, hMSH2) associated with MMR55–57 were not associated with differential FU/LV benefit in our studies; current guidelines by the American Society of Clinical Oncology conclude that there is insufficient evidence to recommend the use of these markers as predictors of response to therapy.23
This report describes our process for identifying genes that can be used to estimate recurrence risk and differential FU/LV chemotherapy benefit on the basis of the relationship of quantitative gene expression at the time of diagnosis and clinical outcome in patients with stage II/III colon cancer treated with surgery or surgery plus FU/LV. The results of these four studies have been used to develop a multigene assay (Figs 3 and 4) for prediction of recurrence risk and differential benefit of adjuvant FU/LV chemotherapy that can be used to divide patients with stage II colon cancer into groups with different likelihoods of recurrence and treatment benefit. The requirement for external validation is being addressed in QUASAR,11 a large, independent study of patients with stage II colon cancer randomly assigned to surgery alone or to surgery followed by adjuvant FU/LV chemotherapy.
Fig 3.
Recurrence score gene panel and algorithm. The recurrence score that is based on 12 genes (seven cancer-related genes and five reference genes) is derived from the reference-normalized expression measurements in four steps and is scaled from 0 to 100. First, expression of each gene is normalized relative to the expression of the five reference genes (ie, ATP5E, GPX1, PGK1, UBB, and VDAC2). Reference-normalized expression measurements range from two to 15, with a 1-unit increase reflecting approximately twice as much input RNA. Expression of individual genes is given a threshold as follows: if FAP, Ki-67 or MYBL2 measurement is less than 6.5 CT, it is considered to be 6.5; if GADD45B measurement is less than 5 CT, then it is considered to be 5. Genes are grouped on the basis of function and/or correlated expression. Second, the stromal and cell cycle group scores are calculated as averages of the reference-normalized, individual gene expression measurements as follows: stromal group score = (BGN + FAP + INHBA) ÷ 3; cell cycle group score = (Ki-67 + C-MYC + MYBL2) ÷ 3. Third, the unscaled recurrence score (RSu) is calculated with the use of coefficients that are defined on the basis of regression analysis of gene expression and recurrence in four development studies: RSu = + 0.15 × stromal group score − 0.30 × cell cycle group score + 0.15 × GADD45B. A plus sign indicates that increased expression is associated with increased risk of recurrence, and a minus sign indicates that increased expression is associated with decreased risk of recurrence. Fourth, the recurrence score (RS) is rescaled as follows: RS = 44 × (RSu + 0.82); if RS is less than 0, then RS = 0, and if RS is greater than 100, then RS = 100.
Fig 4.
Treatment score gene panel and algorithm. The treatment score that is based on 11 genes (six cancer-related genes and the same five reference genes; Fig 3) is derived from the reference-normalized expression measurements in three steps and is scaled from 0 to 100. First, expression of each gene is normalized relative to the expression of the five reference genes (ATP5E, GPX1, PGK1, UBB, and VDAC2). Expression of individual genes is given a threshold as follows: if MAD2L1 or RUNX1 measurement is less than 5.5 CT, it is considered to be 5.5; if BIK measurement is less than 6 CT, then it is considered to be 6; and if EFNB2 measurement is less than 5 CT, then it is considered to be 5. Second, the unscaled treatment score (TSu) is calculated with the use of coefficients that are defined on the basis of regression analysis of gene expression and chemotherapy benefit in four development studies: TSu = −0.3 × EFNB2 − 0.04 × RUNX1 + 0.1 × MAD2L1 + 0.3 × BIK + 0.1 × AXIN2 + 0.1 × HSPE1. A plus sign indicates that increased expression is associated with increased chemotherapy benefit, and a minus sign indicates that increased expression is associated with decreased chemotherapy benefit. Third, the treatment score (TS) is rescaled as follows: TS = 37 × (TSu − 1); if TS is less than 0, then TS = 0, and if TS is greater than 100, then TS = 100.
Acknowledgment
We thank Barbara C. Good, PhD, Director of Scientific Publications for the National Surgical Adjuvant Breast and Bowel Project, for editorial assistance; and Meike Labusch, Angela Chen, Anhthu Nguyen, Bhavin Padhiar, Debjani Dutta, Jayadevi Krishnakumar, Jennie Jeong, Jenny Wu, Hyun Soo Son, Mei-Lan Liu, Mylan Pho, Ranjana Ambannavar, Lauren Intagliata, Freda Lane, James Hackett, and Jeanne Yue from Genomic Health, Inc, for their contributions to sample and data processing and data analysis.
Appendix
Development of the Colon Cancer Recurrence Score and Treatment Score Assays
To develop a tumor gene expression assay for use with tumor blocks that are routinely prepared following surgery, we used a multistep approach.
First, a real-time reverse transcriptase polymerase chain reaction method to quantify the expression of hundreds of genes in RNA isolated from three 10-μm sections of fixed, paraffin-embedded tumor tissue was developed.24
Second, we selected 761 candidate genes from the published literature, genomic databases, pathway analysis, and from microarray-based gene expression profiling experiments performed with fresh frozen tissue.17–21 The 761 candidate genes are listed in Appendix Table A2 (online only).
Third, we performed four independent clinical studies in a total of 1,851 patients with colon cancer to test the relationship between the expression of the candidate genes and time to recurrence. We reasoned that in any single gene expression study a considerable number of genes may correlate with outcome as a result of chance alone. To identify true positives, we performed multiple independent studies to identify whether expression of any of the candidate genes correlated with recurrence across the studies. We hypothesized that the genes most highly correlated with recurrence would survive evaluation across diverse patients and treatments, and we selected heterogeneous populations for development of the gene list. After National Surgical Adjuvant Breast and Bowel Project (NSABP) C-01/C-02 and NSABP C-04 were conducted, the gene list was reduced to 375 genes on the basis of the strength of the relationship between gene expression and recurrence risk as well as the association of gene expression with chemotherapy benefit; and the NSABP C-06 patients and Cleveland Clinic cohorts were studied with 375 genes (Appendix Table A2). There were 48 genes for which expression was associated with recurrence in three of four studies at an unadjusted P value less than .05, and 66 genes had an interaction of gene expression and chemotherapy treatment significant at an unadjusted P < .1.
Fourth, we used the results from the four development studies to select the final gene panels and to design algorithms to compute a recurrence score (RS) and a treatment score (TS) for each tumor sample. The list of gene candidates was narrowed down using a number of considerations that included, but were not limited to, the strength of the associations with recurrence, the consistency of performance across studies, consistency of performance in patients with stage II and stage III disease, and analytic performance. The correlation of the expression of the genes with respect to each other was analyzed by unsupervised cluster analysis and by principal component analysis. Two major gene groups were identified among the prognostic genes: a stromal group (containing extracellular matrix genes, such as BGN, FAP, INHBA, and SPARC; early response genes, such as GADD45B; and invasion genes, such as PAI), and a cell cycle group (genes such as Ki-67, MYBL2, MCM2). Increased expression of stromal genes was associated with increased risk of recurrence, whereas increased expression of cell cycle genes was associated with decreased risk of recurrence. Among the 66 predictive genes, there were several multifunctional transcription factors (RUNX1, TCF1, CREBBP, KLF5) and genes involved in cell cycle and apoptosis (eg, MAD2L1, AURKB, BIK, TOP2A, BUB1, CDC2), hypoxia/stress response (HSPE1, NR4A1, RhoB, HIF1A, CREBBP, EPAS), wnt signaling (AXIN 2 and LEF), mismatch repair (MSH2 and MSH3), and angiogenesis (EFNB2). Analyses were then performed to determine the appropriate number of terms to include in the model and the functional forms of the variables. For this purpose, we used correlation analysis, dimension reduction (including stepwise variable selection and classification and regression trees), Martingale residual analysis, and bootstrap resampling. Multiple analyses across the four studies were conducted to determine whether models with a larger number of genes were better able to predict risk of recurrence and/or chemotherapy benefit and whether having fewer genes resulted in loss of robustness. We observed that relatively parsimonious models with anywhere from six to 10 recurrence-risk genes and five to six chemotherapy-benefit genes were adequate. The selection of the final seven recurrence-risk and six chemotherapy-benefit genes was based primarily on the strength of their performance across all studies and the consistency of primer/probe performance in the assay.
Fig A1.
Outline of the strategy for determining relationships between tumor gene expression, disease recurrence, and differential benefit from fluorouracil (FU) plus leucovorin (LV). NSABP, National Surgical Adjuvant Breast and Bowel Project; QUASAR, Quick and Simple and Reliable.
Fig A2.
(A) Kaplan-Meier plot of recurrence-free interval by nodal status on National Surgical Adjuvant Breast and Bowel Project (NSABP) C-01/C-02: 0 positive nodes and ≥ 12 nodes examined (n = 62), 0 positive nodes and less than 12 nodes examined (n = 64), 1 to 3 positive nodes (n = 94), or ≥ 4 positive nodes (n = 41). (B) Kaplan-Meier plot of recurrence-free interval by nodal status at the Cleveland Clinic: 0 positive nodes and ≥ 12 nodes examined (n = 387), 0 positive nodes and less than 12 nodes examined (n = 117), 1 to 3 positive nodes (n = 201), or ≥ 4 positive nodes (n = 60). (C) Kaplan-Meier plot of recurrence-free interval by nodal status on NSABP C-04: 0 positive nodes and ≥ 12 nodes examined (n = 66), 0 positive nodes and less than 12 nodes examined (n = 68), 1 to 3 positive nodes (n = 114), or ≥ 4 positive nodes (n = 56). (D) Kaplan-Meier plot of recurrence-free interval by nodal status on NSABP C-06: 0 positive nodes and ≥ 12 nodes examined (n = 119), 0 positive nodes and less than 12 nodes examined (n = 116), 1 to 3 positive nodes (n = 189), or ≥ 4 positive nodes (n = 84).
Fig A3.
Agreement of the univariate and multivariate hazard ratios (HRs) for 143 genes significantly related to recurrence-free interval for patients on the National Surgical Adjuvant Breast and Bowel Project C-01/C02 study.
Fig A4.
Agreement of the univariate and multivariate hazard ratios (HRs) for 119 genes significantly related to recurrence-free interval for patients on the Cleveland Clinic study.
Fig A5.
Unsupervised hierarchical clustering of the 66 genes with significant gene by treatment interaction using data from all four studies.
Table A1.
Demographics and Baseline Medical Characteristics of Four Study Cohorts
| Characteristic | No. of Patients | Patients by Study Cohort |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| NSABPC-01/C-02 |
NSABP C-04 |
Cleveland Clinic |
NSABP C-06 |
||||||
| No. | % | No. | % | No. | % | No. | % | ||
| Sex | |||||||||
| Female | 869 | 129 | 47.8 | 144 | 46.8 | 350 | 45.8 | 246 | 48.4 |
| Male | 982 | 141 | 52.2 | 164 | 53.2 | 415 | 54.2 | 262 | 51.6 |
| Age, years | |||||||||
| ≥ 60 | 1,209 | 164 | 60.7 | 144 | 46.8 | 603 | 78.8 | 298 | 58.7 |
| < 60 | 642 | 106 | 39.3 | 164 | 53.2 | 162 | 21.2 | 210 | 41.3 |
| Tumor location | |||||||||
| Left | 405 | 69 | 25.6 | 68 | 22.1 | 153 | 20.0 | 115 | 22.6 |
| Rectosigmoid | 635 | 96 | 35.6 | 110 | 35.7 | 252 | 32.9 | 177 | 34.8 |
| Right | 786 | 95 | 35.2 | 122 | 39.6 | 360 | 47.1 | 209 | 41.1 |
| Multiple or unknown | 25 | 10 | 3.7 | 8 | 2.6 | 0 | 0.0 | 7 | 1.4 |
| Surgical procedure | |||||||||
| Colectomy/hemicolectomy | 1,196 | 184 | 68.1 | 181 | 58.8 | 526 | 68.8 | 305 | 60.0 |
| Segmental/anterior resection | 563 | 56 | 20.7 | 109 | 35.4 | 205 | 26.8 | 193 | 38.0 |
| Other/unknown | 92 | 30 | 11.1 | 18 | 5.8 | 34 | 4.4 | 10 | 2.0 |
| No. of nodes examined | |||||||||
| < 12 | 700 | 129 | 50.2 | 154 | 51.2 | 175 | 22.9 | 242 | 47.7 |
| ≥ 12 | 1,130 | 128 | 49.8 | 147 | 48.8 | 590 | 77.1 | 265 | 52.3 |
| No. of positive nodes | |||||||||
| 0 | 1,007 | 131 | 49.2 | 137 | 44.6 | 504 | 65.9 | 235 | 46.3 |
| 1-3 | 598 | 94 | 35.3 | 114 | 37.1 | 201 | 26.3 | 189 | 37.2 |
| ≥ 4 | 241 | 41 | 15.4 | 56 | 18.2 | 60 | 7.8 | 84 | 16.5 |
| Nodal status of examined nodes | |||||||||
| 0 of < 12 examined | 365 | 64 | 24.5 | 68 | 22.4 | 117 | 15.3 | 116 | 22.8 |
| 0 of ≥ 12 examined | 634 | 62 | 23.8 | 66 | 21.7 | 387 | 50.6 | 119 | 23.4 |
| 1-3 overall | 598 | 94 | 36.0 | 114 | 37.5 | 201 | 26.3 | 189 | 37.2 |
| ≥ 4 overall | 241 | 41 | 15.7 | 56 | 18.4 | 60 | 7.8 | 84 | 16.5 |
| Tumor stage | |||||||||
| II | 1,007 | 131 | 48.5 | 137 | 44.5 | 504 | 65.9 | 235 | 46.3 |
| III | 844 | 139 | 51.5 | 171 | 55.5 | 261 | 34.1 | 273 | 53.7 |
| Tumor grade | |||||||||
| High | 460 | 66 | 24.5 | 48 | 15.6 | 173 | 22.7 | 173 | 34.1 |
| Low | 1,386 | 203 | 75.5 | 260 | 84.4 | 588 | 77.3 | 335 | 65.9 |
| Mucinous status | |||||||||
| Mucinous | 295 | 23 | 8.5 | 23 | 7.5 | 146 | 19.1 | 103 | 20.3 |
| Not mucinous | 1,556 | 247 | 91.5 | 285 | 92.5 | 619 | 80.9 | 405 | 79.7 |
Abbreviations: NSABP, National Surgical Adjuvant Breast and Bowel Project.
Table A2.
Listing of the 761 Candidate Genes
| No. | Gene Name | Sequence ID | 375 Gene Panel | No. | Gene Name | Sequence ID | 375 Gene Panel |
|---|---|---|---|---|---|---|---|
| 1 | ABCB1 | NM_000927.2 | Yes | 384 | IL6ST | NM_002184.2 | Yes |
| 2 | ABCC5 | NM_005688.1 | Yes | 385 | IL-8 | NM_000584.2 | |
| 3 | ABCC6 | NM_001171.2 | Yes | 386 | ILT-2 | NM_006669.1 | |
| 4 | A-Catenin | NM_001903.1 | 387 | IMP-1 | NM_006546.2 | ||
| 5 | ACP1 | NM_004300.2 | 388 | IMP2 | NM_006548.3 | ||
| 6 | ADAM10 | NM_001110.1 | 389 | ING1L | NM_001564.1 | ||
| 7 | ADAM17 | NM_003183.3 | 390 | ING5 | NM_032329.4 | ||
| 8 | ADAMTS12 | NM_030955.2 | Yes | 391 | INHA | NM_002191.2 | |
| 9 | ADPRT | NM_001618.2 | 392 | INHBA | NM_002192.1 | Yes | |
| 10 | AGXT | NM_000030.1 | 393 | INHBB | NM_002193.1 | ||
| 11 | AKAP12 | NM_005100.2 | Yes | 394 | IRS1 | NM_005544.1 | Yes |
| 12 | AKT1 | NM_005163.1 | 395 | ITGA3 | NM_002204.1 | ||
| 13 | AKT2 | NM_001626.2 | 396 | ITGA4 | NM_000885.2 | ||
| 14 | AKT3 | NM_005465.1 | Yes | 397 | ITGA5 | NM_002205.1 | Yes |
| 15 | AL137428 | AL137428.1 | 398 | ITGA6 | NM_000210.1 | ||
| 16 | ALCAM | NM_001627.1 | Yes | 399 | ITGA7 | NM_002206.1 | |
| 17 | ALDH1A1 | NM_000689.1 | 400 | ITGAV | NM_002210.2 | Yes | |
| 18 | ALDOA | NM_000034.2 | 401 | ITGB1 | NM_002211.2 | Yes | |
| 19 | AMFR | NM_001144.2 | Yes | 402 | ITGB3 | NM_000212.1 | Yes |
| 20 | ANGPT2 | NM_001147.1 | Yes | 403 | ITGB4 | NM_000213.2 | Yes |
| 21 | ANTXR1 | NM_032208.1 | Yes | 404 | ITGB5 | NM_002213.3 | |
| 22 | ANXA1 | NM_000700.1 | Yes | 405 | KCNH2 iso a/b | NM_000238.2 | |
| 23 | ANXA2 | NM_004039.1 | Yes | 406 | KCNH2 iso a/c | NM_172057.1 | Yes |
| 24 | ANXA5 | NM_001154.2 | Yes | 407 | KCNK4 | NM_016611.2 | |
| 25 | AP-1 | NM_002228.2 | 408 | KDR | NM_002253.1 | ||
| 26 | APC | NM_000038.1 | Yes | 409 | Ki-67 | NM_002417.1 | Yes |
| 27 | APEX-1 | NM_001641.2 | 410 | KIAA0125 | NM_014792.2 | ||
| 28 | APG-1 | NM_014278.2 | Yes | 411 | KIF22 | NM_007317.1 | Yes |
| 29 | APN | NM_001150.1 | 412 | KIF2C | NM_006845.2 | ||
| 30 | APOC1 | NM_001645.3 | 413 | KIFC1 | XM_371813.1 | Yes | |
| 31 | AREG | NM_001657.1 | Yes | 414 | Kitlng | NM_000899.1 | |
| 32 | ARG | NM_005158.2 | 415 | KLF5 | NM_001730.3 | Yes | |
| 33 | ARHF | NM_019034.2 | 416 | KLF6 | NM_001300.4 | Yes | |
| 34 | ATOH1 | NM_005172.1 | 417 | KLK10 | NM_002776.1 | Yes | |
| 35 | ATP5A1 | NM_004046.3 | Yes | 418 | KLK6 | NM_002774.2 | Yes |
| 36 | ATP5E | NM_006886.2 | Yes | 419 | KLRK1 | NM_007360.1 | Yes |
| 37 | AURKB | NM_004217.1 | Yes | 420 | KNTC2 | NM_006101.1 | |
| 38 | Axin 2 | NM_004655.2 | Yes | 421 | K-ras | NM_033360.2 | Yes |
| 39 | axin1 | NM_003502.2 | Yes | 422 | K-ras mutant 1 | GHI_k-ras_mut1 | Yes |
| 40 | BAD | NM_032989.1 | Yes | 423 | K-ras mutant 2 | GHI_k-ras_mut2 | Yes |
| 41 | BAG1 | NM_004323.2 | 424 | K-rasmutant 3 | GHI_k-ras_mut3 | Yes | |
| 42 | BAG2 | NM_004282.2 | 425 | KRAS2 | NM_004985.3 | ||
| 43 | BAG3 | NM_004281.2 | 426 | KRT19 | NM_002276.1 | ||
| 44 | Bak | NM_001188.1 | 427 | KRT8 | NM_002273.1 | Yes | |
| 45 | Bax | NM_004324.1 | Yes | 428 | LAMA3 | NM_000227.2 | Yes |
| 46 | BBC3 | NM_014417.1 | 429 | LAMB3 | NM_000228.1 | ||
| 47 | BCAS1 | NM_003657.1 | 430 | LAMC2 | NM_005562.1 | Yes | |
| 48 | B-Catenin | NM_001904.1 | Yes | 431 | LAT | NM_014387.2 | Yes |
| 49 | Bcl2 | NM_000633.1 | 432 | LCN2 | NM_005564.2 | ||
| 50 | BCL2L10 | NM_020396.2 | 433 | LDLRAP1 | NM_015627.1 | ||
| 51 | BCL2L11 | NM_138621.1 | Yes | 434 | LEF | NM_016269.2 | Yes |
| 52 | BCL2L12 | NM_138639.1 | 435 | LGALS3 | NM_002306.1 | Yes | |
| 53 | Bclx | NM_001191.1 | 436 | LGMN | NM_001008530 | ||
| 54 | BCRP | NM_004827.1 | 437 | LILRB3 | NM_006864.1 | ||
| 55 | BFGF | NM_007083.1 | 438 | LMNB1 | NM_005573.1 | Yes | |
| 56 | BGN | NM_001711.3 | Yes | 439 | LMYC | NM_012421.1 | Yes |
| 57 | BID | NM_001196.2 | 440 | LOX | NM_002317.3 | Yes | |
| 58 | BIK | NM_001197.3 | Yes | 441 | LOXL2 | NM_002318.1 | Yes |
| 59 | BIN1 | NM_004305.1 | 442 | LRP5 | NM_002335.1 | Yes | |
| 60 | BLMH | NM_000386.2 | Yes | 443 | LRP6 | NM_002336.1 | Yes |
| 61 | BMP2 | NM_001200.1 | 444 | LY6D | NM_003695.2 | ||
| 62 | BMP4 | NM_001202.2 | 445 | MAD | NM_002357.1 | ||
| 63 | BMP7 | NM_001719.1 | 446 | MAD1L1 | NM_003550.1 | Yes | |
| 64 | BMPR1A | NM_004329.2 | 447 | MAD2L1 | NM_002358.2 | Yes | |
| 65 | BRAF | NM_004333.1 | Yes | 448 | MADH2 | NM_005901.2 | Yes |
| 66 | Braf Mutant 1 | GHI_BRAF_mut4 | Yes | 449 | MADH4 | NM_005359.3 | Yes |
| 67 | BRCA1 | NM_007295.1 | Yes | 450 | MADH7 | NM_005904.1 | Yes |
| 68 | BRCA2 | NM_000059.1 | Yes | 451 | MAP2 | NM_031846.1 | |
| 69 | BRK | NM_005975.1 | 452 | MAP2K1 | NM_002755.2 | ||
| 70 | BTF3 | NM_001207.2 | 453 | MAP3K1 | XM_042066.8 | ||
| 71 | BTRC | NM_033637.2 | 454 | MAPK14 | NM_139012.1 | ||
| 72 | BUB1 | NM_004336.1 | Yes | 455 | Maspin | NM_002639.1 | Yes |
| 73 | BUB1B | NM_001211.3 | 456 | MAX | NM_002382.3 | ||
| 74 | BUB3 | NM_004725.1 | 457 | MCM2 | NM_004526.1 | Yes | |
| 75 | C20 orf1 | NM_012112.2 | Yes | 458 | MCM3 | NM_002388.2 | Yes |
| 76 | C20ORF126 | NM_030815.2 | Yes | 459 | MCM6 | NM_005915.2 | Yes |
| 77 | C8orf4 | NM_020130.2 | Yes | 460 | MCP1 | NM_002982.1 | Yes |
| 78 | CA9 | NM_001216.1 | 461 | MDK | NM_002391.2 | ||
| 79 | c-abl | NM_005157.2 | 462 | MDM2 | NM_002392.1 | ||
| 80 | Cad17 | NM_004063.2 | Yes | 463 | MGAT5 | NM_002410.2 | Yes |
| 81 | CALD1 | NM_004342.4 | Yes | 464 | MGMT | NM_002412.1 | |
| 82 | CAPG | NM_001747.1 | Yes | 465 | mGST1 | NM_020300.2 | |
| 83 | CAPN1 | NM_005186.2 | 466 | MMP1 | NM_002421.2 | Yes | |
| 84 | CASP8 | NM_033357.1 | 467 | MMP12 | NM_002426.1 | ||
| 85 | CASP9 | NM_001229.2 | Yes | 468 | MMP2 | NM_004530.1 | Yes |
| 86 | CAT | NM_001752.1 | 469 | MMP7 | NM_002423.2 | Yes | |
| 87 | CAV1 | NM_001753.3 | Yes | 470 | MMP9 | NM_004994.1 | Yes |
| 88 | CBL | NM_005188.1 | 471 | MRP1 | NM_004996.2 | ||
| 89 | CCL20 | NM_004591.1 | 472 | MRP2 | NM_000392.1 | ||
| 90 | CCL3 | NM_002983.1 | 473 | MRP3 | NM_003786.2 | Yes | |
| 91 | CCNA2 | NM_001237.2 | Yes | 474 | MRP4 | NM_005845.1 | |
| 92 | CCNB1 | NM_031966.1 | Yes | 475 | MRPL40 | NM_003776.2 | |
| 93 | CCNB2 | NM_004701.2 | 476 | MSH2 | NM_000251.1 | Yes | |
| 94 | CCND1 | NM_001758.1 | 477 | MSH3 | NM_002439.1 | Yes | |
| 95 | CCND3 | NM_001760.2 | 478 | MSH6 | NM_000179.1 | ||
| 96 | CCNE1 | NM_001238.1 | 479 | MT3 | NM_005954.2 | ||
| 97 | CCNE2 | NM_057749.1 | Yes | 480 | MTA1 | NM_004689.2 | |
| 98 | CCNE2variant 1 | NM_057749var1 | Yes | 481 | MUC1 | NM_002456.1 | Yes |
| 99 | CCR7 | NM_001838.2 | Yes | 482 | MUC2 | NM_002457.1 | Yes |
| 100 | CD105 | NM_000118.1 | 483 | MUC5B | XM_039877.11 | ||
| 101 | CD134 | NM_003327.1 | 484 | MUTYH | NM_012222.1 | ||
| 102 | CD18 | NM_000211.1 | Yes | 485 | MVP | NM_017458.1 | |
| 103 | CD24 | NM_013230.1 | Yes | 486 | MX1 | NM_002462.2 | |
| 104 | CD28 | NM_006139.1 | 487 | MXD4 | NM_006454.2 | ||
| 105 | CD31 | NM_000442.1 | 488 | MYBL2 | NM_002466.1 | Yes | |
| 106 | CD34 | NM_001773.1 | 489 | MYH11 | NM_002474.1 | Yes | |
| 107 | CD3z | NM_000734.1 | Yes | 490 | MYLK | NM_053025.1 | Yes |
| 108 | CD44E | X55150 | Yes | 491 | NAT2 | NM_000015.1 | |
| 109 | CD44s | M59040.1 | Yes | 492 | NAV2 | NM_182964.3 | Yes |
| 110 | CD44v3 | AJ251595v3 | 493 | NCAM1 | NM_000615.1 | Yes | |
| 111 | CD44v6 | AJ251595v6 | Yes | 494 | NDE1 | NM_017668.1 | |
| 112 | CD68 | NM_001251.1 | Yes | 495 | NDRG1 | NM_006096.2 | |
| 113 | CD80 | NM_005191.2 | Yes | 496 | NDUFS3 | NM_004551.1 | |
| 114 | CD82 | NM_002231.2 | 497 | NEDD8 | NM_006156.1 | Yes | |
| 115 | CD8A | NM_171827.1 | 498 | NEK2 | NM_002497.1 | Yes | |
| 116 | CD9 | NM_001769.1 | 499 | NF2 | NM_000268.2 | ||
| 117 | CDC2 | NM_001786.2 | Yes | 500 | NFKBp50 | NM_003998.1 | Yes |
| 118 | CDC20 | NM_001255.1 | Yes | 501 | NFKBp65 | NM_021975.1 | |
| 119 | cdc25A | NM_001789.1 | 502 | NISCH | NM_007184.1 | ||
| 120 | CDC25B | NM_021874.1 | 503 | Nkd-1 | NM_033119.3 | Yes | |
| 121 | CDC25C | NM_001790.2 | Yes | 504 | NMB | NM_021077.1 | |
| 122 | CDC4 | NM_018315.2 | Yes | 505 | NMBR | NM_002511.1 | |
| 123 | CDC42 | NM_001791.2 | 506 | NME1 | NM_000269.1 | Yes | |
| 124 | CDC42BPA | NM_003607.2 | Yes | 507 | NOS3 | NM_000603.2 | |
| 125 | CDC6 | NM_001254.2 | Yes | 508 | NOTCH1 | NM_017617.2 | Yes |
| 126 | CDCA7variant 2 | NM_145810.1 | Yes | 509 | NOTCH2 | NM_024408.2 | |
| 127 | CDH1 | NM_004360.2 | Yes | 510 | NPM1 | NM_002520.2 | |
| 128 | CDH11 | NM_001797.2 | Yes | 511 | NR4A1 | NM_002135.2 | Yes |
| 129 | CDH3 | NM_001793.3 | Yes | 512 | NRG1 | NM_013957.1 | |
| 130 | CDK2 | NM_001798.2 | 513 | NRP1 | NM_003873.1 | Yes | |
| 131 | CDX1 | NM_001804.1 | 514 | NRP2 | NM_003872.1 | Yes | |
| 132 | Cdx2 | NM_001265.2 | Yes | 515 | NTN1 | NM_004822.1 | |
| 133 | CEACAM1 | NM_001712.2 | 516 | NUFIP1 | NM_012345.1 | ||
| 134 | CEACAM6 | NM_002483.2 | 517 | ODC1 | NM_002539.1 | Yes | |
| 135 | CEBPB | NM_005194.2 | Yes | 518 | OPN | NM_000582.1 | Yes |
| 136 | CEGP1 | NM_020974.1 | 519 | ORC1L | NM_004153.2 | ||
| 137 | CENPA | NM_001809.2 | Yes | 520 | OSM | NM_020530.3 | |
| 138 | CENPE | NM_001813.1 | 521 | OSMR | NM_003999.1 | Yes | |
| 139 | CENPF | NM_016343.2 | Yes | 522 | P14ARF | S78535.1 | Yes |
| 140 | CES2 | NM_003869.4 | 523 | p16-INK4 | L27211.1 | Yes | |
| 141 | CGA | NM_001275.2 | 524 | p21 | NM_000389.1 | Yes | |
| 142 | CGB | NM_000737.2 | Yes | 525 | p27 | NM_004064.1 | |
| 143 | CHAF1B | NM_005441.1 | 526 | P53 | NM_000546.2 | ||
| 144 | CHD2 | NM_001271.1 | 527 | p53R2 | AB036063.1 | Yes | |
| 145 | CHFR | NM_018223.1 | Yes | 528 | PADI4 | NM_012387.1 | |
| 146 | Chk1 | NM_001274.1 | Yes | 529 | PAI1 | NM_000602.1 | Yes |
| 147 | Chk2 | NM_007194.1 | 530 | Pak1 | NM_002576.3 | ||
| 148 | CIAP1 | NM_001166.2 | 531 | PARC | NM_015089.1 | ||
| 149 | cIAP2 | NM_001165.2 | Yes | 532 | PCAF | NM_003884.3 | |
| 150 | c-kit | NM_000222.1 | 533 | PCNA | NM_002592.1 | Yes | |
| 151 | CKS1B | NM_001826.1 | 534 | PDGFA | NM_002607.2 | Yes | |
| 152 | CKS2 | NM_001827.1 | Yes | 535 | PDGFB | NM_002608.1 | Yes |
| 153 | Claudin 4 | NM_001305.2 | Yes | 536 | PDGFC | NM_016205.1 | Yes |
| 154 | CLDN1 | NM_021101.3 | Yes | 537 | PDGFD | NM_025208.2 | Yes |
| 155 | CLDN7 | NM_001307.3 | Yes | 538 | PDGFRa | NM_006206.2 | Yes |
| 156 | CLIC1 | NM_001288.3 | Yes | 539 | PDGFRb | NM_002609.2 | |
| 157 | CLTC | NM_004859.1 | Yes | 540 | PFN1 | NM_005022.2 | |
| 158 | CLU | NM_001831.1 | 541 | PFN2 | NM_053024.1 | Yes | |
| 159 | cMet | NM_000245.1 | Yes | 542 | PGK1 | NM_000291.1 | Yes |
| 160 | c-myb | NM_005375.1 | Yes | 543 | PI3K | NM_002646.2 | Yes |
| 161 | cMYC | NM_002467.1 | Yes | 544 | PI3KC2A | NM_002645.1 | |
| 162 | CNN | NM_001299.2 | 545 | PIK3CA | NM_006218.1 | ||
| 163 | COL1A1 | NM_000088.2 | Yes | 546 | PIM1 | NM_002648.2 | |
| 164 | COL1A2 | NM_000089.2 | Yes | 547 | Pin1 | NM_006221.1 | |
| 165 | COPS3 | NM_003653.2 | 548 | PKD1 | NM_000296.2 | ||
| 166 | COX2 | NM_000963.1 | 549 | PKR2 | NM_002654.3 | Yes | |
| 167 | COX3 | MITO_COX3 | 550 | PLA2G2A | NM_000300.2 | ||
| 168 | CP | NM_000096.1 | 551 | PLAUR | NM_002659.1 | ||
| 169 | CRBP | NM_002899.2 | 552 | PLK | NM_005030.2 | Yes | |
| 170 | CREBBP | NM_004380.1 | Yes | 553 | PLK3 | NM_004073.2 | Yes |
| 171 | CRIP2 | NM_001312.1 | 554 | PLOD2 | NM_000935.2 | ||
| 172 | cripto | NM_003212.1 | Yes | 555 | PMS1 | NM_000534.2 | |
| 173 | CRK(a) | NM_016823.2 | 556 | PMS2 | NM_000535.2 | ||
| 174 | CRMP1 | NM_001313.1 | 557 | PPARG | NM_005037.3 | ||
| 175 | CRYAB | NM_001885.1 | Yes | 558 | PPID | NM_005038.1 | |
| 176 | CSEL1 | NM_001316.2 | Yes | 559 | PPM1D | NM_003620.1 | Yes |
| 177 | CSF1 | NM_000757.3 | Yes | 560 | PPP2R4 | NM_178001.1 | |
| 178 | CSK (SRC) | NM_004383.1 | 561 | PR | NM_000926.2 | ||
| 179 | c-Src | NM_005417.3 | Yes | 562 | PRDX2 | NM_005809 | Yes |
| 180 | CTAG1B | NM_001327.1 | 563 | PRDX3 | NM_006793.2 | ||
| 181 | CTGF | NM_001901.1 | Yes | 564 | PRDX4 | NM_006406.1 | Yes |
| 182 | CTHRC1 | NM_138455.2 | Yes | 565 | PRDX6 | NM_004905.2 | |
| 183 | CTLA4 | NM_005214.2 | 566 | PRKCA | NM_002737.1 | Yes | |
| 184 | CTNNBIP1 | NM_020248.2 | 567 | PRKCB1 | NM_002738.5 | Yes | |
| 185 | CTSB | NM_001908.1 | Yes | 568 | PRKCD | NM_006254.1 | |
| 186 | CTSD | NM_001909.1 | 569 | PRKR | NM_002759.1 | ||
| 187 | CTSH | NM_004390.1 | 570 | pS2 | NM_003225.1 | Yes | |
| 188 | CTSL | NM_001912.1 | Yes | 571 | PTCH | NM_000264.2 | Yes |
| 189 | CTSL2 | NM_001333.2 | 572 | PTEN | NM_000314.1 | Yes | |
| 190 | CUL1 | NM_003592.2 | 573 | PTGER3 | NM_000957.2 | Yes | |
| 191 | CUL4A | NM_003589.1 | Yes | 574 | PTHLH | NM_002820.1 | |
| 192 | CXCL12 | NM_000609.3 | Yes | 575 | PTHR1 | NM_000316.1 | |
| 193 | CXCR4 | NM_003467.1 | Yes | 576 | PTK2 | NM_005607.3 | |
| 194 | CYBA | NM_000101.1 | 577 | PTK2B | NM_004103.3 | ||
| 195 | CYP1B1 | NM_000104.2 | Yes | 578 | PTP4A3 | NM_007079.2 | |
| 196 | CYP2C8 | NM_000770.2 | Yes | 579 | PTP4A3 v2 | NM_032611.1 | Yes |
| 197 | CYP3A4 | NM_017460.3 | Yes | 580 | PTPD1 | NM_007039.2 | |
| 198 | CYR61 | NM_001554.3 | Yes | 581 | PTPN1 | NM_002827.2 | |
| 199 | DAPK1 | NM_004938.1 | Yes | 582 | PTPRF | NM_002840.2 | |
| 200 | DCC | NM_005215.1 | 583 | PTPRJ | NM_002843.2 | Yes | |
| 201 | DCC_exons18-23 | X76132_18-23 | 584 | PTPRO | NM_030667.1 | ||
| 202 | DCC_exons6-7 | X76132_6-7 | 585 | PTTG1 | NM_004219.2 | ||
| 203 | DCK | NM_000788.1 | 586 | RAB32 | NM_006834.2 | Yes | |
| 204 | DDB1 | NM_001923.2 | 587 | RAB6C | NM_032144.1 | ||
| 205 | DET1 | NM_017996.2 | 588 | RAC1 | NM_006908.3 | ||
| 206 | DHFR | NM_000791.2 | Yes | 589 | RAD51C | NM_058216.1 | |
| 207 | DHPS | NM_013407.1 | 590 | RAD54L | NM_003579.2 | Yes | |
| 208 | DIABLO | NM_019887.1 | 591 | RAF1 | NM_002880.1 | Yes | |
| 209 | DIAPH1 | NM_005219.2 | 592 | RALBP1 | NM_006788.2 | Yes | |
| 210 | DICER1 | NM_177438.1 | 593 | RANBP2 | NM_006267.3 | Yes | |
| 211 | DKK1 | NM_012242.1 | Yes | 594 | ranBP7 | NM_006391.1 | |
| 212 | DLC1 | NM_006094.3 | Yes | 595 | RANBP9 | NM_005493.2 | |
| 213 | DPYD | NM_000110.2 | Yes | 596 | RAP1GDS1 | NM_021159.3 | |
| 214 | DR4 | NM_003844.1 | Yes | 597 | RARA | NM_000964.1 | |
| 215 | DR5 | NM_003842.2 | 598 | RARB | NM_016152.2 | ||
| 216 | DRG1 | NM_004147.3 | 599 | RASSF1 | NM_007182.3 | ||
| 217 | DSP | NM_004415.1 | 600 | RBM5 | NM_005778.1 | ||
| 218 | DTYMK | NM_012145.1 | 601 | RBX1 | NM_014248.2 | Yes | |
| 219 | DUSP1 | NM_004417.2 | Yes | 602 | RCC1 | NM_001269.2 | Yes |
| 220 | DUSP2 | NM_004418.2 | 603 | REG4 | NM_032044.2 | Yes | |
| 221 | DUT | NM_001948.2 | Yes | 604 | RFC | NM_003056.1 | |
| 222 | DYRK1B | NM_004714.1 | 605 | RhoB | NM_004040.2 | Yes | |
| 223 | E2F1 | NM_005225.1 | Yes | 606 | rhoC | NM_175744.1 | Yes |
| 224 | EDN1 | NM_001955.1 | 607 | RIZ1 | NM_012231.1 | ||
| 225 | EFNA1 | NM_004428.2 | Yes | 608 | RNF11 | NM_014372.3 | |
| 226 | EFNA3 | NM_004952.3 | 609 | ROCK1 | NM_005406.1 | Yes | |
| 227 | EFNB1 | NM_004429.3 | 610 | ROCK2 | NM_004850.3 | Yes | |
| 228 | EFNB2 | NM_004093.2 | Yes | 611 | RPLPO | NM_001002.2 | |
| 229 | EFP | NM_005082.2 | Yes | 612 | RPS13 | NM_001017.2 | Yes |
| 230 | EGFR | NM_005228.1 | 613 | RRM1 | NM_001033.1 | Yes | |
| 231 | EGLN1 | NM_022051.1 | 614 | RRM2 | NM_001034.1 | Yes | |
| 232 | EGLN3 | NM_022073.2 | Yes | 615 | RTN4 | NM_007008.1 | |
| 233 | EGR1 | NM_001964.2 | Yes | 616 | RUNX1 | NM_001754.2 | Yes |
| 234 | EGR3 | NM_004430.2 | Yes | 617 | RXRA | NM_002957.3 | |
| 235 | EI24 | NM_004879.2 | Yes | 618 | S100A1 | NM_006271.1 | Yes |
| 236 | EIF4E | NM_001968.1 | Yes | 619 | S100A2 | NM_005978.2 | |
| 237 | EIF4EL3 | NM_004846.1 | Yes | 620 | S100A4 | NM_002961.2 | Yes |
| 238 | ELAVL1 | NM_001419.2 | Yes | 621 | S100A8 | NM_002964.3 | |
| 239 | EMP1 | NM_001423.1 | Yes | 622 | S100A9 | NM_002965.2 | |
| 240 | EMR3 | NM_032571.2 | 623 | S100P | NM_005980.2 | Yes | |
| 241 | EMS1 | NM_005231.2 | 624 | SAT | NM_002970.1 | Yes | |
| 242 | ENO1 | NM_001428.2 | Yes | 625 | SBA2 | NM_018639.3 | Yes |
| 243 | EP300 | NM_001429.1 | 626 | SDC1 | NM_002997.1 | ||
| 244 | EPAS1 | NM_001430.3 | Yes | 627 | SEMA3B | NM_004636.1 | |
| 245 | EpCAM | NM_002354.1 | 628 | SEMA3F | NM_004186.1 | ||
| 246 | EPHA2 | NM_004431.2 | 629 | SEMA4B | NM_020210.1 | Yes | |
| 247 | EPHB2 | NM_004442.4 | Yes | 630 | SFRP2 | NM_003013.2 | Yes |
| 248 | EPHB4 | NM_004444.3 | 631 | SFRP4 | NM_003014.2 | Yes | |
| 249 | EphB6 | NM_004445.1 | Yes | 632 | SGCB | NM_000232.1 | Yes |
| 250 | EPM2A | NM_005670.2 | 633 | SHC1 | NM_003029.3 | Yes | |
| 251 | ErbB3 | NM_001982.1 | 634 | SHH | NM_000193.2 | ||
| 252 | ERCC1 | NM_001983.1 | 635 | SI | NM_001041.1 | Yes | |
| 253 | ERCC2 | NM_000400.2 | 636 | Siah-1 | NM_003031.2 | ||
| 254 | EREG | NM_001432.1 | Yes | 637 | SIAT4A | NM_003033.2 | Yes |
| 255 | ERK1 | Z11696.1 | 638 | SIAT7B | NM_006456.1 | ||
| 256 | ERK2 | NM_002745.1 | 639 | SIM2 | NM_005069.2 | Yes | |
| 257 | ESPL1 | NM_012291.1 | Yes | 640 | SIN3A | NM_015477.1 | |
| 258 | EstR1 | NM_000125.1 | 641 | SIR2 | NM_012238.3 | Yes | |
| 259 | ETV4 | NM_001986.1 | 642 | SKP1A | NM_006930.2 | ||
| 260 | F3 | NM_001993.2 | Yes | 643 | SKP2 | NM_005983.2 | Yes |
| 261 | FABP4 | NM_001442.1 | Yes | 644 | SLC25A3 | NM_213611.1 | Yes |
| 262 | FAP | NM_004460.2 | Yes | 645 | SLC2A1 | NM_006516.1 | |
| 263 | fas | NM_000043.1 | 646 | SLC31A1 | NM_001859.2 | Yes | |
| 264 | fasl | NM_000639.1 | 647 | SLC5A8 | NM_145913.2 | ||
| 265 | FASN | NM_004104.4 | Yes | 648 | SLC7A5 | NM_003486.4 | |
| 266 | FBXO5 | NM_012177.2 | Yes | 649 | SLPI | NM_003064.2 | Yes |
| 267 | FBXW7 | NM_033632.1 | 650 | SMARCA3 | NM_003071.2 | Yes | |
| 268 | FDXR | NM_004110.2 | 651 | SNAI1 | NM_005985.2 | ||
| 269 | FES | NM_002005.2 | 652 | SNAI2 | NM_003068.3 | Yes | |
| 270 | FGF18 | NM_003862.1 | Yes | 653 | SNRPF | NM_003095.1 | Yes |
| 271 | FGF2 | NM_002006.2 | Yes | 654 | SOD1 | NM_000454.3 | Yes |
| 272 | FGFR1 | NM_023109.1 | 655 | SOD2 | NM_000636.1 | Yes | |
| 273 | FGFR2 isoform 1 | NM_000141.2 | 656 | SOS1 | NM_005633.2 | Yes | |
| 274 | FHIT | NM_002012.1 | 657 | SOX17 | NM_022454.2 | ||
| 275 | FIGF | NM_004469.2 | 658 | SPARC | NM_003118.1 | Yes | |
| 276 | FLJ12455 | NM_022078.1 | 659 | SPINT2 | NM_021102.1 | Yes | |
| 277 | FLJ20712 | AK000719.1 | 660 | SPRY1 | AK026960.1 | Yes | |
| 278 | FLT1 | NM_002019.1 | 661 | SPRY2 | NM_005842.1 | Yes | |
| 279 | FLT4 | NM_002020.1 | 662 | SR-A1 | NM_021228.1 | ||
| 280 | FOS | NM_005252.2 | Yes | 663 | ST14 | NM_021978.2 | Yes |
| 281 | FOXO3A | NM_001455.1 | Yes | 664 | STAT1 | NM_007315.1 | |
| 282 | FPGS | NM_004957.3 | Yes | 665 | STAT3 | NM_003150.1 | |
| 283 | FRP1 | NM_003012.2 | 666 | STAT5A | NM_003152.1 | ||
| 284 | FST | NM_006350.2 | Yes | 667 | STAT5B | NM_012448.1 | Yes |
| 285 | Furin | NM_002569.1 | 668 | STC1 | NM_003155.1 | Yes | |
| 286 | FUS | NM_004960.1 | 669 | STK11 | NM_000455.3 | ||
| 287 | FUT1 | NM_000148.1 | 670 | STK15 | NM_003600.1 | Yes | |
| 288 | FUT3 | NM_000149.1 | 671 | STMN1 | NM_005563.2 | ||
| 289 | FUT6 | NM_000150.1 | Yes | 672 | STMY3 | NM_005940.2 | Yes |
| 290 | FXYD5 | NM_014164.4 | 673 | STS | NM_000351.2 | ||
| 291 | FYN | NM_002037.3 | Yes | 674 | SURV | NM_001168.1 | Yes |
| 292 | FZD1 | NM_003505.1 | Yes | 675 | TAGLN | NM_003186.2 | Yes |
| 293 | FZD2 | NM_001466.2 | 676 | TBP | NM_003194.1 | ||
| 294 | FZD6 | NM_003506.2 | 677 | TCF-1 | NM_000545.3 | Yes | |
| 295 | G1P2 | NM_005101.1 | 678 | TCF-7 | NM_003202.2 | ||
| 296 | GADD45 | NM_001924.2 | 679 | TCF7L1 | NM_031283.1 | ||
| 297 | GADD45B | NM_015675.1 | Yes | 680 | TCF7L2 | NM_030756.1 | |
| 298 | GADD45G | NM_006705.2 | 681 | TCFL4 | NM_170607.2 | ||
| 299 | GAGE4 | NM_001474.1 | 682 | TEK | NM_000459.1 | ||
| 300 | GBP1 | NM_002053.1 | 683 | TERC | U86046.1 | Yes | |
| 301 | GBP2 | NM_004120.2 | Yes | 684 | TERT | NM_003219.1 | |
| 302 | G-Catenin | NM_002230.1 | Yes | 685 | TFF3 | NM_003226.1 | Yes |
| 303 | GCLC | NM_001498.1 | 686 | TGFA | NM_003236.1 | ||
| 304 | GCLM | NM_002061.1 | 687 | TGFB2 | NM_003238.1 | Yes | |
| 305 | GCNT1 | NM_001490.3 | Yes | 688 | TGFB3 | NM_003239.1 | Yes |
| 306 | GDF15 | NM_004864.1 | 689 | TGFBI | NM_000358.1 | Yes | |
| 307 | GIT1 | NM_014030.2 | Yes | 690 | TGFBR1 | NM_004612.1 | Yes |
| 308 | GJA1 | NM_000165.2 | Yes | 691 | TGFBR2 | NM_003242.2 | Yes |
| 309 | GJB2 | NM_004004.3 | Yes | 692 | THBS1 | NM_003246.1 | Yes |
| 310 | GPX1 | NM_000581.2 | Yes | 693 | THY1 | NM_006288.2 | Yes |
| 311 | GPX2 | NM_002083.1 | 694 | TIMP1 | NM_003254.1 | Yes | |
| 312 | Grb10 | NM_005311.2 | Yes | 695 | TIMP2 | NM_003255.2 | Yes |
| 313 | GRB14 | NM_004490.1 | 696 | TIMP3 | NM_000362.2 | Yes | |
| 314 | GRB2 | NM_002086.2 | 697 | TJP1 | NM_003257.1 | ||
| 315 | GRB7 | NM_005310.1 | 698 | TK1 | NM_003258.1 | Yes | |
| 316 | GRIK1 | NM_000830.2 | 699 | TLN1 | NM_006289.2 | Yes | |
| 317 | GRO1 | NM_001511.1 | 700 | TMEPAI | NM_020182.3 | Yes | |
| 318 | GRP | NM_002091.1 | 701 | TMSB10 | NM_021103.2 | Yes | |
| 319 | GRPR | NM_005314.1 | Yes | 702 | TMSB4X | NM_021109.2 | Yes |
| 320 | GSK3B | NM_002093.2 | Yes | 703 | TNC | NM_002160.1 | |
| 321 | GSTA3 | NM_000847.3 | 704 | TNF | NM_000594.1 | ||
| 322 | GSTM1 | NM_000561.1 | 705 | TNFRSF5 | NM_001250.3 | ||
| 323 | GSTM3 | NM_000849.3 | 706 | TNFRSF6B | NM_003823.2 | ||
| 324 | GSTp | NM_000852.2 | Yes | 707 | TNFSF4 | NM_003326.2 | |
| 325 | GSTT1 | NM_000853.1 | Yes | 708 | TOP2A | NM_001067.1 | Yes |
| 326 | H2AFZ | NM_002106.2 | Yes | 709 | TOP2B | NM_001068.1 | |
| 327 | HB-EGF | NM_001945.1 | Yes | 710 | TP | NM_001953.2 | Yes |
| 328 | hCRA a | U78556.1 | Yes | 711 | TP53BP1 | NM_005657.1 | Yes |
| 329 | HDAC1 | NM_004964.2 | Yes | 712 | TP53BP2 | NM_005426.1 | Yes |
| 330 | HDAC2 | NM_001527.1 | 713 | TP53I3 | NM_004881.2 | ||
| 331 | HDGF | NM_004494.1 | 714 | TRAG3 | NM_004909.1 | Yes | |
| 332 | hENT1 | NM_004955.1 | 715 | TRAIL | NM_003810.1 | Yes | |
| 333 | Hepsin | NM_002151.1 | 716 | TS | NM_001071.1 | Yes | |
| 334 | HER2 | NM_004448.1 | Yes | 717 | TST | NM_003312.4 | |
| 335 | Herstatin | AF177761.2 | 718 | TUBA1 | NM_006000.1 | Yes | |
| 336 | HES6 | NM_018645.3 | Yes | 719 | TUBB | NM_001069.1 | |
| 337 | HGF | M29145.1 | 720 | TUFM | NM_003321.3 | Yes | |
| 338 | HIF1A | NM_001530.1 | Yes | 721 | TULP3 | NM_003324.2 | |
| 339 | HK1 | NM_000188.1 | 722 | tusc4 | NM_006545.4 | ||
| 340 | HLA-DPB1 | NM_002121.4 | 723 | UBB | NM_018955.1 | Yes | |
| 341 | HLA-DRA | NM_019111.3 | 724 | UBC | NM_021009.2 | ||
| 342 | HLA-DRB1 | NM_002124.1 | 725 | UBE2C | NM_007019.2 | Yes | |
| 343 | HLA-G | NM_002127.2 | Yes | 726 | UBE2M | NM_003969.1 | Yes |
| 344 | HMGB1 | NM_002128.3 | 727 | UBL1 | NM_003352.3 | ||
| 345 | hMLH | NM_000249.2 | 728 | UCP2 | NM_003355.2 | ||
| 346 | HNRPAB | NM_004499.2 | Yes | 729 | UGT1A1 | NM_000463.2 | |
| 347 | HNRPD | NM_031370.2 | Yes | 730 | UMPS | NM_000373.1 | Yes |
| 348 | HoxA1 | NM_005522.3 | 731 | UNC5A | XM_030300.7 | ||
| 349 | HoxA5 | NM_019102.2 | Yes | 732 | UNC5B | NM_170744.2 | Yes |
| 350 | HOXB13 | NM_006361.2 | Yes | 733 | UNC5C | NM_003728.2 | |
| 351 | HOXB7 | NM_004502.2 | Yes | 734 | upa | NM_002658.1 | Yes |
| 352 | HRAS | NM_005343.2 | Yes | 735 | UPP1 | NM_003364.2 | Yes |
| 353 | HSBP1 | NM_001537.1 | 736 | VCAM1 | NM_001078.2 | ||
| 354 | HSD17B1 | NM_000413.1 | 737 | VCL | NM_003373.2 | Yes | |
| 355 | HSD17B2 | NM_002153.1 | Yes | 738 | VCP | NM_007126.2 | Yes |
| 356 | HSPA1A | NM_005345.4 | Yes | 739 | VDAC1 | NM_003374.1 | |
| 357 | HSPA1B | NM_005346.3 | Yes | 740 | VDAC2 | NM_003375.2 | Yes |
| 358 | HSPA4 | NM_002154.3 | 741 | VDR | NM_000376.1 | ||
| 359 | HSPA5 | NM_005347.2 | 742 | VEGF | NM_003376.3 | Yes | |
| 360 | HSPA8 | NM_006597.3 | Yes | 743 | VEGF_altsplice1 | AF486837.1 | Yes |
| 361 | HSPB1 | NM_001540.2 | 744 | VEGF_altsplice2 | AF214570.1 | Yes | |
| 362 | HSPCA | NM_005348.2 | 745 | VEGFB | NM_003377.2 | Yes | |
| 363 | HSPE1 | NM_002157.1 | Yes | 746 | VEGFC | NM_005429.2 | Yes |
| 364 | HSPG2 | NM_005529.2 | Yes | 747 | VIM | NM_003380.1 | Yes |
| 365 | ICAM1 | NM_000201.1 | 748 | WIF | NM_007191.2 | Yes | |
| 366 | ICAM2 | NM_000873.2 | Yes | 749 | WISP1 | NM_003882.2 | Yes |
| 367 | ID1 | NM_002165.1 | 750 | WNT2 | NM_003391.1 | Yes | |
| 368 | ID2 | NM_002166.1 | 751 | Wnt-3a | NM_033131.2 | ||
| 369 | ID3 | NM_002167.2 | Yes | 752 | Wnt-5a | NM_003392.2 | |
| 370 | ID4 | NM_001546.2 | Yes | 753 | Wnt-5b | NM_032642.2 | |
| 371 | IFIT1 | NM_001548.1 | 754 | WWOX | NM_016373.1 | ||
| 372 | IGF1 | NM_000618.1 | Yes | 755 | XPA | NM_000380.2 | |
| 373 | IGF1R | NM_000875.2 | 756 | XPC | NM_004628.2 | ||
| 374 | IGF2 | NM_000612.2 | 757 | XRCC1 | NM_006297.1 | ||
| 375 | IGFBP2 | NM_000597.1 | 758 | YB-1 | NM_004559.1 | ||
| 376 | IGFBP3 | NM_000598.1 | Yes | 759 | YWHAH | NM_003405.2 | |
| 377 | IGFBP5 | NM_000599.1 | Yes | 760 | zbtb7 | NM_015898.2 | |
| 378 | IGFBP6 | NM_002178.1 | 761 | ZG16 | NM_152338.1 | ||
| 379 | IGFBP7 | NM_001553 | Yes | ||||
| 380 | IHH | NM_002181.1 | |||||
| 381 | IL10 | NM_000572.1 | |||||
| 382 | IL1B | NM_000576.2 | |||||
| 383 | IL6 | NM_000600.1 |
Table A3.
Definitions of the Chemotherapy Benefit Groups
| Chemotherapy Benefit Group | X = 0.859exp[1.839×RSu+3.526+1.781xTSu]−0.859exp[1.839×RSu] |
|---|---|
| Low | X less than 2% |
| Intermediate | X greater than or equal to 2% and less than 6% |
| High | X greater than or equal to 6% |
NOTE. The unscaled recurrence score and unscaled treatment score were used in combination to determine a chemotherapy benefit group for each patient, as the absolute chemotherapy benefit was also a function of baseline recurrence risk.
Table A4.
Relationship Between Baseline Patient Characteristics and RFI in NSABP C-01/C-02 According to Cox Regression Analysis
| Variable | No. of Patients | HR | 95% CI | P |
|---|---|---|---|---|
| Female v male | 270 | 0.82 | 0.57 to 1.19 | .299 |
| Age, per 1 year increase | 270 | 1.00 | 0.99 to 1.02 | .702 |
| Age ≥ 60 v < 60 years | 270 | 1.02 | 0.70 to 1.48 | .929 |
| Location of tumor | 270 | .006 | ||
| Right v left | 2.46 | 1.45 to 4.19 | ||
| Rectosigmoid v left | 1.76 | 1.02 to 3.04 | ||
| Multiple/unknown v left | 2.21 | 0.82 to 5.93 | ||
| Protocol (C-02 v C-01) | 270 | 0.65 | 0.42 to 1.02 | .052 |
| Surgery + BCG v surgery only | 270 | 1.37 | 0.94 to 2.00 | .107 |
| No. of nodes examined | 270 | .335 | ||
| < 12 v ≥ 12 | 1.18 | 0.81 to 1.73 | ||
| Unknown v≥ 12 | 1.81 | 0.82 to 4.00 | ||
| No. of positive nodes | 266 | < .001 | ||
| 1-3 v 0 | 1.73 | 1.13 to 2.65 | ||
| ≥ 4 v 0 | 2.94 | 1.80 to 4.78 | ||
| Nodal involvement status | 261 | < .001 | ||
| 0 of < 12 examined v 0 of ≥ 12 examined | 2.45 | 1.27 to 4.72 | ||
| 1-3 v 0 of ≥ 12 examined | 2.79 | 1.50 to 5.19 | ||
| ≥ 4 v 0 of ≥ 12 examined | 4.74 | 2.44 to 9.20 | ||
| Stage III v II | 270 | 2.10 | 1.43 to 3.08 | < .001 |
| Tumor grade at GHI | ||||
| First reading high v low | 270 | 1.35 | 0.88 to 2.09 | .180 |
| Second reading high v low | 269 | 1.51 | 1.01 to 2.26 | .054 |
| Surgery | 270 | .346 | ||
| Segmental/anterior resection v colectomy/hemicolectomy | 1.27 | 0.82 to 1.96 | ||
| Other/unknown v colectomy/hemicolectomy | 0.78 | 0.42 to 1.48 | ||
| Mucinous tumor | 270 | 1.64 | 0.92 to 2.93 | .115 |
| Surgery year | 270 | 0.95 | 0.88 to 1.02 | .151 |
Abbreviations: RFI, recurrence-free interval; NSABP, National Surgical Adjuvant Breast and Bowel Project; HR, hazard ratio; BCG, Bacillus Calmette-Guérin; GHI, Genomic Health, Inc.
Table A5.
Relationship Between Baseline Patient Characteristics and RFI in CC Study According to Cox Regression Analysis
| Variable | No. of Patients | HR | 95% CI | P |
|---|---|---|---|---|
| Female v male | 765 | 0.93 | 0.67 to 1.28 | .653 |
| Age, per 1 year increase | 765 | 1.02 | 1.00 to 1.03 | .010 |
| Age ≥ 60 v < 60 years | 765 | 1.49 | 0.98 to 2.26 | .054 |
| Location of tumor | 765 | .609 | ||
| Right v left | 1.25 | 0.80 to 1.95 | ||
| Rectosigmoid v left | 1.15 | 0.71 to 1.85 | ||
| No. of nodes examined | ||||
| < 12 v ≥ 12 | 765 | 1.66 | 1.17 to 2.36 | .006 |
| No. of positive nodes | 765 | < .001 | ||
| 1-3 v 0 | 2.03 | 1.42 to 2.91 | ||
| ≥ 4 v 0 | 4.22 | 2.67 to 6.67 | ||
| Nodal involvement status | 765 | < .001 | ||
| 0 of < 12 examined v 0 of ≥ 12 examined | 1.50 | 0.90 to 2.50 | ||
| 1-3 v 0 of ≥ 12 examined | 2.26 | 1.53 to 3.33 | ||
| ≥ 4 v0 of ≥ 12 examined | 4.69 | 2.89 to 7.60) | ||
| Stage III v II | 765 | 2.43 | 1.76 to 3.36 | < .001 |
| GHI tumor grade high v Low | 765 | 0.87 | 0.61 to 1.26 | .460 |
| CC tumor grade high vLow | 761 | 1.28 | 0.89 to 1.86 | .195 |
| Surgery | 765 | .947 | ||
| Segmental/anterior resection v colectomy/hemicolectomy | 0.94 | 0.65 to 1.36 | ||
| Other/unknown v colectomy/hemicolectomy | 0.94 | 0.41 to 2.15 | ||
| Mucinous tumor | 765 | 0.64 | 0.40 to 1.02 | .049 |
| Surgery year | 765 | .131 | ||
| 1986-1990 v 1981-1985 | 0.90 | 0.59 to 1.38 | ||
| 1991-1995 v 1981-1985 | 0.56 | 0.34 to 0.94 | ||
| 1996-2000 v 1981-1985 | 0.88 | 0.57 to 1.35 | ||
| T stage T4 v T1, T2, and T3 | 765 | 1.74 | 1.22 to 2.48 | .003 |
| Fixative | 765 | .090 | ||
| Hollandes v formalin | 1.53 | 0.91 to 2.59 | ||
| Zenkers or Bouins v formalin | 2.06 | 1.06 to 4.02 | ||
| MMR deficient v proficient | 712 | 0.75 | 0.45 to 1.27 | .266 |
| Surgeon | 765 | .742 |
Abbreviations: RFI, recurrence-free interval; CC, Cleveland Clinic; HR, hazard ratio; GHI, Genomic Health, Inc; MMR, mismatch repair.
Table A6.
Relationship Between Baseline Patient Characteristics and RFI in NSABP C-04 Study According to Cox Regression Analysis
| Variable | No. of Patients | HR | 95% CI | P |
|---|---|---|---|---|
| Age ≥ 60 v < 60 years | 308 | 0.76 | 0.50 to 1.15 | .191 |
| Age, per 1 year increase | 308 | 0.99 | 0.97 to 1.01 | .240 |
| Female v male | 308 | 0.81 | 0.53 to 1.23 | .322 |
| Race | 308 | .470 | ||
| Black v white | 0.65 | 0.28 to 1.49 | ||
| Other v white | 1.26 | 0.51 to 3.12 | ||
| Location of tumor | 308 | .972 | ||
| Right v left | 1.13 | 0.65 to 1.97 | ||
| Rectosigmoid v left | 1.11 | 0.63 to 1.95 | ||
| Multiple/unknown v left | 0.95 | 0.22 to 4.09 | ||
| Surgery | 308 | .836 | ||
| Segmental/anterior resection v colectomy/hemicolectomy | 1.13 | 0.73 to 1.75 | ||
| Other/unknown v colectomy/hemicolectomy | 0.96 | 0.38 to 2.40 | ||
| Tumor grade by GHI | ||||
| First reading | 308 | 1.37 | 0.81 to 2.32 | .260 |
| Second reading | 308 | 1.04 | 0.64 to 1.68 | .884 |
| No. of nodes examined | 308 | .245 | ||
| < 12 v ≥ 12 | 1.37 | 0.90 to 2.10 | ||
| Unknown v ≥ 12 | 1.97 | 0.61 to 6.39 | ||
| Tumor stage III v II | 308 | 1.44 | 0.94 to 2.21 | .089 |
| No. of positive nodes | 307 | .002 | ||
| 1-3 v 0 | 1.06 | 0.64 to 1.73 | ||
| ≥ 4 v 0 | 2.43 | 1.47 to 4.04 | ||
| Nodes involvement | 304 | < .001 | ||
| 0 of < 12 examined v 0 of ≥ 12 examined | 2.27 | 1.11 to 4.66 | ||
| 1-3 v 0 of ≥ 12 examined | 1.66 | 0.83 to 3.32 | ||
| ≥ 4 v 0 of ≥ 12 examined | 3.83 | 1.90 to 7.72 | ||
| Mucinous tumor | 308 | 1.53 | 0.77 to 3.04 | .255 |
| Surgery date, quarters | 308 | 1.00 | 0.88 to 1.13 | .975 |
| Surgery year | 308 | 0.98 | 0.63 to 1.53 | .924 |
Abbreviations: RFI, recurrence-free interval; NSABP, National Surgical Adjuvant Breast and Bowel Project; HR, hazard ratio; GHI, Genomic Health, Inc.
Table A7.
Relationship Between Baseline Patient Characteristics and RFI in NSABP C-06 Study According to Cox Regression Analysis
| Variable | No. of Patients | HR | 95% CI | P |
|---|---|---|---|---|
| Female v male | 508 | 1.14 | 0.80 to 1.61 | .463 |
| Age, per 1 year increase | 508 | 1.00 | 0.99 to 1.02 | .632 |
| Age ≥ 60 v < 60 years | 508 | 1.13 | 0.79 to 1.62 | .485 |
| Ethnicity | 508 | .492 | ||
| Black v white | 0.98 | 0.53 to 1.82 | ||
| Other/unknown v white | 0.53 | 0.17 to 1.68 | ||
| Location of tumor | 508 | .805 | ||
| Right v left | 1.10 | 0.70 to 1.73 | ||
| Rectosigmoid v left | 0.95 | 0.59 to 1.53 | ||
| Multiple/unknown v left | 0.55 | 0.08 to 4.07 | ||
| No. of nodes examined | ||||
| < 12 v ≥ 12 | 507 | 1.09 | 0.77 to 1.30 | .632 |
| Tumor stage III v II | 508 | 2.84 | 1.91 to 4.22 | < .001 |
| T stage* | 508 | .241 | ||
| T1 v T3 | 1.51 | 0.37 to 6.11 | ||
| T2 v T3 | 0.83 | 0.41 to 1.70 | ||
| No. of positive nodes | 508 | < .001 | ||
| 1-3 v 0 | 2.19 | 1.41 to 3.38 | ||
| ≥ 4 v 0 | 4.49 | 2.84 to 7.09 | ||
| Nodal involvement | 508 | <.001 | ||
| 0 of < 12 examined v 0 of ≥ 12 examined | 1.70 | 0.84 to 3.41 | ||
| 1-3 v 0 of ≥ 12 examined | 2.91 | 1.59 to 5.35 | ||
| ≥ 4 v 0 of ≥ 12 examined | 5.98 | 3.21 to 11.1 | ||
| GHI tumor grade | ||||
| First reading | 508 | 0.95 | 0.63 to 1.42 | .794 |
| Second reading | 508 | 1.08 | 0.75 to 1.56 | .668 |
| Surgery | 508 | .424 | ||
| Segmental/anterior resection v resection v colectomy/hemicolectomy | 0.89 | 0.61 to 1.28 | ||
| Other/unknown v colectomy/hemicolectomy | 1.80 | 0.66 to 4.92 | ||
| Mucinous tumor | 508 | 0.71 | 0.44 to 1.14 | .144 |
| Surgery date, quarters | 508 | 0.97 | 0.90 to 1.05 | .427 |
| Surgery year | 508 | 0.88 | 0.66 to 1.17 | .367 |
Abbreviations: RFI, recurrence-free interval; NSABP, National Surgical Adjuvant Breast and Bowel Project; HR, hazard ratio; GHI, Genomic Health, Inc.
Data insufficient to estimate T4 effect.
Footnotes
See accompanying editorial on page 3904
Supported by Public Health Service Grants No. U10-CA-37377, U10-CA-69974, U10-CA-12027, and U10-CA-69651 from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, and by Genomic Health, Inc.
Presented in part at the 42nd Annual Meeting of the American Society of Clinical Oncology, June 2-6, 2006, Atlanta, GA, and at the Annual Gastrointestinal Cancers Symposium of the American Society of Clinical Oncology, January 25-27, 2008, Orlando, FL.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00427570.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Kim M. Clark-Langone, Genomic Health, Inc (C); Margarita Lopatin, Genomic Health, Inc (C); Drew Watson, Genomic Health, Inc (C); Frederick L. Baehner, Genomic Health, Inc (C); Steven Shak, Genomic Health, Inc (C); Joffre Baker, Genomic Health, Inc (C); J. Wayne Cowens, Genomic Health, Inc (C) Consultant or Advisory Role: Michael J. O'Connell, Genomic Health, Inc (U) Stock Ownership: Kim M. Clark-Langone, Genomic Health, Inc; Margarita Lopatin, Genomic Health, Inc; Drew Watson, Genomic Health, Inc; Steven Shak, Genomic Health, Inc; Joffre Baker, Genomic Health, Inc; J. Wayne Cowens, Genomic Health, Inc Honoraria: Greg Yothers, Genomic Health, Inc Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Michael J. O'Connell, Ian Lavery, Soonmyung Paik, Margarita Lopatin, Frederick L. Baehner, Steven Shak, Joffre Baker, J. Wayne Cowens, Norman Wolmark
Financial support: Steven Shak
Administrative support: Soonmyung Paik, J. Wayne Cowens, Norman Wolmark
Provision of study materials or patients: Ian Lavery, Soonmyung Paik
Collection and assembly of data: Ian Lavery, Greg Yothers, Soonmyung Paik, Kim M. Clark-Langone, Drew Watson, Frederick L. Baehner, Steven Shak, J. Wayne Cowens
Data analysis and interpretation: Michael J. O'Connell, Ian Lavery, Greg Yothers, Soonmyung Paik, Kim M. Clark-Langone, Margarita Lopatin, Drew Watson, Steven Shak, Joffre Baker, J. Wayne Cowens
Manuscript writing: Michael J. O'Connell, Greg Yothers, Soonmyung Paik, Kim M. Clark-Langone, Margarita Lopatin, Frederick L. Baehner, Steven Shak, Joffre Baker, J. Wayne Cowens
Final approval of manuscript: Michael J. O'Connell, Ian Lavery, Greg Yothers, Soonmyung Paik, Kim M. Clark-Langone, Margarita Lopatin, Drew Watson, Frederick L. Baehner, Steven Shak, Joffre Baker, J. Wayne Cowens, Norman Wolmark
REFERENCES
- 1.Moertel CG, Fleming TR, Macdonald JS, et al. Fluorouracil plus levamisole as effective adjuvant treatment after resection of stage III colon carcinoma: A final report. Ann Intern Med. 1995;122:321–326. doi: 10.7326/0003-4819-122-5-199503010-00001. [DOI] [PubMed] [Google Scholar]
- 2.O'Connell MJ, Mailliard JA, Kahn MJ, et al. Controlled trial of fluorouracil and low-dose leucovorin given for 6 months as postoperative adjuvant therapy for colon cancer. J Clin Oncol. 1997;15:246–250. doi: 10.1200/JCO.1997.15.1.246. [DOI] [PubMed] [Google Scholar]
- 3.Wolmark N, Rockette H, Fisher B, et al. The benefit of leucovorin-modulated fluorouracil as postoperative adjuvant therapy for primary colon cancer: Results from NSABP protocol C-03. J Clin Oncol. 1993;11:1879–1887. doi: 10.1200/JCO.1993.11.10.1879. [DOI] [PubMed] [Google Scholar]
- 4.Moertel CG, Fleming TR, Macdonald JS, et al. Intergroup study of fluorouracil plus levamisole as adjuvant therapy for stage II/Dukes' B2 colon cancer. J Clin Oncol. 1995;13:2936–2943. doi: 10.1200/JCO.1995.13.12.2936. [DOI] [PubMed] [Google Scholar]
- 5.Mamounas E, Wieand S, Wolmark N, et al. Comparative efficacy of adjuvant chemotherapy in patients with Dukes' B versus Dukes' C colon cancer: Results from four National Surgical Adjuvant Breast and Bowel Project adjuvant studies (C-01, C-02, C-03, C-04) J Clin Oncol. 1999;17:1349–1355. doi: 10.1200/JCO.1999.17.5.1349. [DOI] [PubMed] [Google Scholar]
- 6.Efficacy of adjuvant fluorouracil and folinic acid in B2 colon cancer: International multicenter pooled analysis of B2 colon cancer trials (IMPACT B2) investigators. J Clin Oncol. 1999;17:1356–1363. [PubMed] [Google Scholar]
- 7.Benson AB, 3rd, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 8.André T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343–2351. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 9.Kuebler JP, Wieand HS, O'Connell MJ, et al. Oxaliplatin combined with weekly bolus 5-fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: Results from NSABP Protocol C-07. J Clin Oncol. 2007;25:2198–2204. doi: 10.1200/JCO.2006.08.2974. [DOI] [PubMed] [Google Scholar]
- 10.André T, Boni C, Navarro M, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC Trial. J Clin Oncol. doi: 10.1200/JCO.2008.20.6771. [epub ahead of print on May 8, 2009] [DOI] [PubMed] [Google Scholar]
- 11.QUASAR Collaborative Group. Adjuvant chemotherapy versus observation in patients with colorectal cancer: A randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 12.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 13.Ogunbiyi OA, Goodfellow PJ, Herfarth K, et al. Confirmation that chromosome 18q allelic loss in colon cancer is a prognostic indicator. J Clin Oncol. 1998;16:427–433. doi: 10.1200/JCO.1998.16.2.427. [DOI] [PubMed] [Google Scholar]
- 14.Jen J, Kim H, Piantadosi S, et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331:213–221. doi: 10.1056/NEJM199407283310401. [DOI] [PubMed] [Google Scholar]
- 15.Barratt PL, Seymour MT, Stenning SP, et al. DNA markers predicting benefit from adjuvant fluorouracil in patients with colon cancer: A molecular study. Lancet. 2002;360:1381–1391. doi: 10.1016/s0140-6736(02)11402-4. [DOI] [PubMed] [Google Scholar]
- 16.Krajewska M, Kim H, Kim C, et al. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin Cancer Res. 2005;11:5451–5461. doi: 10.1158/1078-0432.CCR-05-0094. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Jatkoe T, Zhang Y, et al. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol. 2004;22:1564–1571. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 18.Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 19.Bertucci F, Salas S, Eysteries S, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 20.Arango D, Laiho P, Kokko A, et al. Gene-expression profiling predicts recurrence in Dukes' C colorectal cancer. Gastroenterology. 2005;129:874–884. doi: 10.1053/j.gastro.2005.06.066. [DOI] [PubMed] [Google Scholar]
- 21.Barrier A, Boelle PY, Roser F, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24:4685–4691. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 22.Sargent DJ, Marsini S, Thibodeau SN, et al. Confirmation of deficient mismatched repair (dMMR) as a predictive marker for lack of benefit from 5-FU based chemotherapy in stage II and III colon cancer (CC): A pooled molecular reanalysis of randomized chemotherapy trials. J Clin Oncol. 2008;26(suppl):180s. abstr 4008. [Google Scholar]
- 23.Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 24.Cronin M, Pho M, Dutta D, et al. Measurement of gene expression in archival paraffin-embedded tissues: Development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 26.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 27.Wolmark N, Fisher B, Rockette H, et al. Postoperative adjuvant chemotherapy or BCG for colon cancer: Results from NSABP protocol C-01. J Natl Cancer Inst. 1988;80:30–36. doi: 10.1093/jnci/80.1.30. [DOI] [PubMed] [Google Scholar]
- 28.Wolmark N, Rockette H, Wickerham DL, et al. Adjuvant therapy of Dukes' A, B, and C adenocarcinoma of the colon with portal-vein fluorouracil infusion: Preliminary results of National Surgical Adjuvant Breast and Bowel Project Protocol C-02. J Clin Oncol. 1990;8:1466–1475. doi: 10.1200/JCO.1990.8.9.1466. [DOI] [PubMed] [Google Scholar]
- 29.Wolmark N, Rockette H, Mamounas E. Clinical trial to assess the relative efficacy of fluorouracil and leucovorin, fluorouracil and levamisole, and fluorouracil, leucovorin, and levamisole in patients with Dukes' B and C carcinoma of the colon: Results from National Surgical Adjuvant Breast and Bowel Project C-04. J Clin Oncol. 1999;17:3553–3559. doi: 10.1200/JCO.1999.17.11.3553. [DOI] [PubMed] [Google Scholar]
- 30.Lembersky BC, Wieand HS, Petrelli NJ, et al. Oral uracil and tegafur plus leucovorin compared with intravenous fluorouracil and leucovorin in stage II and III carcinoma of the colon: Results from National Surgical Adjuvant Breast and Bowel Project Protocol C-06. J Clin Oncol. 2006;24:2059–2064. doi: 10.1200/JCO.2005.04.7498. [DOI] [PubMed] [Google Scholar]
- 31.Hamilton SR, Aaltonen LA. Lyon, France: IARC Press; 2000. World Health Organization Classification of Tumors: Pathology and Genetics of Tumours of the Digestive System. [Google Scholar]
- 32.Clark-Langone KM, Wu JY, Sangli C, et al. Biomarker discovery for colon cancer using a 761 gene RT-PCR assay. BMC Genomics. 2007;8:279. doi: 10.1186/1471-2164-8-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giardiello FM, Brensinger JD, Petersen GM. AGA technical review on hereditary colorectal cancer and genetic testing. Gastroenterology. 2001;121:198–213. doi: 10.1053/gast.2001.25581. [DOI] [PubMed] [Google Scholar]
- 34.Cox D. R. Regression Models and Life-Tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 35.Anderberg MR. New York, NY: Academic Press; 1973. Cluster Analysis for Applications; pp. 131–140. [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 37.Good P. New York, NY: Springer-Verlag; 2000. Permutation Tests: A Practical Guide to Resampling Methods for Testing Hypotheses. [Google Scholar]
- 38.Efron B, Tibshirani RJ. London, United Kingdom: Chapman and Hall; 1993. An Introduction to the Bootstrap. [Google Scholar]
- 39.Chernick MR. New York, NY: Wiley; 1999. Bootstrap Methods: A Practitioner's Guide—Wiley Series in Probability and Statistics. [Google Scholar]
- 40.Simon R. A checklist for evaluating reports of expression profiling for treatment selection. Clin Adv Hematol Oncol. 2006;4:219–224. [PubMed] [Google Scholar]
- 41.Alonzo TA. Standards for reporting prognostic tumor marker studies. J Clin Oncol. 2005;23:9053–9054. doi: 10.1200/JCO.2005.04.3778. [DOI] [PubMed] [Google Scholar]
- 42.Esteban J, Baker J, Cronin M, et al. Tumor gene expression and prognosis in breast cancer: Multi-gene RT-PCR assay of paraffin-embedded tissue. J Clin Oncol. 2003;22(suppl):850. abstr 3416. [Google Scholar]
- 43.Allegra CJ, Paik S, Colangelo LH, et al. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes' B and C colon cancer: A National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project collaborative study. J Clin Oncol. 2003;21:241–250. doi: 10.1200/JCO.2003.05.044. [DOI] [PubMed] [Google Scholar]
- 44.Garrity MM, Burgart LJ, Mahoney MR, et al. Prognostic value of proliferation, apoptosis, defective DNA mismatch repair, and p53 overexpression in patients with resected Dukes' B2 or C colon cancer: A North Central Cancer Treatment Group Study. J Clin Oncol. 2004;22:1572–1582. doi: 10.1200/JCO.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 45.Dukes CE. The classification of cancer of the rectum. J Pathol Bact. 1932;35:323–332. [Google Scholar]
- 46.Dukes CE, Bussey HJ. The spread of rectal cancer and its effect on prognosis. Br J Cancer. 1958;12:309–320. doi: 10.1038/bjc.1958.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaupel P, Mayer A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 48.Tajima A, Hess MT, Cabrera BL, et al. The mismatch repair complex hMutS alpha recognizes 5-fluorouracil-modified DNA: Implications for chemosensitivity and resistance. Gastroenterology. 2004;127:1678–1684. doi: 10.1053/j.gastro.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 49.de Bruin M, van Capel T, Van der Born K, et al. Role of platelet-derived endothelial cell growth factor/thymidine phosphorylase in fluoropyrimidine sensitivity. Br J Cancer. 2003;88:957–964. doi: 10.1038/sj.bjc.6600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Triest B, Pinedo HM, Blaauwgeers JL, et al. Prognostic role of thymidine synthase, thymidine phosphorylase/platelet-derived endothelial cell growth factor and proliferation markers in colorectal cancer. Clin Cancer Res. 2000;6:1063–1072. [PubMed] [Google Scholar]
- 51.van Kuilenburg AB. Screening for dihyrodropyrimidine dehydrogenase deficiency: To do or not to do, that's the question. Cancer Invest. 2006;24:215–217. doi: 10.1080/07357900500524702. [DOI] [PubMed] [Google Scholar]
- 52.Peters GJ, Smorenburg CH, Van Groeningen CJ, et al. Prospective clinical trials using a pharmacogenetic/pharmacogenomic approach. J Chemother. 2004;16(suppl 4):25–30. doi: 10.1179/joc.2004.16.Supplement-1.25. [DOI] [PubMed] [Google Scholar]
- 53.Salonga D, Danenberg KD, Johnson M, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 54.Ichikawa W, Uetake H, Shirota Y, et al. Combination of dihydropyrimidine dehydrogenase and thymidylate synthase gene expression in primary tumors as predictive parameters for the efficacy of fluoropyrimidine-based chemotherapy for metastatic colorectal cancer. Clin Cancer Res. 2003;9:786–791. [PubMed] [Google Scholar]
- 55.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Storojeva I, Boulay JL, Heinimann K, et al. Prognostic and predictive relevance of microsatellite instability in colorectal cancer. Oncol Rep. 2005;14:241–249. [PubMed] [Google Scholar]
- 57.Carethers JM, Smith EJ, Behling CA, et al. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]