Abstract
The aryl hydrocarbon (dioxin) receptor (AhR) is a ligand-dependent transcription factor that produces a wide range of biological and toxic effects in many species and tissues. Whereas the best-characterized high-affinity ligands include structurally related halogenated aromatic hydrocarbons (HAHs) and polycyclic aromatic hydrocarbons (PAHs), the AhR is promiscuous and can also be activated by structurally diverse exogenous and endogenous chemicals. However, little is known about how these diverse ligands actually bind to and activate the AhR. Utilizing AhR ligand binding, DNA binding, and reporter gene expression assays, we have identified a novel ligand-selective antagonist (CH223191) that preferentially inhibits the ability of some classes of AhR agonists (2,3,7,8-tetrachlorodibenzo-p-dioxin and related HAHs), but not others (PAHs, flavonoids, or indirubin), to bind to and/or activate the AhR and AhR signal transduction. HAH-specific antagonism of AhR-dependent reporter gene expression by CH223191 was observed with mouse, rat, human, and guinea pig cell lines. Ligand- and species-selective antagonism was also observed with the AhR antagonists 3′-methoxy-4′-nitroflavone and 6,2′,4′,-trimethoxyflavone. Our results suggest that the differences in the binding by various ligands to the AhR contribute to the observed structural diversity of AhR ligands and could contribute in ligand-specific variation in AhR functionality and the toxic and biological effects of various classes of AhR agonists.
Keywords: 2,3,7,8-tetrachlorodibenzo-p-dioxin; TCDD; Ah receptor; beta-naphthoflavone; CH223191
The aryl hydrocarbon (dioxin) receptor (AhR) is a ligand-dependent basic helix-loop-helix-Per-Arnt-Sim (PAS)–containing transcription factor that responds to exogenous and endogenous chemicals with the induction/repression of expression of a diverse battery of genes in a wide range of species and tissues (reviewed in Beischlag et al., 2008; Bradshaw and Bell, 2009; Kewley et al., 2004; Ma, 2001). The best-characterized high-affinity ligands for the AhR include a variety of toxic halogenated aromatic hydrocarbons (HAHs), such as the polychlorinated dibenzo-p-dioxins, dibenzofurans, and biphenyls, and numerous polycyclic aromatic hydrocarbons (PAHs) and PAH-like chemicals, such as benzo(a)pyrene, 3-methylcholanthrene, and beta-naphthoflavone (BNF) (Denison et al., 1998; Poland and Knutson, 1982; Safe, 1990). However, a relatively large number of natural, endogenous, and synthetic AhR agonists have also been identified in recent years whose structures and physicochemical characteristics are dramatically different from the prototypical HAH and PAH AhR ligands (Denison and Heath-Pagliuso, 1998; Denison and Nagy, 2003; Denison et al., 1998; Nguyen and Bradfield, 2008), which suggests that the AhR has an extremely promiscuous ligand-binding pocket.
Exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, dioxin), the protypical and most potent AhR ligand/agonist, and related dioxin-like halogenated aromatic hydrocarbons (dl-HAHs), results in a wide variety of toxic and biological responses, many of which have been shown to require AhR-dependent gene expression (Bradshaw and Bell, 2009; Furness and Whelan, 2009; Poland and Knutson, 1982; Safe, 1990). Although the spectrum of biochemical and toxicological responses mediated by the AhR are species and tissue specific, ligand-specific differences have also been observed, with TCDD and other dl-HAHs, but not PAHs or other non-HAH AhR agonists, producing a characteristic spectrum of AhR-dependent toxic effects (Bradshaw and Bell, 2009; Denison et al., 1998; Poland and Knutson, 1982; Safe, 1990). Although the greater toxic potency of HAHs appears to be related to their resistance to metabolism and persistent AhR activation (Bradshaw and Bell, 2009; Denison et al., 1998; Poland and Glover, 1980; Poland and Knutson, 1982; Safe, 1990), ligand-dependent differences in the structure and function of the AhR protein and/or AhR protein complex may also play a role (Gouédard et al., 2004; Matikainen et al., 2001).
Although the extreme structural diversity of AhR ligands is analogous to the ligand promiscuity reported for some members of the steroid, thyroid, and retinoic acid receptor superfamily (Ghosh et al., 2003; Ngan et al., 2009; Noy, 2007), a mechanistic understanding of AhR ligand diversity is lacking due to the absence of a three-dimensional (3D) structure of the AhR ligand–binding domain (LBD). However, several studies examining the ability of low- and high-affinity ligands to bind to AhRs containing mutations within the ligand-binding pocket have provided some evidence for differential binding of structurally diverse ligands within the AhR LBD (Backlund and Ingelman-Sundberg, 2004; Goryo et al., 2007; Whelan et al., 2010). These results suggested to us that if structurally diverse ligands can bind to different amino acids within the AhR ligand–binding pocket, then it should be possible to identify an antagonist that would preferentially affect one ligand, or a class of ligands, over another. Accordingly, we examined the ability of a variety of chemicals to differently inhibit the ability of the AhR to be activated by the prototypical agonists TCDD (an HAH) and BNF (a PAH-like chemical). Here we report that the compound CH223191, previously shown to be a pure AhR antagonist that competes with TCDD for binding to the AhR (Kim et al., 2006), is actually an HAH-selective AhR antagonist, producing little or no antagonism of AhR activation by maximally inducing concentrations of PAHs or other non-HAH AhR agonists. Not only is CH223191 the first reported ligand-selective antagonist of the AhR but also our results provide strong supporting evidence for the differential interaction of HAHs and non-HAH agonists within the AhR LBD and provide novel insights into the mechanisms contributing to the observed structural promiscuity of AhR ligands.
MATERIALS AND METHODS
Chemicals.
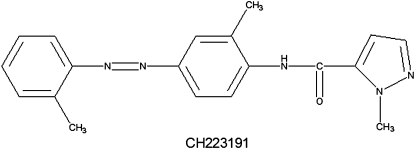
TCDD, [3H]TCDD, and 3′-methoxy-4′-nitroflavone (MNF) were generously provided by Dr Steven Safe (Texas A&M University) and 6,2′,4′,-trimethoxyflavone (TMF) by Dr Gary Perdew (Pennsylvania State University). 3,3′,4,4′-Tetrachlorobiphenyl (PCB77), 3,3′,4,4′,5-pentachlorobiphenyl (PCB126), and 2,3,7,8-tetrachlorodibenzofuran (TCDF) were from Accustandard (New Haven, CT) and BNF, alpha-naphthoflavone (ANF), benzo(a)anthracene (BAA), dibenz(a,h)anthracene (DBA), benzo(k)fluoranthene (BKF), indirubin, and dimethyl sulfoxide (DMSO) from Aldrich Chemicals (St Louis, MO). CH223191 was from Chembridge Corporation (San Diego, CA), and its structure (Fig. 1) was confirmed by nuclear magnetic resonance (NMR). Flavonoids (T7, AU1, AJ9, and AY9) were from Dr Michael Nantz (University of Louisville; Springsteel et al., 2003), and we previously confirmed their AhR agonist activity and relative potency (unpublished data).
FIG. 1.
Structure of CH223191.
Cell culture, chemical treatment, and reporter gene expression.
Recombinant guinea pig intestinal adenocarcinoma (G16L1.1c8) and rat (H4L1.1c4), mouse (H1L1.1c2), and human (HG2L6.1c3) hepatoma cells containing a stably transfected AhR-responsive dioxin-responsive element (DRE)-driven luciferase reporter plasmid were grown as described (Garrison et al., 1996; Han et al., 2004). Cells were plated into white, clear-bottomed 96-well culture plates (75,000 cells per well), incubated with DMSO (1% final concentration) or the indicated concentration of TCDD, BNF, or other agonist in the absence or presence of CH223191 for the indicated time at 37°C and luciferase activity determined (Han et al., 2004).
DNA- and ligand-binding analysis.
Male Hartley guinea pig hepatic cytosol (8 mg protein/ml) was incubated with 20nM TCDD, 1μM BNF, or carrier solvent (DMSO) in the absence or presence of the indicated concentration of CH223191 for 2 h at 20°C, and binding of an in vitro transformed AhR complex to a 32P-labeled oligonucleotide containing the dioxin-responsive element (DRE) oligonucleotide was determined using gel retardation analysis as described in detail (Denison et al., 2002). The amount of ligand-induced AhR:Arnt:[32P]-DRE complex was determined by phosphorimager analysis and expressed relative to the amount of complex produced by maximal activating concentrations of TCDD or BNF. For ligand binding, cytosol (2 mg protein/ml) was incubated with 10nM [3H]TCDD in the absence or presence of 1μM TCDF or 10μM of CH223191 and [3H]TCDD-specific binding determined by sucrose density centrifugation (Denison et al., 2002).
Nuclear localization analysis.
Ligand-dependent AhR nuclear localization was determined using mouse hepatoma (TAOc1BPrc1) cells that have been stably transfected with an N-terminal yellow fluorescent protein (YFP)-AhR chimera (yAhR) expression plasmid. These cells (referred to as yAHAYc6 cells) respond to AhR agonists with nuclear accumulation of yAhR that occurs in a chemical-, dose-, and time-dependent manner (Hayashi and Denison, in preparation). One day before microscopy, yAHAYc6 cells were plated onto 4-well Lab-Tek chambered #1.0 borosilicate cover glass (Nunc) in 600 μl of media. Prior to chemical treatment, cells were rinsed with PBS and treated with the desired chemicals in Opti-MEM reduced serum media (Invitrogen). After a 1-h incubation at 37°C, cells were visualized using an Olympus IX71 fluorescence microscope with a YFP filter (excitation: HQ500/20, emission: HQ520lp, beam splitter: Q515lp; Chroma Technology Corp.). Live cell images were captured and processed using a Hamamatsu Orca camera and Slidebook 4.2 imaging software (Intelligent Imaging Innovations).
RESULTS
CH223191 Preferentially Inhibits AhR-Dependent Reporter Gene Expression in Cells from Various Species by TCDD
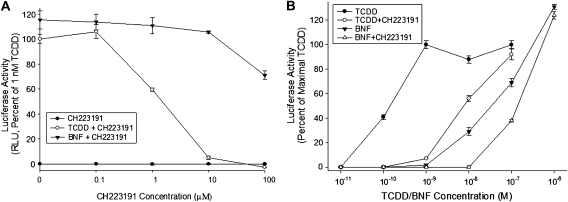
Although CH223191 has been previously shown to antagonize the ability of TCDD to stimulate AhR-dependent gene expression in mice in vivo and in cells in culture (Kim et al., 2006), its effect on other AhR agonists was not examined. To determine whether CH223191 can antagonize the induction of AhR-dependent gene expression by different ligands, we first examined its effects on guinea pig intestinal adenocarcinoma (G16L1.1c8) and mouse hepatoma (H1L1.1c2) cells that contain a stably transfected DRE-luciferase reporter plasmid. Co-incubation of G16L1.1c8 cells with increasing concentrations of CH223191 and equally effective/maximally inducing concentrations of BNF (1μM) or TCDD (1nM) revealed that whereas CH223191 decreased TCDD-dependent induction of luciferase in a concentration-dependent manner (Fig. 2A), BNF-induced luciferase activity was only partially inhibited at the highest CH223191 concentration (100μM). Co-incubation of mouse hepatoma (H1L1.1c2 cells) with a constant concentration of CH223191 (10μM) and increasing concentrations of BNF or TCDD revealed complete inhibition of induction by TCDD and only partial (< 30%) inhibition of induction by BNF; no inhibition of BNF was observed with a maximally inducing concentration of BNF (Fig. 2B). The CH223191-dependent antagonism of induction by TCDD was overcome with higher concentrations of TCDD, indicating that inhibition by CH223191 was competitive in nature. These results demonstrate that CH223191 is a preferential and more effective antagonist of AhR-dependent gene activation by TCDD than that by BNF. These results indicate a preferential effect of CH223191 on TCDD-stimulated AhR-dependent gene expression. We also confirmed the preferential ability of CH223191 to antagonize TCDD- but not BNF-inducible AhR-dependent reporter gene expression in stably transfected rat (H4L1.1c4), and human (HG2L6.1c3) hepatoma cell lines (Table 1). The relative inhibitory potency (IC50) of CH223191 against TCDD-dependent gene induction in each cell line, determined from this concentration-response analysis, was 1.1μM for guinea pig cells, 1.5μM for mouse cells, 3.1μM for rat cells, and 0.2μM for human cells (data not shown). In addition to luciferase, CH223191 preferentially inhibited TCDD-dependent induction of expression of an AhR-responsive green fluorescent protein reporter gene that had been stably integrated into rat hepatoma (H4IIe) cells (data not shown). Accordingly, we expect CH223191 to inhibit expression of other AhR-responsive genes (endogenous and transfected). Taken together, the above results demonstrate that CH223191 is a preferential inhibitor of TCDD-induced AhR-dependent gene expression in various species.
FIG. 2.
Differential antagonistic effects of CH223191 on TCDD- or BNF-induced luciferase reporter gene expression in a stably transfected cell line. (A) Guinea pig (G16L1.1c8) cells were incubated with TCDD (1nM) or BNF (1μM) in the absence or presence of increasing concentrations of CH223191 (0.1–100μM) for 4 h and luciferase activity determined. (B) Mouse hepatoma (H1L1.1c2) cells were incubated with increasing concentrations of TCDD or BNF in the absence or presence of 10μM CH223191 for 4 h at 37°C and luciferase activity determined. Values are expressed as a percentage of the maximal induction of luciferase by 1nM TCDD and represent the mean ± SD of triplicate determinations.
TABLE 1.
The Effect of CH223191 on TCDD- and BNF-Induced AhR-Dependent Luciferase Reporter Gene Expression in Guinea Pig (G16L1.1c8), Mouse (H1L1.1c2), Rat (H4L1.1c4), and Human (HG2L6.1c3) Cell Lines
| Treatment |
Luciferase activity in different cell lines (relative light units) |
||||
| Chemical | CH223191 (10μM) | G16L1.1c8 | H1L1.1c2 | H4L1.1c4 | HG2L6.1c3 |
| DMSO | − | 19 ± 2a | 12 ± 1 | 40 ± 14 | 12 ± 1 |
| + | 4 ± 3 | 3 ± 1 | 17 ± 5 | 9 ± 1 | |
| TCDD (1nM) | − | 81 ± 5 | 545 ± 61 | 4173 ± 555 | 73 ± 12 |
| + | 17 ± 1b | 13 ± 1b | 131 ± 36b | 10 ± 1b | |
| BNF (1μM) | − | 94 ± 7 | 550 ± 64 | 4883 ± 181 | 53 ± 15 |
| + | 86 ± 2 | 511 ± 40 | 4681 ± 679 | 64 ± 14 | |
Values represent the mean ± SD of triplicate determinations of luciferase activity in cells incubated with the indicated chemicals for 4 h at 37°C.
Values are significantly different from the luciferase activity of TCDD-treated cells incubated in the absence of CH223191 at p < 0.01 as determined by the student's t-test.
CH223191 Inhibits TCDD- but Not BNF-Dependent AhR Nuclear Translocation
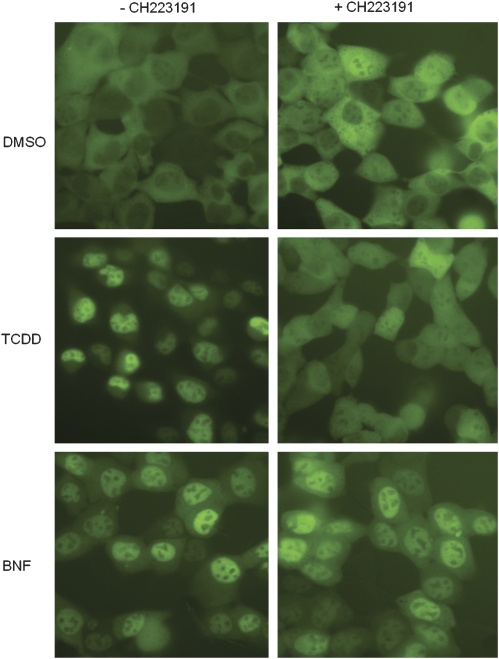
The TCDD-selective inhibition of AhR-dependent gene expression by CH223191 can result from effects on any step in the AhR signaling pathway. To determine whether CH223191 can selectively affect ligand-dependent AhR nuclear accumulation, we examined its effect on BNF- and TCDD-dependent nuclear translocation of a yAhR stably expressed in mouse hepatoma (yAHAYc6) cells. As expected, equally effective concentrations of BNF and TCDD treatment of yAHAYc6 cells resulted in nuclear accumulation of yAhR compared with the DMSO solvent control (Fig. 3, left panels). Co-incubation of yAHAYc6 cells with CH223191 dramatically reduced TCDD-dependent nuclear accumulation of yAhR, with little reduction in BNF-dependent nuclear accumulation of yAhR (Fig. 3, right panels). These results suggest that the CH223191-dependent reduction in TCDD-inducible luciferase activity results from a decrease in AhR nuclear accumulation.
FIG. 3.
CH223191 inhibits TCDD- but not BNF-stimulated AhR nuclear translocation in mouse hepatoma cells. Recombinant mouse hepatoma (yAHAYc6) cells containing a stably transfected yAhR chimera expression vector were treated with DMSO (solvent control), TCDD (1nM), or BNF (1μM) in the absence or presence of 10μM CH223191. After 1 h of incubation at 37°C, YFP fluorescence in the cells was visualized using an Olympus IX71 fluorescence microscope and live cell images were captured and processed as described under the “Materials and Methods” section.
CH223191 Preferentially Inhibits TCDD- but Not BNF-Stimulated In Vitro AhR Transformation and DNA Binding
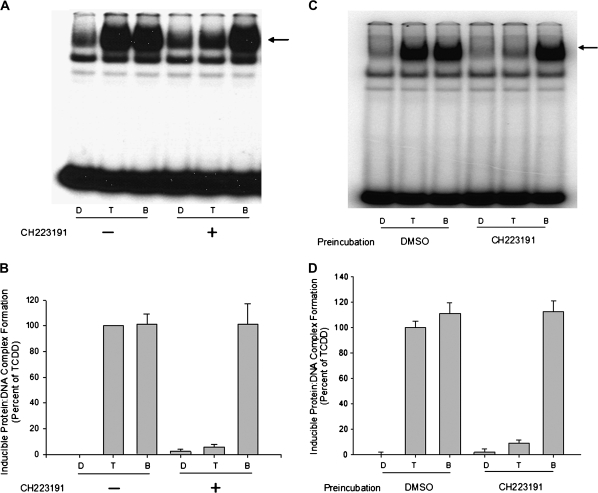
Inhibition of TCDD-dependent nuclear accumulation of the AhR by CH223191 could result from an inhibition of any step in the AhR signaling pathway prior to nuclear translocation of the AhR. We next assessed the ligand-selective inhibitory effect of CH223191 on the ability of TCDD and BNF to stimulate AhR transformation (i.e., ligand-dependent conversion of the AhR into a form that dimerizes with the nuclear AhR nuclear translocator [Arnt] protein) and binding of the liganded AhR:Arnt heterodimer to DNA containing its specific DNA recognition site, the DRE. Whereas incubation of guinea pig hepatic cytosol with maximally inducing concentrations of BNF or TCDD results in AhR transformation into its DNA-binding form, co-incubation with CH223191 inhibited TCDD- but not BNF-dependent AhR transformation/DNA binding (Figs. 4A and 4B).
FIG. 4.
Differential inhibitory effects of CH223191 on TCDD- or BNF-stimulated AhR transformation and DNA binding in vitro. Guinea pig hepatic cytosol (8 mg protein/ml) was (A, B) incubated with DMSO (20 μl/ml), TCDD (20nM), or BNF (2μM) in the absence or presence of 10μM CH223191 for 2 h at 20°C or (C, D) preincubated with 10μM CH223191 for 1 h at 20°C followed by the addition of DMSO (20 μl/ml), TCDD (20nM), or BNF (1μM) and further incubated for 2 h at 20°C. Protein-DNA complexes were resolved by gel retardation analysis (A, C) and the amount of induced protein-DNA complex formation determined by phosphorimager analysis (B, D). Values are expressed as a percent of maximal TCDD-induced protein-DNA complex formation and represent the mean ± SD of triplicate determinations. The arrow indicates the position of the induced protein-DNA (ligand:AhR:Arnt:DRE) complex.
To eliminate the possibility that the lack of antagonism of BNF by CH223191 was not simply due to the higher concentrations of BNF used in the incubations relative to that of TCDD (1μM versus 20nM, respectively), but was the result of ligand-selective antagonism, we examined the effect of CH223191 preincubation on ligand-dependent AhR transformation and DNA binding. In these studies, cytosol was preincubated with CH223191 for 1 h prior to the addition of TCDD or BNF, followed by further incubation for 2 h and measurement of AhR transformation/DNA binding. Similar to the co-incubation results, CH223191 preincubation only inhibited TCDD-dependent AhR transformation and DNA binding (Figs. 4C and 4D). These results, combined with the inability of CH223191 to reduce DNA binding of AhR complexes already transformed by TCDD or BNF (Fig. 5), demonstrated that antagonism by CH223191 results from an inhibition of AhR transformation events rather than affecting DNA binding by TCDD-transformed AhR complex. Increasing the concentration of TCDD in TCDD/CH223191 co-incubation experiments (Supplementary fig. S1) not only led to an increase in TCDD-inducible AhR DNA binding at the highest TCDD concentration (20nM) but also led to an increase in AhR-dependent gene expression (Fig. 2B), consistent with a competitive mechanism of inhibition by CH223191. The ability of CH223191 to competitively reduce [3H]TCDD-specific binding to the mouse AhR (Kim et al., 2006) or to guinea pig hepatic cytosolic AhR (Supplementary fig. S2) also confirmed that CH223191 is a competitive AhR ligand. These results, combined with the lack of any agonist activity of CH223191, demonstrate that CH223191 is a novel TCDD-selective pure AhR antagonist.
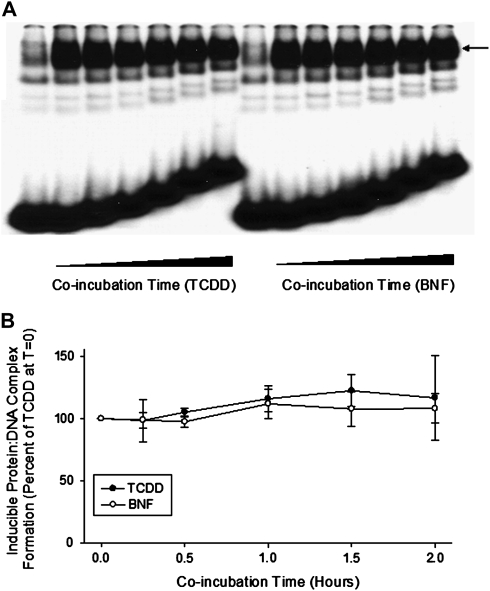
FIG. 5.
CH223191 has no effect on DNA binding of AhR that has already undergone TCDD- or BNF-dependent transformation in vitro. Guinea pig hepatic cytosol (8 mg protein/ml) was incubated with DMSO (20 μl/ml), TCDD (2nM), or BNF (2μM) for 2 h, followed by the addition of CH223191 (to 10μM) and aliquots removed at the indicated times and induced protein-DNA complex formation determined by gel retardation analysis (A) and the amount of induced protein-DNA complex formation determined by phosphorimager analysis (B). Values are expressed as a percent of the maximal TCDD-induced protein-DNA complex formation observed at time “0” and represent the mean ± SD of triplicate determinations. The arrow in (A) indicates the position of the induced protein-DNA (ligand:AhR:Arnt:DRE) complex.
CH223191 Is an HAH-Selective Antagonist of the AhR
The above results demonstrate that CH223191 preferentially antagonizes TCDD compared with that of BNF; however, whether this ligand selectivity extends to other AhR agonists or classes of agonists is unknown. Accordingly, we examined the ability of CH223191 to inhibit AhR-dependent luciferase reporter gene induction by equally effective concentrations of 12 other AhR agonists, including several HAHs (TCDF, PCB77, and PCB126), PAHs (BAA, BKF, and DBA), synthetic flavonoids (ANF, T7, AU1, AJ9, and AY9 [structures shown in Supplementary fig. S3]), and indirubin (Denison and Nagy, 2003; Denison et al., 1998, 2002; Knockaert et al., 2004). CH223191 antagonized only the ability of the HAHs, but not the non-HAH agonists (PAHs, flavonoids, or indirubin), to stimulate AhR-dependent gene expression in rat hepatoma cells (Table 2 and Supplementary fig. S3). Interestingly, induction of luciferase activity by AY9, a dihalogen-substituted flavonoid, was inhibited by 50% when co-incubated with 10μM CH223191 (Table 2), and this inhibition occurred in a CH223191 concentration-dependent manner (Supplementary fig. S3). Overall, the results of our analysis of 14 selected AhR agonists are consistent with the hypothesis that CH223191 is an HAH-selective AhR antagonist.
TABLE 2.
The Effect of CH223191 on AhR-Dependent Reporter Gene Expression Induced by Equally Effective Inducing Concentrations Selected AhR Agonists in Rat Hepatoma (H4L1.1c4) Cells
| AhR agonist |
Luciferase activity (percent of 1nM TCDD) |
|||
| Class | Chemical | Concentration | − CH223191 | + CH223191 |
| HAHs | TCDD | 1nM | 100 ± 6a | 3 ± 1b |
| TCDF | 10nM | 104 ± 1 | 4 ± 1b | |
| PCB126 | 10nM | 96 ± 5 | 2 ± 2b | |
| PCB77 | 10nM | 86 ± 2 | 1 ± 1b | |
| PAHs | BAA | 1μM | 100 ± 4 | 102 ± 4 |
| BKF | 100nM | 100 ± 1 | 99 ± 1 | |
| DBA | 100nM | 94 ± 7 | 102 ± 2 | |
| Flavonoids | BNF | 1μM | 114 ± 3 | 122 ± 5 |
| T7 | 1μM | 98 ± 4 | 95 ± 2 | |
| AU1 | 1μM | 93 ± 11 | 106 ± 3 | |
| ANF | 10μM | 105 ± 4 | 129 ± 4 | |
| AJ9 | 1μM | 76 ± 4 | 83 ± 1 | |
| AY9 | 1μM | 107 ± 1 | 52 ± 4b | |
| Other | Indirubin | 1μM | 76 ± 2 | 81 ± 4 |
Values represent the mean ± SD of triplicate determinations of luciferase activity expressed as a percent of that induced by 1nM TCDD.
Values are significantly reduced in the presence of 10μM CH223191 at p < 0.01 as determined by the student's t-test.
TMF and MNF Exhibit Species-Specific, HAH-Selective AhR Antagonist Activity
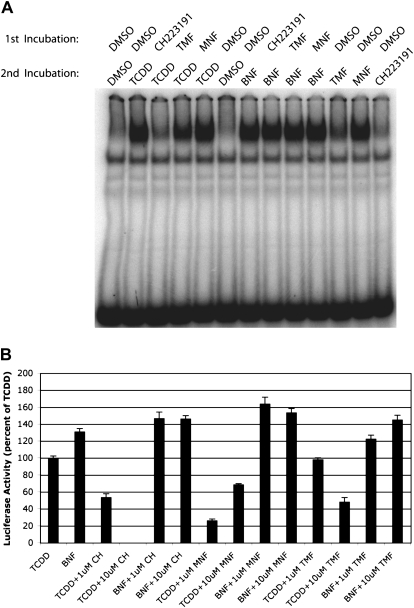
The results of the above analyses demonstrate the ligand selectivity of CH223191; however, few studies have directly compared the ligand selectivity of other AhR antagonists in the same experimental system. To determine whether CH223191 is unique in its ligand selectivity, we examined the ability of two previously reported pure AhR flavonoid antagonists TMF and MNF (Lu et al., 1995; Murray et al., 2010a) to inhibit TCDD- and BNF-dependent transformation and DNA binding of guinea pig hepatic cytosol AhR using the antagonist preincubation approach described for the CH223191 studies in Figure 4. Although TMF reduced the ability of TCDD to stimulate AhR DNA binding, MNF had little effect; CH223191 completely inhibited TCDD-dependent AhR DNA binding (Fig. 6A). The ability of MNF itself to stimulate AhR DNA binding indicates that it is a full agonist in this experimental system. TMF, like CH223191, failed to inhibit the ability of BNF to stimulate AhR DNA binding, and thus it exhibits some ligand specificity in its antagonist activity. The action of these antagonists on AhR-dependent gene expression was also examined by determining their effect on the induction of luciferase activity in recombinant mouse hepatoma (H1L1.1c2) cells co-incubated with TCDD (1nM) or BNF (1μM) in the absence or presence of 1 or 10μM of antagonist (CH223191, MNF, or TMF) for 4 h (Fig. 6B). Although these results revealed antagonism of TCDD induction of luciferase gene induction by CH223191, TMF, and MNF (the higher levels of induction at 10μM MNF result from its partial agonist activity; Henry and Gasiewicz, 2008; Zhou and Gasiewicz, 2003), all three antagonists did not inhibit BNF-dependent luciferase gene induction in these cells but actually enhanced it to a small degree. The observed species differences in agonist/antagonist activity of MNF observed between the rat (gene induction) and guinea pig (DNA binding) AhR bioassays are consistent with those previously reported differences in MNF effects on mouse (antagonist activity) and guinea pig (agonist activity) AhR activity that were shown to result from a single amino acid difference in the LBD of the AhR of these species (Henry and Gasiewicz, 2008). Together, the above results indicate the ability of other AhR antagonists to also exhibit ligand selectivity.
FIG. 6.
Differential inhibitory effects of the AhR antagonists TMF and MNF on TCDD- or BNF-stimulated AhR DNA binding and gene expression. (A) Guinea pig hepatic cytosol (8 mg protein/ml) was preincubated with DMSO (20 μl/ml), 10μM CH223191, 10μM TMF, or 1μM MNF for 1 h at 20°C followed by the addition of DMSO (20 μl/ml), TCDD (20nM), or BNF (1μM) and further incubation for 2 h at 20°C. Protein-DNA complexes were resolved by gel retardation analysis as described in the “Materials and Methods” section. (B) Recombinant rat hepatoma (H4L1.1c4) cells were incubated with 1nM TCDD or 1μM BNF in the absence or presence of DMSO, of the indicated concentration of CH223191, MNF, or TMF for 4 h at 37°C and luciferase activity determined. Values are expressed as a percentage of the maximal induction of luciferase by 1nM TCDD and represent the mean ± SD of triplicate determinations.
DISCUSSION
Activation of the AhR can result in a wide variety of toxic and biological effects, and the specific spectrum of observed effects is ligand dependent, with metabolically stable ligands (HAHs) producing the prototypical spectrum of TCDD-like AhR-dependent toxicity (Bradshaw and Bell, 2009; Denison et al., 1998; Furness and Whelan, 2009; Poland and Glover, 1980; Poland and Knutson, 1982; Safe, 1990). More recently, the AhR has been shown to bind and be activated by an extremely wide variety of structurally diverse exogenous and endogenous chemicals that have little similarity to the prototypical HAH/PAH AhR ligands (Bradshaw and Bell, 2009; Denison and Heath-Pagliuso, 1998; Denison et al., 1998; Nguyen and Bradfield, 2008). Attempts to model all known AhR agonists by structure-activity relationship approaches have been unsuccessful and are complicated by the dramatic diversity in AhR ligand structure and physiochemical properties (Denison et al., 1998; Petko et al., 2010). Although the reported diversity in AhR ligand structure has continued to rapidly expand, our understanding of the molecular mechanisms responsible for the binding by and activation of the AhR by these structurally diverse ligands remains to be elucidated.
CH223191 exerts its antagonist effect against TCDD through its ability to act as a competitive ligand, and this is demonstrated through both ligand-binding analysis (Kim et al., 2006; Supplementary fig. 2) and the increase in agonist activity (i.e., AhR-dependent DNA binding and gene expression) that occurs with increasing concentrations of TCDD in the presence of a constant concentration of CH223191 (Fig. 2B and Supplementary fig. 1). Although the competitive nature of CH223191 binding to the AhR initially suggested to us that the lack of inhibitory effect on BNF or other non-HAH AhR ligands might simply be due to the fact that these ligands are used at a significantly higher concentration than that of TCDD or other HAHs, due to their lower relative affinity/potency, our results indicate that this is not the case. The preincubation experiments (Figs. 4C and 4D) indicated that even though CH223191 is bound to the AhR/AhR LBD (supported by its ability to block binding and activation by TCDD), BNF can still bind and stimulate AhR transformation and DNA binding. Thus, binding of CH223191 must occur with residues that do not affect the ability of non-HAH ligands to bind and stimulate AhR transformation and DNA binding. Although CH223191 can partially inhibit BNF-dependent AhR activation, the degree of inhibition (< 30%) was dramatically less than that observed with TCDD (Fig. 2). When BNF or other non-HAH agonists were used at concentrations that are equally effective as that of a maximally inducing concentration of TCDD, no inhibition was observed. Additionally, when we consider the significantly lower inducing potency of BNF compared with TCDD (a 1000-fold more BNF is required for maximal AhR-dependent gene expression [compare equally effective inducing concentrations of 1μM BNF with 1nM TCDD]) and that a 10,000-fold molar excess of CH223191 (10μM) completely antagonized induction by 1nM TCDD (Fig. 2), if both TCDD and BNF were interacting with the AhR identically, then 10μM CH223191 should have produced significant inhibition of gene induction by 1μM BNF, but this was not observed. The ability of 10μM CH223191 to significantly inhibit induction by the dihalogen-substituted flavonoid AY9 not only indicates that this concentration of CH223191 is sufficient to inhibit the induction by 1μM of AY9, but considering the above results, CH223191 appears to have a preference for halogen-substituted AhR agonists.
Similar to CH223191, we observed that the flavonoid antagonists TMF and MNF also exhibited ligand-specific antagonism, suggesting that all three compounds can exert antagonism through a common mechanism. Although several reports have indicated that these antagonists can inhibit the activity of some non-HAH AhR agonists (Goergens et al., 2009; Han et al., 2009; Murray et al., 2010a; van Grevenynghe et al., 2005; Veldhoen et al., 2009), the high degree of variability in the potency and efficacy of a specific AhR antagonist between experimental systems suggests that AhR antagonist activity occurs in a ligand-, species-, and context-specific manner. Accordingly, accurate comparative analysis of the mechanism of action of AhR antagonists requires their analysis in a common experimental system. The preferential antagonism of HAHs by CH223191, TMF, and MNF reported here is consistent with the hypothesis that there are significant differences in the binding of the HAHs and non-HAH AhR agonists within the ligand-binding pocket, and they reveal two distinct classes of AhR agonists (based on their response to CH223191). Although it is clear from previous [3H]TCDD competitive binding studies that HAH and non-HAH AhR ligands can directly compete with each other for binding to the AhR ligand–binding pocket (reviewed in Denison et al., 1998), these antagonists must interact with the AhR in such a way to preferentially inhibit the binding of HAHs, but not non-HAH ligands, to the AhR LBD. Alternatively, it is possible that CH223191 and other AhR antagonists could act as a selective AhR modulator (SAhRM) in that their binding to the AhR LBD could alter the structure of the protein and/or LBD in such a way to exclude some ligands but not others. This is supported by previous studies demonstrating ligand-specific differences in steroid hormone receptor structure (Connor et al., 2001; De Bosscher, 2010; Dusell et al., 2008; Ellmann et al., 2009; Kazmin et al., 2006). Alternatively, CH223191 could bind to the AhR outside of the LBD and act as an allosteric modulator, thereby affecting overall AhR structure that leads to preferential inhibition of HAH access/binding to the LBD. Mechanistically, it is not clear how CH233191 or other AhR antagonists specifically inhibit the binding of TCDD and dioxin-like HAHs, and given the lack of a 3D crystal/NMR structure of the AhR ligand–binding pocket, the exact nature of AhR ligand interactions within the binding pocket remain to be established. However, our recently developed homology model of the mouse AhR LBD (Pandini et al., 2007, 2009) has allowed us to identify the specific amino acid residues whose side chains lie within or are adjacent to the ligand-binding pocket and which are candidate residues for these differential interactions. We are currently using site-directed mutagenesis approaches to specifically identify the amino acids within the AhR LBD involved in agonist- and antagonist-specific interactions. However, the halogenated side chains of the HAHs appear to play a key role in this differential ligand binding. This is supported by the observed inhibition of induction of AhR-dependent gene expression by the only flavonoid (AY9) that contains multiple halogens in its structure and suggests that AY9 might actually bind within the AhR LBD in a manner more similar to that of HAHs than the non-HAH AhR ligands. This could suggest that the residue(s) targeted by CH223191 may be involved in preferential interactions with halogens present on these ligands.
Differential binding of structurally diverse ligands within the AhR LBD is also indirectly supported by previous mutagenesis/ligand-binding analysis studies (Backlund and Ingelman-Sundberg, 2004; Goryo et al., 2007; Whelan et al., 2010). Mutation of a conserved tyrosine residue into phenylalanine (Y320F) in the human AhR LBD (comparable with residue 316 in the mouse AhR) reportedly resulted in the selective loss of binding and/or activation by several low-affinity non-HAH AhR ligands (i.e., 2-mercapto-5-methoxybenzimidazole, primaquine, and omeprazole) but not by the high-affinity ligand TCDD (Backlund and Ingelman-Sundberg, 2004). In contrast, insertion of a single mutation in a closely associated position into the mouse AhR LBD (F318L) was reported to produce a receptor that could be activated by the PAH ligand 3-methylcholanthrene but not by BNF or TCDD (Goryo et al., 2007). Based on our mouse AhR LBD homology model (Pandini et al., 2007, 2009), the mutations F318L and Y316F (Y320 in the human AhR) both reside in the same small alpha helix (Eα) present at the top of the ligand-binding cavity. Interestingly, whereas the loss of the aromatic group in the side chain with the Y318L mutation eliminates binding by TCDD/BNF (Goryo et al., 2007), the Y316F mutation, which retains an aromatic ring in this position, eliminates binding by low-affinity non-HAH ligands but not TCDD (Backlund and Ingelman-Sundberg, 2004). These results, combined with our previous mutagenesis results (Pandini et al., 2007, 2009), indicate that the presence of an aromatic residue in these positions is critical for AhR ligand binding/functional activity and that changes in the specific residues within this helix differentially affect binding of HAH and non-HAH ligands. Interestingly, whereas mutation of histidine 285 to alanine in the LBD eliminates binding and activation of the AhR by TCDD, mutation to phenylalanine only reduced the potency/affinity of TCDD (Pandini et al., 2009). In contrast to the above mutations, histidine 285 is contained within a central strand of the beta sheet (Aβ) of the LBD with its side chain pointing into the center of the binding cavity, and previous studies (Pandini et al., 2009) suggest that it plays a role in stabilizing TCDD binding through aromatic interactions. The results of Whelan et al. (2010) indicate that the presence of a tyrosine residue in this position allowed binding and activation of the AhR by the novel agonist YH439 but not TCDD, consistent with differences in the ability of these two agonists to bind within the ligand-binding pocket and demonstrating the key nature of the aromatic side chain of this amino acid in binding specificity. Although the effect of the specific mutations described above on the overall 3D structure of the AhR LBD and whether they result in an altered conformation of the LBD that contributes to and/or is responsible for these observed differential ligand-specific effects is not known. However, these studies are consistent with the results presented here that support differential binding by ligands or classes of ligands within the AhR ligand–binding pocket, and these differences could contribute to the observed structural promiscuity of AhR ligands. Future mutagenesis, protein modeling, and docking analysis using the AhR LBD model will provide further insights into these unique aspects of AhR ligand binding.
The structural diversity and differential binding of AhR ligands also suggest the existence of selective modulators of the AhR, similar to that reported for nuclear steroid hormone receptor. Previous studies have shown that the functional activity of nuclear hormone receptors can be altered in a ligand-selective manner, and these functional changes appear to be directly related to ligand-specific changes in the overall structure of the receptor and consequently impact the specific proteins (i.e., coactivators) to which it interacts (Connor et al., 2001; De Bosscher, 2010; Dusell et al., 2008; Ellmann et al., 2009; Kazmin et al., 2006). Similarly, given the ligand promiscuity of the AhR and differences in ligand binding, one can envision the existence of ligand-specific differences in AhR structure and function resulting from specific differences in the binding of ligands to the AhR (i.e., the existence of SAhRMs). In fact, several SAhRMs have already been identified based on their ability to selectively produce some AhR-dependent responses and not others (i.e., inhibition of inflammatory gene expression without induction of CYP1A1 [Murray et al., 2010b] and ligand-selective inhibition of estrogen receptor action [Zhang et al., 2008, 2009]). Like that observed for selective modulators of nuclear hormone receptors, ligand-specific differences in AhR co-activator recruitment have been observed (Zhang et al., 2008) and likely result from ligand-specific differences in AhR structure. Additionally, ligand-dependent differences in nucleotide-specific DNA binding of the AhR have been reported (Gouédard et al., 2004; Matikainen et al., 2001), and this could contribute to the observed diversity in ligand-specific AhR responses. Given these aspects, one can envision dramatic differences in the overall spectrum of AhR-depended responses observed in a given cell type or in vivo that is entirely driven by the specific ligand and its mechanism of interaction with the AhR. This could contribute to the wide spectrum of AhR-dependent toxic and biological effects observed following exposure to various AhR agonists.
Although metabolically persistent AhR ligands, such as that of TCDD and related dl-HAHs, can produce adverse AhR-dependent responses (Denison and Heath-Pagliuso, 1998; Denison and Nagy, 2003; Denison et al., 1998; Safe, 1990), the demonstration of a critical role for ligand-dependent activation of the AhR in aspects of immune function (Esser et al., 2009; Marshall and Kerkvliet, 2010) and tissue-specific developmental effects (Bradshaw and Bell, 2009; Furness and Whelan, 2009), combined with documented effects of AhR ligands on cell cycle, inflammation, and cancer cell proliferation, support the potential therapeutic application for activators/inhibitors of the AhR/AhR signaling pathway (Bradshaw and Bell, 2009; DuSell et al., 2010; Furness and Whelan, 2009; Hall et al., 2010). Although a variety of AhR antagonists have been identified, including ANF, resveratrol, curcumin, luteolin, and others (Bradshaw and Bell, 2009; Denison and Nagy, 2003; Denison et al., 1998, 2002), the partial agonist activity associated with these compounds complicate clear analysis of their inhibitory effects. Only three pure AhR antagonists have been reported, namely CH223191 (Kim et al., 2006) and the synthetic flavonoids TMF (Murray et al., 2010a) and MNF (Lu et al., 1995). Whereas TMF can antagonize HAH and non-HAH AhR agonist activity in human cells and cytosol in vitro (Murray et al., 2010a), only CH223191 has been shown to be a potent antagonist of TCDD both in vitro and in vivo and it can inhibit TCDD-dependent toxicity in vivo (Kim et al., 2006). The agonist activity of MNF is unique in that it occurs in a species- and context-specific manner (Henry and Gasiewicz, 2008; Lu et al., 1995; Zhou and Gasiewicz, 2003). Accordingly, CH223191 might be a useful therapeutic agent itself and/or a unique lead compound for the development of more potent antagonists of AhR-dependent toxic effects produced by TCDD and related dl-HAHs. Preliminary structure-activity analysis suggests that the HAH antagonistic activity of CH223191 requires the 1-methyl-1H-pyrazole-5-carboxamide portion of the molecule because its removal results in a compound (o-aminoazotoluene) with relative potent agonist activity (data not shown). Further structure-activity relationship analysis of CH223191 and its derivatives will provide important insights into aspects of the molecular responsible for its selective inhibitory function and AhR ligand–binding specificity. Although it remains to be determined whether CH223191 will inhibit the binding and activation of the AhR by endogenous compounds, we envision that the endogenous ligands are more likely to bind to the AhR in a manner more similar to the non-HAH AhR agonists and expect that they would not be inhibited by CH223191. Examination of the ability of CH223191 to inhibit the toxic/biological effects of TCDD without antagonizing endogenous AhR-dependent developmental effects would be important to determine in future studies. Overall, we have identified and characterized a novel AhR antagonist that has not only revealed significant differences in the binding of HAHs and non-HAHs to the AhR, but this reagent will allow for a more in-depth analysis of AhR ligand–binding specificity, the effects of TCDD, and AhR biology.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Environmental Health Sciences of the National Institutes of Health (ES012498, ES007685); a Superfund Research Grant (ES004699); startup funds from the 100 Talents Program of the Chinese Academy of Sciences; the California Agricultural Experiment Station and the American taxpayers.
Supplementary Material
Acknowledgments
We thank Dr Steven Safe for the TCDD, [3H]TCDD, and MNF, Dr Gary Perdew for the TMF, Dr Mike Nantz (University of Louisville) for the flavonoids, and Christopher Dicus (University of California, Davis) for the NMR analysis of CH223191 structure.
References
- Backlund M, Ingelman-Sundberg M. Different structural requirements of the ligand binding domain of the aryl hydrocarbon receptor for high- and low-affinity ligand binding and receptor activation. Mol. Pharmacol. 2004;65:416–425. doi: 10.1124/mol.65.2.416. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw TD, Bell DR. Relevance of the aryl hydrocarbon receptor (AhR) for clinical toxicology. Clin. Toxicol. 2009;47:632–642. doi: 10.1080/15563650903140423. [DOI] [PubMed] [Google Scholar]
- Connor CE, Norris JD, Broadwater G, Willson TM, Gottardis MM, Dewhirst MW, McDonnell DP. Circumventing tamoxifen resistance in breast cancer cells using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer Res. 2001;61:2917–2922. [PubMed] [Google Scholar]
- De Bosscher K. Selective glucocorticoid receptor modulators. J. Steroid Biochem. Mol. Biol. 2010;120:96–104. doi: 10.1016/j.jsbmb.2010.02.027. [DOI] [PubMed] [Google Scholar]
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull. Environ. Contam. Toxicol. 1998;61:557–568. doi: 10.1007/pl00002973. [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Denison MS, Pandini A, Nagy SR, Baldwin EP, Bonati L. Ligand binding and activation of the Ah receptor. Chem. Biol. Int. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- Denison MS, Rogers JM, Rushing SR, Jones CL, Tetangco SC, Heath-Pagliuso S. Analysis of the Ah receptor signal transduction pathway. In: Maines M, Costa LG, Reed DJ, Sassa S, Sipes IG, editors. Current Protocols in Toxicology. New York, NY: John Wiley and Sons; 2002. pp. 4.8.1–4.8.45. [DOI] [PubMed] [Google Scholar]
- Denison MS, Seidel SD, Rogers WJ, Ziccardi M, Winter GM, Heath-Pagliuso S. Natural and synthetic ligands for the Ah receptor. In: Puga A, Wallace KB, editors. Molecular Biology Approaches to Toxicology. Philadelphia, PA: Taylor & Francis; 1998. pp. 393–410. [Google Scholar]
- DuSell CD, Nelson ER, Wittmann BM, Fretz JA, Thomas RS, Pike JW, McDonnell DP. Regulation of aryl hydrocarbon receptor function by selective estrogen receptor modulators. Mol. Endocrinol. 2010;24:33–46. doi: 10.1210/me.2009-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuSell CD, Umetani M, Shaul PW, Manglesdorf DJ, McDonnell DP. 27-Hydroxycholesterol is an endogenous selective estrogen receptor modulator. Mol. Endocrinol. 2008;22:65–77. doi: 10.1210/me.2007-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmann S, Sticht H, Thiel F, Beckmann MW, Strick R, Strissel PL. Estrogen and progesterone receptor: from molecular structures to clinical targets. Cell Mol. Life Sci. 2009;66:2405–2426. doi: 10.1007/s00018-009-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30:447–454. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Furness SG, Whelan F. The pleiotropy of dioxin toxicity—xenobiotic misappropriation of the aryl hydrocarbon receptor's alternative physiological roles. Pharmacol. Ther. 2009;124:336–354. doi: 10.1016/j.pharmthera.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Garrison PM, Tullis K, Aarts JM, Brouwer A, Giesy JP, Denison MS. Species-specific recombinant cell lines as bioassay systems for the detection of 2,3,7,8-tetrachlorodibenzo-p-dioxin-like chemicals. Fundam. Appl. Toxicol. 1996;30:194–203. doi: 10.1006/faat.1996.0056. [DOI] [PubMed] [Google Scholar]
- Ghosh U, Ganessunker D, Sattigeri VJ, Carlson KE, Mortensen DJ, Katzenellenbogen BS, Katzenellenbogen JA. Estrogenic diazenes: heterocyclic non-steroidal estrogens of unusual structure with selectivity for estrogen receptor subtypes. Bioorg. Med. Chem. 2003;11:629–657. doi: 10.1016/s0968-0896(02)00309-7. [DOI] [PubMed] [Google Scholar]
- Goergens A, Frericks M, Esser C. The aryl hydrocarbon receptor is only marginally involved in the antileukemic effects of its ligand curcumin. Anticancer Res. 2009;29:4657–4664. [PubMed] [Google Scholar]
- Goryo K, Suzuki A, Del Carpio CA, Siizaki K, Kuriyama E, Mikami Y, Kinoshita K, Yasumoto K, Rannug A, Miyamoto A, et al. Identification of amino aid residues in the Ah receptor involved in ligand binding. Biochem. Biophys. Res. Comm. 2007;354:396–402. doi: 10.1016/j.bbrc.2006.12.227. [DOI] [PubMed] [Google Scholar]
- Gouédard C, Barouki R, Morel Y. Dietary polyphenols increase paraoxonase 1 gene expression by an aryl hydrocarbon receptor-dependent mechanism. Mol. Cell. Biol. 2004;24:5209–5222. doi: 10.1128/MCB.24.12.5209-5222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Barhoover MA, Kazmin D, McDonnell DP, Greenlee WF, Thomas RS. Activation of the aryl hydrocarbon receptor inhibits invasive and metastatic feature of human breast cancer cells and promotes breast cancer cell differentiation. Mol. Endocrinol. 2010;24:359–369. doi: 10.1210/me.2009-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Nagy SR, Denison MS. Comparison of recombinant cell bioassays for the detection of Ah receptor agonists. Biofactors. 2004;20:11–22. doi: 10.1002/biof.5520200102. [DOI] [PubMed] [Google Scholar]
- Han EH, Kim HG, Im JH, Jeong TC, Jeong HG. Up-regulation of CYP1A1 by rutaecarpine is dependent on aryl hydrocarbon receptor and calcium. Toxicology. 2009;266:38–47. doi: 10.1016/j.tox.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Henry EC, Gasiewicz TA. Molecular determinants of species-specific agonist and antagonist activity of a substituted flavone towards the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 2008;472:77–88. doi: 10.1016/j.abb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmin D, Prytkova T, Cook CE, Wolfinger R, Chu TM, Beratan D, Norris JD, Chang CY, McDonnell DP. Linking ligand-induced alterations in androgen receptor structure to differential gene expression: a first step in the rational design of selective androgen receptor modulators. Mol. Endocrinol. 2006;20:1201–1207. doi: 10.1210/me.2005-0309. [DOI] [PubMed] [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell. Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Kim SH, Henry EC, Kim DK, Kim YH, Shin KJ, Han MS, Lee TG, Kang JK, Gasiewicz TA, Ryu SH, et al. Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-4-o-tolylazo-phenyl)amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol. Pharmacol. 2006;69:1871–1878. doi: 10.1124/mol.105.021832. [DOI] [PubMed] [Google Scholar]
- Knockaert M, Blondel M, Bach S, Leost M, Elbi C, Hager GL, Nagy SR, Han D, Denison M, French M, et al. Independent actions on cyclin-dependent kinases and aryl hydrocarbon receptor mediate the antiproliferative effects of indirubins. Oncogene. 2004;23:4400–4412. doi: 10.1038/sj.onc.1207535. [DOI] [PubMed] [Google Scholar]
- Lu YF, Santostefano M, Cunningham BD, Threadgill MD, Safe S. Identification of 3′-methoxy-4′-nitroflavone as a pure aryl hydrocarbon (Ah) receptor antagonist and evidence for more than one form of the nuclear Ah receptor in MCF-7 human breast cancer cells. Arch. Biochem. Biophys. 1995;316:470–477. doi: 10.1006/abbi.1995.1062. [DOI] [PubMed] [Google Scholar]
- Ma Q. Induction of CYP1A1. The AhR/DRE paradigm: transcription, receptor regulation, and expanding biological roles. Curr. Drug Metabol. 2001;2:149–164. doi: 10.2174/1389200013338603. [DOI] [PubMed] [Google Scholar]
- Marshall NB, Kerkvliet NI. Dioxin and immune regulation: emerging role of aryl hydrocarbon receptor in the generation of regulatory T cells. Ann. N. Y. Acad. Sci. 2010;1183:25–37. doi: 10.1111/j.1749-6632.2009.05125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu HY, Laine J, Sakai T, Korsmeyer SJ, Casper RF, et al. Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat. Genet. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- Murray IA, Flaveny CA, DiNatale BC, Chairo CR, Schroeder JC, Kusnadi A, Perdew GH. Antagonism of aryl hydrocarbon receptor signalling by 6,2′,4′-trimethoxyflavone. J. Pharmacol. Exp. Ther. 2010a;332:135–144. doi: 10.1124/jpet.109.158261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Morales JL, Flaveny CA, Dinatale BC, Chiaro C, Gowdahalli K, Amin S, Perdew GH. Evidence for ligand-mediated selective modulation of aryl hydrocarbon receptor activity. Mol. Pharmacol. 2010b;77:247–254. doi: 10.1124/mol.109.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan CH, Beglov D, Rudnitskaya AN, Kozakov D, Waxman DJ, Vajda S. The structural basis of pregnane X receptor binding promiscuity. Biochemistry. 2009;48:11572–11581. doi: 10.1021/bi901578n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 2008;21:102–106. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy N. Ligand specificity of nuclear hormone receptors: sifting through promiscuity. Biochemistry. 2007;46:13461–13467. doi: 10.1021/bi7018699. [DOI] [PubMed] [Google Scholar]
- Pandini A, Denison MS, Song Y, Soshilov AA, Bonati L. Structural and functional characterization of the Aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis. Biochemistry. 2007;46:696–708. doi: 10.1021/bi061460t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandini A, Soshilov AA, Song Y, Zhao J, Bonati L, Denison MS. Detection of the TCDD binding fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis. Biochemistry. 2009;48:5972–5983. doi: 10.1021/bi900259z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petko PI, Rowlands JC, Budinsky R, Zhao B, Denison MS, Mekenyan O. Mechanism-based common reactivity pattern (COREPA) modelling of aryl hydrocarbon receptor binding affinity. SAR QSAR Environ. Res. 2010;21:187–214. doi: 10.1080/10629360903570933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Glover E. 2,3,7,8-Tetrachlorodibenzo-p-dioxin: segregation of toxicity with the Ah locus. Mol. Pharmacol. 1980;17:86–94. [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit. Rev. Toxicol. 1990;21:51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- Springsteel MF, Galietta LJ, Ma T, By K, Berger GO, Yang H, Dicus CW, Choung W, Quan C, Shelat AA, et al. Benzoflavone activators of the cystic fibrosis transmembrane conductance regulator: towards a pharmacophore model for the nucleotide-binding domain. Bioorg. Med. Chem. 2003;11:4113–4120. doi: 10.1016/s0968-0896(03)00435-8. [DOI] [PubMed] [Google Scholar]
- van Grevenynghe J, Bernard M, Langouet S, Le Berre C, Fest T, Fardel O. Human CD34-positive hematopoietic stem cells constitute targets for carcinogenic polycyclic aromatic hydrocarbons. J. Pharmacol. Exp. Ther. 2005;314:693–702. doi: 10.1124/jpet.105.084780. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture media are essential for optimal differentiation of Th17 T cells. J. Exp. Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan F, Hao N, Furness SG, Whitelaw ML, Chapman-Smith A. Amino acid substitutions in the aryl hydrocarbon receptor (AhR) ligand binding domain reveal YH439 as a atypical AhR activator. Mol. Pharmacol. 2010;77:1037–1046. doi: 10.1124/mol.109.062927. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lei P, Liu X, Li X, Walker K, Kotha L, Rowlands C, Safe S. The aryl hydrocarbon receptor as a target for estrogen receptor-negative breast cancer chemotherapy. Endocr. Relat. Cancer. 2009;16:835–844. doi: 10.1677/ERC-09-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Rowlands C, Safe S. Ligand-dependent interactions of the Ah receptor with coactivators in a mammalian two-hybrid assay. Toxicol. Appl. Pharmacol. 2008;227:196–206. doi: 10.1016/j.taap.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gasiewicz TA. 3′-Methyl-4′-nitroflavone, a reported aryl hydrocarbon receptor antagonist, enhances Cyp1a1 transcription by a dioxin response element-dependent mechanism. Arch. Biochem. Biophys. 2003;416:68–80. doi: 10.1016/s0003-9861(03)00274-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.