Abstract
Brazil is the largest producer and consumer of common bean (Phaseolus vulgaris L.), which is the most important source of human dietary protein in that country. This study assessed the genetic diversity and the structure of a sample of 279 geo-referenced common bean landraces from Brazil, using molecular markers. Sixty-seven microsatellite markers spread over the 11 linkage groups of the common bean genome, as well as Phaseolin, PvTFL1y, APA and four SCAR markers were used. As expected, the sample showed lower genetic diversity compared to the diversity in the primary center of diversification. Andean and Mesoamerican gene pools were both present but the latter gene pool was four times more frequent than the former. The two gene pools could be clearly distinguished; limited admixture was observed between these groups. The Mesoamerican group consisted of two sub-populations, with a high level of admixture between them leading to a large proportion of stabilized hybrids not observed in the centers of domestication. Thus, Brazil can be considered a secondary center of diversification of common bean. A high degree of genome-wide multilocus associations even among unlinked loci was observed, confirming the high level of structure in the sample and suggesting that association mapping should be conducted in separate Andean and Mesoamerican Brazilian samples.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1350-5) contains supplementary material, which is available to authorized users.
Introduction
Common bean (Phaseolus vulgaris L.) is the most important source of protein in the Brazilian diet. In combination with rice, the crop makes up the basic daily meal for most Brazilians throughout the country. Brazil has ranked, over the last decades, as the largest producer of common bean in the world and also as the main consumer (Yokoyama and Stone 2000). Although Brazil is not the primary center of diversity of common bean, it is expected that a large diversity of the domesticated gene pool is represented in Brazil, considering the history of the crop’s cultivation in the country. Common bean was cultivated by Brazilian native populations before the European conquest, based on historical reports (Hoehne 1937) and archeological data (Freitas 2006). The process of assimilation of common bean cultivation into the new culture—the one resulting after the European conquest—has not been studied extensively. However, it is recognized that the cultivation of the crop became very popular in most of the regions within the country, in a diversity of environments, and predominantly in smallholder-farmer systems (Vieira 1988; Borém and Carneiro 1998). Vieira (1972) reported there were several hundreds of common bean landraces that were cultivated in Brazil in this period, emphasizing the importance of such reservoir (predominantly, landraces) as sources of resistance to diseases. Common bean cropping systems are until now also very diverse in Brazil (e.g., single crop vs. in association; different levels of inputs, etc.) (Vieira 1988; Borém and Carneiro 1998).
Common bean is a diploid (2n = 2x = 22) and predominantly selfing species, with average outcrossing rates estimated at under 3% (Ramalho and Abreu 2006) although occasionally higher values are obtained (Ibarra-Pérez et al. 1997). The species has been domesticated independently in Mesoamerica and the southern Andes, based on several kinds of data [distribution of wild populations, archeological remains, historical texts, and evolutionary studies based on several types of molecular markers, including phaseolin (the major seed protein in P. vulgaris] (reviewed in Gepts 1998; Gepts et al. 2008; McClean et al. 2008; Kwak et al. 2009). This species occurs in its wild form in different countries of Latin America, from the northern region of Mexico up to Northeastern Argentina, but not in Brazil (Debouck 1986). As a result of this bi-centric process of domestication, the domesticated common bean presents two distinct major gene pools, an Andean and a Mesoamerican one (Gepts 1998; Gepts et al. 2008). The distinction between those two gene pools is usually very clear in common bean collections, either by different kinds of molecular data (Gepts 1988; Koenig and Gepts 1989; Emydgio et al. 2003; Pallottini et al. 2004) or by morphological characters (Singh et al. 1991). These two gene pools are also separated by partial reproductive isolation, both in wild and domesticated populations (Gepts and Bliss 1985; Koinange and Gepts 1992), which leads to hybrid weakness in the F1 (Gepts and Bliss 1985) and later generations (Singh and Molina 1996).
The first attempts to study the organization of diversity of Brazilian common bean landraces used electrophoretic types of phaseolin seed protein (Gepts et al. 1988; Pereira and Souza 1992). These studies showed that the majority of market classes among domesticated beans had an ‘S’ type, characteristic of the Mesoamerican gene pool, while other classes showed the ‘T’ type, characteristic of the Andean gene pool. Later on, other studies assessed the genetic diversity of common bean landraces from the Southern region of Brazil with RAPD (Maciel et al. 2001) and AFLP markers (Maciel et al. 2003). The studies of Maciel et al. (2001, 2003) confirmed the overall distinction between Andean and Mesoamerican accessions of domesticated P. vulgaris in Brazilian samples. However, the distinction between the two major gene pools was not as clear in the study of Maciel et al. (2003), in which some of the landraces showed a ‘T’ phaseolin type but clustered in the Mesoamerican group, suggesting some admixture between these gene pools. Maciel et al. (2003) also identified a larger diversity within the landraces stratum than within the commercial cultivar group, emphasizing the importance of the landraces as sources of genetic variation for common bean in Brazil. Fonseca and da Silva (1977) and Chiorato et al. (2006) assessed the diversity of Brazilian common bean landraces using morphological descriptors.
Multilocus associations (MAs) are an important aspect of the organization of genetic diversity within and among genomes, particularly in highly structured populations, such as in common bean (Kwak and Gepts 2009; Rossi et al. 2009). Understanding the nature of MA within a genome is a pre-requisite for the identification of associations between genome polymorphisms and qualitative or quantitative traits, such as in association analysis methods (Flint-Garcia et al. 2003). Kwak and Gepts (2009) performed a genome-wide MA analysis in common bean and identified a high percentage of loci in MA when the whole sample (including both Andean and Mesoamerican gene pools) was analyzed, while a reduction in MA was observed by analyzing separate gene pools.
There is a need for a more comprehensive analysis of genetic diversity and population structure in Brazilian P. vulgaris based on a larger sample representative of the major bean growing areas of the crop and a genome-wide sample of markers. This aspect is particularly important for the landrace group, which could be an important reservoir of genetic diversity and rusticity, considering the history of this crop in Brazil. Moreover, the availability of a large number of microsatellite markers developed and mapped for the species (Yu et al. 2000; Gaitán-Solís et al. 2002; Blair et al. 2003; Grisi et al. 2007), in addition to the availability of new statistical tools that can improve the population genetic analysis with the visualization of admixture processes (Pritchard et al. 2000) and MA analysis, facilitates a more complete study of the genetic diversity of the domesticated pool of P. vulgaris in Brazil.
Materials and methods
Sampling of the bean collection
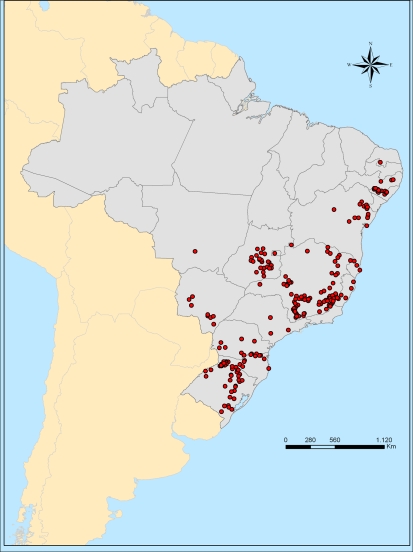
Common bean (P. vulgaris L.) landraces accessions used in this study were obtained from the Common Bean Gene Bank at the Empresa Brasileira de Pesquisa Agropecuária (EMBRAPA) Arroz e Feijão. Based on passport data, one randomly chosen accession per Brazilian municipality was included in the study sample to maximize the geographic representation of the sample. The preliminary list of accessions in the study sample was reviewed by one of us, Jaime Fonseca, as the former EMBRAPA germplasm explorer, to verify that all the selected accessions were landraces, to replace accessions considered as mixtures, and to ascertain that the most important landraces within each region were represented in the study sample. Thus, a total of 279 landraces accessions of common bean were included (Supplementary Table 1; Fig. 1). As standard genotypes, two other accessions of common bean were also included: BAT93 as a breeding line typical of the Mesoamerican gene pool, and Jalo EEP553 as a representative Andean cultivar (and, furthermore, a cultivar in Brazil; Voysest 1983). The BAT93 × Jalo EEP558 recombinant inbred population is the core mapping populations in P. vulgaris (Freyre et al. 1998; Gepts et al. 2008). This sample did not include wild P. vulgaris as wild beans are absent in Brazil (Freytag and Debouck 2002).
Fig. 1.
Collecting sites of common bean landraces in Brazil
For DNA extraction, small, young leaves were collected from one plant per accession, around 30 days after planting, and immediately placed on ice, followed by storage at −80°C. Genomic DNA was extracted following the cetyl trimethyl-ammonium bromide (CTAB) procedure described by Doyle and Doyle (1987).
Marker analyses
Sixty-seven microsatellite markers (distributed over all 11 linkage groups of the P. vulgaris gene map) were used here (Yu et al. 2000; Gaitán-Solís et al. 2002; Blair et al. 2003; Grisi et al. 2007; Table 1). Microsatellite analysis was conducted as described by Kwak et al. (2009), including an economic method of fluorescent labeling of microsatellite fragments amplified by PCR (Schuelke 2000).
Table 1.
Genetic and mapping information for microsatellites and other markers used in this study
| Marker | Linkage group | Relative position within linkage group (cM) | Referencea | Type | SSR sequence | No. of alleles | Gene diversity | H o | PIC |
|---|---|---|---|---|---|---|---|---|---|
| BMd45 | 1 | 0 | 1 | Gene-based | (AG)5 | 5 | 0.43 | 0 | 0.34 |
| PVag003 | 1 | 146 | 4 | Gene-based | (AG)8 | 3 | 0.54 | 0 | 0.43 |
| BMd10 | 1 | 152 | 1 | Gene-based | (GA)8 | 5 | 0.55 | 0.012 | 0.46 |
| PVBR139 | 1 | 174 | 3 | Genomic | (AG)7 | 2 | 0.21 | 0.004 | 0.18 |
| BM157 | 1 | NA | 2 | Genomic | (GA)16 | 5 | 0.35 | 0 | 0.33 |
| BM146 | 1 | NA | 2 | Genomic | (CTGTTG)4(CTG)4(TTG)3(CTG)3(CTG)4 | 4 | 0.17 | 0 | 0.15 |
| PvTFL1y | 1 | NA | 5 | Not applicable | 2 | 0.23 | 0 | 0.21 | |
| BM156 | 2 | 0 | 2 | Genomic | (CT)32 | 8 | 0.43 | 0.004 | 0.41 |
| PVBR243 | 2 | 14 | 3 | Genomic | (CT)20 | 4 | 0.42 | 0 | 0.37 |
| GATS91 | 2 | 21 | 2 | Genomic | (GA)17 | 16 | 0.85 | 0.004 | 0.83 |
| PVgccacc001 | 2 | 37 | 4 | Gene-based | (GCCACC)5 | 3 | 0.64 | 0 | 0.56 |
| BM143 | 2 | 53 | 2 | Genomic | (GA)35 | 14 | 0.81 | 0.019 | 0.79 |
| BM139 | 2 | 101 | 2 | Genomic | (CT)25 | 8 | 0.38 | 0 | 0.36 |
| PVBR106 | 2 | NA | 3 | Genomic | (CTT)2(CT)5 | 5 | 0.35 | 0 | 0.3 |
| PVBR125 | 2 | NA | 3 | Genomic | (GA)6A(GA)3 | 4 | 0.55 | 0.007 | 0.49 |
| SW13 | 2 | NA | 6 | Not applicable | 2 | 0.45 | 0 | 0.35 | |
| AG1 | 3 | 0 | 2 | Genomic | (GA)8GGTA(GA)5GGGGACG(AG)4 | 3 | 0.09 | 0.004 | 0.09 |
| BMd1 | 3 | 8 | 1 | Gene-based | (AT)9 | 6 | 0.62 | 0 | 0.56 |
| PVat008 | 3 | 8 | 4 | Gene-based | (AT)9 | 12 | 0.71 | 0 | 0.67 |
| BM159 | 3 | 41 | 2 | Genomic | (CT)9(CA)8 | 5 | 0.42 | 0 | 0.38 |
| BMd36 | 3 | 72 | 1 | Genomic | (TA)8 | 7 | 0.74 | 0.011 | 0.7 |
| BM172 | 3 | 96 | 2 | Genomic | (GA)23 | 8 | 0.37 | 0 | 0.35 |
| PVBR131 | 3 | NA | 3 | Genomic | (TGA)5 | 3 | 0.47 | 0.004 | 0.39 |
| PVBR87 | 3 | NA | 3 | Genomic | (GA)16 | 11 | 0.76 | 0.004 | 0.73 |
| PVctt001 | 4 | 0 | 4 | Gene-based | (CTT)3(T)3(CTT)6 | 6 | 0.7 | 0.011 | 0.64 |
| PVat001 | 4 | 23 | 4 | Gene-based | (TA)22 | 37 | 0.96 | 0.008 | 0.96 |
| PVat003 | 4 | 49 | 4 | Gene-based | (AT)6 | 4 | 0.43 | 0 | 0.34 |
| BMd26 | 4 | 82 | 1 | Gene-based | (GAT)6 | 6 | 0.38 | 0.022 | 0.32 |
| PVatgc002 | 4 | 133 | 4 | Gene-based | (ATGC)4 | 5 | 0.58 | 0 | 0.54 |
| PVatct001 | 4 | NA | 4 | Gene-based | (ATCT)3 | 1 | 0 | 0 | 0 |
| PVBR112 | 4 | NA | 3 | Genomic | (TC)13 | 4 | 0.4 | 0 | 0.36 |
| PVBR182 | 4 | NA | 3 | Genomic | (AG)12 | 6 | 0.44 | 0.007 | 0.38 |
| SB12 | 4 | NA | 7 | Not applicable | 2 | 0.47 | 0 | 0.36 | |
| SF10 | 4 | NA | 8 | Not applicable | 2 | 0.4 | 0 | 0.32 | |
| APA | 4 | NA | 9 | Not applicable | 2 | 0.31 | 0 | 0.26 | |
| BMd53 | 5 | 0 | 1 | Gene-based | (GTA)5 | 5 | 0.36 | 0.004 | 0.31 |
| BM175 | 5 | 42 | 2 | Genomic | (AT)5(GA)19 | 8 | 0.41 | 0.004 | 0.38 |
| BMd20 | 5 | 72 | 1 | Gene-based | (TA)5 | 4 | 0.51 | 0.018 | 0.46 |
| BMd28a | 5 | 94 | 1 | Gene-based | (GT)4 | 2 | 0.15 | 0 | 0.14 |
| BMd28b | NA | 4 | 0.53 | 0.009 | 0.43 | ||||
| BMd12 | 6 | 0 | 1 | Genomic | (AGC)7 | 6 | 0.16 | 0.046 | 0.16 |
| PVBR163 | 6 | 16 | 3 | Genomic | (AG)16 | 15 | 0.84 | 0.009 | 0.82 |
| PVBR198 | 6 | 29 | 3 | Genomic | (AG)10 | 4 | 0.5 | 0.008 | 0.47 |
| ROC11 | 6 | NA | 10 | Not applicable | 2 | 0.36 | 0 | 0.3 | |
| BM160 | 7 | 0 | 2 | Genomic | (GA)15(GAA)5 | 7 | 0.4 | 0 | 0.37 |
| BM201 | 7 | 76 | 2 | Genomic | (GA)15 | 6 | 0.75 | 0.018 | 0.71 |
| BM210 | 7 | 80 | 2 | Genomic | (CT)15 | 13 | 0.75 | 0.012 | 0.71 |
| BMd40 | 7 | 90 | 1 | Genomic | (AT)6 | 7 | 0.6 | 0.004 | 0.55 |
| BM209 | 7 | 157 | 2 | Genomic | (TA)4(TG)16 | 5 | 0.6 | 0 | 0.52 |
| PVBR35 | 7 | NA | 3 | Genomic | (TC)10 | 8 | 0.54 | 0.018 | 0.51 |
| PVatcc001 | 7 | NA | 4 | Gene-based | (ATCC)3(AG)2(TAC)3 | 5 | 0.34 | 0.004 | 0.3 |
| PVatcc002 | 7 | NA | 4 | Gene-based | (ATCC)3(AG)2(TAC)3T(CTA)3 | 4 | 0.13 | 0.004 | 0.13 |
| PVatcc003 | 7 | NA | 4 | Gene-based | (ATCC)3 | 2 | 0.01 | 0 | 0.01 |
| Phaseolin | 7 | NA | 11 | Not applicable | 2 | 0.33 | 0 | 0.28 | |
| BMd25 | 8 | 0 | 1 | Gene-based | (GAT)6 | 2 | 0.26 | 0.004 | 0.22 |
| BM189 | 8 | 44 | 2 | Genomic | (CT)13 | 11 | 0.81 | 0.019 | 0.78 |
| BM151 | 8 | 84 | 2 | Genomic | (CT)14 | 6 | 0.67 | 0.015 | 0.62 |
| PVBR173 | 8 | NA | 3 | Genomic | (GA)21 | 1 | 0 | 0 | 0 |
| PVBR83 | 8 | NA | 3 | Genomic | (GA)9 | 5 | 0.47 | 0 | 0.37 |
| BM188 | 9 | 0 | 2 | Genomic | (CA)18(TA)7 | 2 | 0.4 | 0 | 0.32 |
| BM141 | 9 | 66 | 2 | Genomic | (GA)29 | 13 | 0.68 | 0.011 | 0.64 |
| BM114 | 9 | 103 | 2 | Genomic | (TA)8(GT)10 | 16 | 0.8 | 0.008 | 0.77 |
| PVat007 | 9 | NA | 4 | Gene-based | (AT)12 | 14 | 0.87 | 0.02 | 0.85 |
| PVBR60 | 9 | NA | 3 | Genomic | (CT)9 | 10 | 0.64 | 0 | 0.59 |
| GATS11B | 10 | 0 | 2 | Genomic | (CT)8 | 6 | 0.46 | 0.007 | 0.42 |
| BMd42 | 10 | 30 | 1 | Genomic | (AT)5 | 7 | 0.67 | 0.014 | 0.62 |
| PVBR181 | 10 | NA | 3 | Genomic | (AG)11 | 5 | 0.4 | 0.004 | 0.36 |
| PVBR185 | 10 | NA | 3 | Genomic | (TC)11 | 5 | 0.57 | 0 | 0.53 |
| BM212 | 10 | NA | 2 | Genomic | (CA)13 | 3 | 0.32 | 0 | 0.28 |
| BMd22 | 11 | 0 | 1 | Gene-based | (TC)6 | 5 | 0.55 | 0.008 | 0.46 |
| BMd33 | 11 | 7 | 1 | Genomic | (ATT)9 | 3 | 0.34 | 0 | 0.28 |
| BMd41 | 11 | 11 | 1 | Genomic | (ATT)9 | 6 | 0.37 | 0.008 | 0.34 |
| BM205b | 11 | 53 | 2 | Genomic | (GT)11 | 5 | 0.43 | 0.011 | 0.39 |
| PVag001 | 11 | 108 | 4 | Gene-based | (GA)11 | 4 | 0.33 | 0 | 0.28 |
| PVBR215 | 11 | NA | 3 | Genomic | (CT)6 | 2 | 0.28 | 0 | 0.24 |
| Mean | 6 | 0.46 | 0.005 | 0.42 | |||||
PIC polymorphism information content, NA not available
a1, Blair et al. (2003); 2, Gaitán-Solís et al. (2002); 3, Grisi et al. (2007); 4 Yu et al. (2000); 5, Kwak et al. (2009); 6, Melotto et al. (1996); 7, Mendez de Vigo et al. (2002); 8, Corrêa et al. (2000); 9, Kami et al. (2006); 10, Johnson et al. (1997); 11, Kami et al. (1995)
The following P. vulgaris SCAR markers were used in this study. Markers SW13 and ROC11 map to linkage groups 2 and 6, respectively, and are linked to the I and bc-3 genes, respectively, both genes conferring resistance to Bean Common Mosaic Virus and Bean Common Mosaic Necrosis Virus (Melotto et al. 1996; Johnson et al. 1997). Markers SB12 and SF10 tag the Co-9 and Co-10 genes (linkage group 4), which confer resistance to anthracnose (Mendez de Vigo et al. 2002; Corrêa et al. 2000). Primers for each SCAR marker were obtained from http://www.css.msu.edu/bic/PDF/SCAR_Markers_2009.pdf. The respective PCRs were conducted as described in the original articles describing the SCAR markers. PCR products were loaded on a vertical, non-denaturing polyacrylamide gel electrophoresis system: MEGA-GEL High Throughput Vertical Unit model C-DASG-400-50, CBS Scientific Co. Gels consisted of 6% (w/v) of acrylamide/bis-acrylamide (19:1), 0.5× TBE buffer, 0.07% (w/v) ammonium persulfate, and 0.08% (w/v) TEMED. PCR products were run for 2 h at 350 V. The DNA marker ladder was All-Purpose Hi-Lo™ DNA marker (Bionexus).
To evaluate the phaseolin type in each accession, a PCR assay was used that had been designed specifically to amplify a region surrounding the 15-bp tandem direct repeat of the phaseolin gene family (Kami et al. 1995). Polymerase chain reaction conditions and primers are described in Kami et al. (1995). PCR products were loaded in a vertical polyacrylamide gel electrophoresis system and ran for 2 h at 350 V.
To genotype the accessions for the PvTFL1y gene, a candidate gene for the determinacy trait in P. vulgaris (Kwak et al. 2008), the primers TFL1y-1a and TFL1y-F4 and PCR conditions developed by Kwak et al. (2008) were used. PCR products were run in 1.5% agarose electrophoresis for 1 h and 40 min, at 117 V.
The APA (Arcelin–phytohemagglutinin–α-amylase inhibitor) locus encodes a multigene family of seed proteins in common bean and is associated with resistance to bruchid insects in this crop. To genotype the accessions for the polymorphism related to this locus, primers designed to amplify fragments ranging from 750 to 900 bp (including the different members of the multi-gene family) (Kami et al. 2006) were used. Polymerase chain reactions contained reagents in the same concentrations as used for SCAR markers; PCR cycles were: 3 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 50°C and 1 min at 72°C; final extension time of 5 min at 72°C. PCR products were loaded in a vertical polyacrylamide gel electrophoresis system and ran for 2 h at 350 V.
Data analyses
The raw marker data are included in Supplementary Table 2. Major allele frequency, allele number, gene diversity (or expected heterozygosity) and observed heterozygosity were calculated according to Weir (1996), while polymorphism information content (PIC) followed Botstein et al. (1980). The above-mentioned parameters were calculated using Powermarker 3.25 software (Liu and Muse 2005).
To evaluate genome-wide multi-locus associations, the microsatellite data were transformed to haplotype data after the heterozygote genotype was treated as missing. The Tassel software (http://www.maizegenetics.net/tassel) was used to calculate the weighted average of the linkage disequilibrium coefficients D′ (standardized disequilibrium coefficient) and r 2 (correlation between alleles at two loci), according to Farnir et al. (2000). Analyses were conducted for the entire plant sample and within the Andean and Mesoamerican samples. To assess whether MA existed primarily among linked markers within linkage groups, a subset of 48 microsatellite markers with known map positions (Table 1; Blair et al. 2003, Grisi et al. 2007) were also analyzed with Tassel. For estimation of experiment-wise P values for linkage disequilibrium tests, 1,000 permutations were conducted as implemented in Tassel (Weir 1996).
The Structure 2.1 software (Pritchard et al. 2000) was used to define the population structure and to assign individuals to populations. The program was run with a preset number of populations (K) ranging from 1 to 10. Twenty independent simulations were performed for each K, using the admixture model, correlated allele frequencies, a running length of 5,000 burn-in and 50,000 Markov chain Monte Carlo (MCMC) repetitions. Results from simulations with the highest likelihood within each number of different K simulations were chosen to assign accessions to populations. Accessions with population membership coefficient of less than 0.8 were identified as potential hybrids. A Structure graphical bar plot of membership coefficients was generated using the Distruct program (Rosenberg 2004). To identify the number of populations that best reflects the structure in the study sample, the following parameters were calculated using an R-script (Structure-Sum) available at http://www.nhm.uio.no/ncb: the likelihoods (posterior probabilities) of simulations for each preset K; the standard deviations of likelihoods; Delta K (Evanno et al. 2005); and the average similarity coefficients for different simulations within each preset K (Nordborg et al. (2005). A Wilcoxon two-sample test was used to compare the mean likelihoods of each preset K. The posterior membership coefficients obtained with Structure for K = 3 are listed in Supplementary Table 3. A neighbor-joining tree was reconstructed based on C.S. Chord distance (Cavalli-Sforza and Edwards 1967), using PowerMarker.
Results
Polymorphism and diversity: microsatellites
Of 80 microsatellite markers tested in this study, 67 produced reliable results when applied to the whole study sample. The reliability of microsatellite markers was based on the presence of peaks of expected size and with consistent shapes over the whole study sample. All the microsatellite markers used produced a single, clear peak of the expected size for each sample, except marker BMd28, which produced two clear peaks of the expected sizes over all the samples. BMd28 was, therefore, scored as a multi-locus marker, BMd28a and BMd28b. Of the 67 microsatellite markers applied to the whole study sample, two markers were monomorphic, PVatct001 and PVBR173 (Table 1).
For the remaining 65 microsatellite markers, gene diversity of individual microsatellites varied from 0.01 (PVatcc003) to 0.96 (PVat001), both gene-based microsatellite markers. The number of alleles identified for each microsatellite varied from 2 (genomic markers PVBR139, PVBR215, and BM188, and gene-based markers PVatcc003, BMd25 and BMd28a) to 37 (PVat001, a gene-based marker). The mean number of alleles over all microsatellite loci was 7. Polymorphism information content varied from 0.09 to 0.85, for the genomic marker AG1 and the gene-based marker PVat007, respectively. The microsatellite markers that presented the highest gene diversity (higher than 0.8) were PVat001, PVat007 (both gene-based markers), GATS91, PVBR163, BM143 and BM189 (all genomic markers). These high diversity markers were located on linkage groups 11, 9, 2, 1, 2 and 8, respectively. The microsatellite markers that showed the lowest gene diversity (lower than 0.3) were PVatct001, BM137, PVBR173, PVatcc003, AG1, PVatcc002, BMd28a, BMd12, BM146, PVBR139, BMd-25 and PVBR215, located on linkage groups 4, 6, 3, 7, 3, 7, 5, 6, 1, 2, 8 and 6, respectively. From these 12 lowest diverse markers, seven were genomic and five were gene-based markers.
Mean gene diversity for genomic markers was 0.49, while the average number of alleles for this type of markers was 6.7. For gene-based markers, mean gene diversity and average allele number were 0.46 and 6.3, respectively. The mean gene diversity in the Brazilian sample was 0.48.
Polymorphisms among other markers
According to the assay of Kami et al. (1995), two types of phaseolin were identified in the Brazilian sample, the “S” and “T” types. Two hundred and twenty-two accessions (79% of the sample) presented an ‘S’ phaseolin type (characteristic of the Mesoamerican gene pool), while the other 59 accessions showed a ‘T’ phaseolin type (characteristic of the Andean gene pool). No other phaseolin types, such as the “C” or “H” types, which are observed in a small fraction of common bean domesticated accessions elsewhere, were present in this sample. This confirmed earlier observations of Gepts et al. (1988) in the case of Brazil.
At the PvTFL1y gene locus, two alleles were identified in the study sample: the 4.1 and the 1.3 kbp alleles. Thirty-seven accessions showed the 4.1 kbp insertion at the PvTFL1y locus, a molecular feature associated with the determinate growth habit in P. vulgaris (Kwak et al. 2008; M. Kwak and P. Gepts, unpublished results). Two hundred and thirty-seven accessions presented the 1.3 kbp allele at this locus, generally associated with the indeterminate growth habit in common bean. Seven accessions could not be scored for this marker, due to failure to amplification. We cannot determine whether the lack of amplification in this and other markers are due to the absence of the gene (null allele) or a technical difficulty such as a deficient DNA extraction or failed PCR amplification. At the APA gene locus, two alleles were identified: a single band allele (48 accessions in the study sample) and a double band allele (206 accessions). Twenty-seven accessions could not be scored for this marker.
With regard to SCAR markers, a high percentage of accessions within the Brazilian germplasm collection of landraces presented the marker allele associated with resistance for the respective diseases. Seventy-two percent of the evaluated accessions showed the amplicon of the SF10 marker (linked to the Co-10 anthracnose resistance allele) whereas 62% presented the amplicon of the SB12 marker (linked to the Co-9 anthracnose resistance allele). Thirty-four percent of the accessions presented the amplicon for the SW13 marker, linked to the I gene, which confers resistance to Bean Common Mosaic Virus. Seventy-seven percent of the accessions showed no band for the ROC11 marker, which is linked to the bc-3 gene, also responsible for resistance to Bean Common Mosaic Virus.
Mean gene diversity in the Brazilian sample considering all the molecular markers together was 0.46 and mean observed heterozygosity was 0.0052.
Identifying membership in the two major gene pools
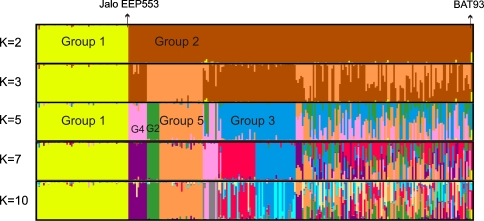
Based on previous studies of genetic diversity of common bean with different types of markers, we hypothesized that the Brazilian collection of common bean landraces is composed of accessions of both Mesoamerican and Andean gene pools and that there is a strong differentiation between these two gene pools. Hence, a Structure analysis was conducted for K = 2 to identify these two major gene pools in the study sample (Fig. 2). Fifty-nine accessions were grouped in cluster 1. This cluster included the control genotype Jalo EEP553. A total of 221 accessions were grouped in cluster 2, including the BAT93 Mesoamerican control accession. A single accession (labeled as 271; gene bank identification CF920002; “feijão de cores”; Supplementary Table 1) was classified as a hybrid between these two major groups, since it its posterior population membership coefficient in the Mesoamerican gene pool was lower than the 0.8 threshold chosen.
Fig. 2.
Structure bar plot of membership coefficients for all the accessions of common bean in the study sample sorted in the same order and classified according to successive selected preset K values ranging from 2 to 10. For K = 2 and K = 5, the groups are identified. G2 and G4: groups 2 and 4, respectively. Hybrid represents a large group of accessions resulting from hybridization mostly among Mesoamerican groups
Cluster 1 contained most of the accessions with a “T” phaseolin type, characteristic of the Andean gene pool (Table 2). Cluster 2 included most of the accessions with an “S” phaseolin type, characteristic of the Mesoamerican gene pool. Nevertheless, 6 of 59 accessions in cluster 1 showed a Mesoamerican, “S” phaseolin type and 6 of 221 accessions in cluster 2 showed an Andean, ‘T’ phaseolin, which suggest introgression between the two gene pools.
Table 2.
Estimate of the number hybrid accessions between the Andean and Mesoamerican gene pools based on phaseolin and Structure analyses
| Status | Andean | Mesoamerican |
|---|---|---|
| Phaseolin analysisa | ||
| Non-hybrid | 53 | 216 |
| Hybrid | 6 | 6 |
| Structure analysisb | ||
| Non-hybrid | 58 | 221 |
| Hybrid | 1 | 1 |
The F ST value for the Andean versus Mesoamerican sub-division was estimated at 0.60. Considering all 75 molecular markers, gene diversity for the Mesoamerican group of accessions was 0.33, while gene diversity of the Andean group was 0.30.
Further definition of the organization of genetic diversity
Table 3 summarizes some parameters for the Structure simulations performed for each preset K value (from K = 2 to 10). Mean likelihoods for the simulations increased with higher preset K values. The differences in the likelihoods among successive preset Ks were significant based on a Wilcoxon two-sample test. Delta K, an ad hoc statistic that has been recommended to help the identification of the best-fitting number of populations within a sample (Evanno et al. 2005), was highest at K = 2 (Table 3). Furthermore, the standard deviations of likelihoods were smallest at K = 3 and largest at K = 4. The standard deviations of the likelihoods were also larger at higher Ks (K = 8 and 9) (Table 3). The aspect of consistency among different simulations within each preset K can also be visualized through the similarity coefficient between different runs for each preset K according to Nordborg et al. (2005) (Table 3). At K = 2 and 3, the similarity coefficients among different simulations were almost 1 and the standard deviations for these coefficients among different simulations were very low. At K = 4, the mean similarity coefficient dropped to 0.62, and the standard deviation was 0.26. At K = 5, the mean similarity coefficient was 0.70 (higher than for preset K = 4) and the standard deviation was 0.24. The mean similarity coefficients among different simulations decreased for K larger than 5.
Table 3.
Mean likelihoods of models and their standard deviations, Delta K for simulations for different K values, mean similarity coefficients and their standard deviations for different Structure simulations within each preset K
| K | Ln P (D)a | Delta K | Similarity coefficientb | ||
|---|---|---|---|---|---|
| Mean | Standard deviation | Mean | Standard deviation | ||
| 2 | −25559.83c | 62.91 | 208.16c | 1.00 | 0.00030 |
| 3 | −23300.03 | 23.07 | 89.66 | 0.99 | 0.00098 |
| 4 | −23108.59 | 1515.67 | 0.86 | 0.62 | 0.26332 |
| 5 | −21603.28 | 283.127 | 2.96 | 0.70 | 0.23525 |
| 6 | −20944.23 | 133.70 | 1.86 | 0.60 | 0.26913 |
| 7 | −20534.81 | 299.00 | 0.666 | 0.51 | 0.18273 |
| 8 | −20323.73 | 400.98 | 0.56 | 0.4 | 0.17342 |
| 9 | −19908.24 | 407.93 | 0.06 | 0.36 | 0.17416 |
| 10 | −19520.35 | 207.81 | NA | 0.43 | 0.16021 |
a20 iterations for each K level
bSimilarity coefficients between different runs (20) within each preset K, according to Nordborg et al. (2005)
cA different set of Structure simulations was run for the estimation of Delta K for K = 2; these simulations used the same parameters (admixture model, correlated allele frequencies, running length of 5,000 burn-in and 50,000 MCMC repetitions)
The separation of Andean and Mesoamerican groups was confirmed in all the different Structure simulations at preset K values >2. Figure 2 showed that the presumably Andean group (yellow group or cluster 1 for preset K = 2) was always preserved as the same separate cluster (yellow color) and without significant evidence of admixture with other groups, in all the different Structure simulations with preset Ks ranging from 2 to 10. The Structure bar graphics also provide information on the level of admixture in the study sample. At K = 2, assuming that posterior membership coefficients between 0.50 and 0.80 may indicate hybridity, only one introgressant in each direction could be observed (Table 2). A Chi-squared test shows no significant differences in the number of introgressants as assessed by the phaseolin and Structure tests (κ 2 = 0.06 < 7.82, P = 0.05 with 3 degrees of freedom). At K = 3, there was a large number of accessions that fell into a hybrid classification between the two clusters inside the Mesoamerican group. The number of accessions in this presumed hybrid group was 61. Accession BAT93, a breeding line resulting from a four-way cross, was a member of this presumed Mesoamerican hybrid group (Fig. 2). At higher preset Ks, larger numbers of individuals were classified as hybrids. Those hybrids always resulted from hybridizations between different subgroups within the presumably Mesoamerican group but not with the Andean group. At K = 5, there was a minor peak for the Delta K. For this number of sub-populations, two Mesoamerican groups observed at K = 3 were each subdivided into two sub-groups. The light brown group of K = 3 was divided into groups 2 and 5, whereas the dark-brown group of K = 3 was divided into groups 3 and 4 at K = 5 (Fig. 2). While there is no obvious explanation for this split at this stage, morpho-agronomic or adaptation data could provide an explanation, pending further analyses.
Neighbor-joining diversity analysis
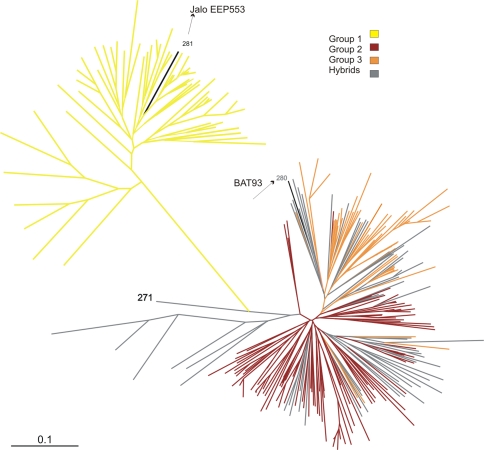
Relationships among accessions were also visualized by a neighbor-joining (NJ) tree based on all 73 polymorphic molecular markers (Fig. 3). In order to compare the results of this diversity analysis with the assignment of individuals to groups using the Structure software, the branches of the tree were colored according to Structure simulations for preset K = 3 (same colors as the Structure bar plot of membership coefficients for K = 3 in Fig. 3).
Fig. 3.
Neighbor-joining tree reconstructed for the Brazilian germplasm collection of landraces based on the C.S. Chord distances and all 73 polymorphic molecular markers. Branches are colored according to the Structure simulation for K = 3. 271: hybrid accession identified at K = 2 (posterior probability < 0.80); gray branches hybrid accessions identified at K = 3 (see Fig. 2 and text)
The presumably Andean gene pool identified with Structure constituted a separated single cluster with exactly the same 59 accessions in the NJ tree (yellow cluster in Figs. 2, 3). The accession labeled as 271, which was the only accession identified as a potential hybrid when Structure simulations were preset to K = 2 (and is, therefore, a potential hybrid between the Andean and Mesoamerican gene pools), also clustered closer to the presumably Andean gene pool than any other accession not identified as a member of the Andean gene pool group.
The two groups identified within the presumably Mesoamerican group when Structure was preset to K = 3 (groups 2 and 3) clustered predominantly separated in the NJ tree (Fig. 3). Just a single accession from group 2 clustered with accessions from group 3, while four accessions from group 3 clustered with accessions from group 2. The accessions classified with Structure simulations as hybrid between the two Mesoamerican groups were spread throughout the Mesoamerican cluster in the NJ tree, not just between groups 2 and 3, but also within each one of these groups.
Genome-wide MAs
When the entire study sample was analyzed for genome-wide MAs, a large fraction (80%) of loci pairs presented significant LD (Table 4). MAs were not restricted to pairs of markers located in the same linkage group, but also occurred between markers mapped on different linkage groups. For the loci pairs that presented significant LD, D′ ranged from 0.16 to 1, with a 0.64 mean, and r 2 ranged from 0.003 to 0.92, with a 0.17 mean.
Table 4.
Counts for comparisons between pair of loci according to significance (P < 0.01) for genome-wide linkage disequilibrium test in the whole study sample and in Mesoamerican and Andean groups separately
| Whole study sample | Mesoamerican group | Andean group | ||||
|---|---|---|---|---|---|---|
| Counts | % | Counts | % | Counts | % | |
| In LD (P < 0.01) | 1,337 | 80 | 360 | 23 | 96 | 8 |
| No. of comparisons evaluated | 1,676 | 1,568 | 1,166 | |||
A genome-wide analysis of separate Andean and Mesoamerican sub-populations, resulted in significantly reduced MA when compared to the MA observed in the whole sample (Table 4). MA in the Andean group (measured as the percentage of marker pairs in disequilibrium) involved just 8% of loci pairs. In the Mesoamerican group, 23% of loci pairs showed a significant MA; they had D′ values ranging from 0.09 to 1, with a 0.44 mean. For these same loci r 2 ranged from 0.002 to 1, with a 0.05 mean. In the Andean group, the loci pairs that presented significant LD had D′ values ranging from 0.20 to 1, with a 0.68 mean, and r 2 values ranging from 0.015 to 1, with a 0.16 mean.
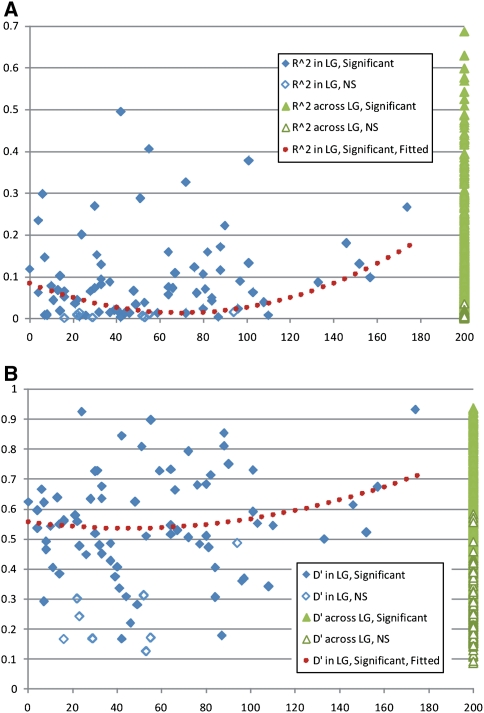
To examine whether MA existed primarily within chromosomes, MA relationships were analyzed in a subset of 48 Simple Sequence repeats (SSRs) with known map locations within the 11 linkage groups (Fig. 4). Statistically significant associations (P < 0.01; experiment-wise) were observed in 966 locus pairs (out of 1,035). Seventy-seven of these significant interactions were among loci located on the same chromosome; 889 involved loci on different chromosomes. Among the 69 non-significant interactions, 5 were on the same chromosome and 64 on different chromosomes. The proportion of significant versus non-significant interactions was the same among loci on the same or different chromosome (Chi-squared test: κ 2 = 0.99, 1 df (0.25 < P < 0.50). Thus, statistically significant MA does not only occur among loci on the same chromosome but also occur in similar proportions among loci within and across chromosomes. Within linkage groups there was no strong relationship between genetic distance and MA intensity (Fig. 4).
Fig. 4.
Magnitude of multilocus associations as measured by r 2 (a) and D′ (b) among Brazilian landraces. The abscissa represents the genetic distance expressed in cM for locus pairs within linkage groups (LG). Along the same axis, “Across LG” represents the r 2 and D′ values for locus pairs distributed among linkage groups
Discussion
Our analysis of the Brazilian common bean landraces reveals several features of this germplasm. First, we confirm that the two major gene pools of domesticated common bean are present in the country, confirming earlier studies (Gepts et al. 1988; Pereira and Souza 1992). This study, however, shows clearly that the Mesoamerican gene pool represents a large majority of the country’s bean landraces. This may be surprising given the closer proximity of the Andes compared to Mesoamerica. This predominance of the latter gene pool may be due to multiple introductions of Mesoamerican germplasm, in pre- and post-conquest times (Gepts et al. 1988). Similarities in climate and soil between Brazil and the Mesoamerican area may help explain the wide distribution of Mesoamerican bean germplasm in that country.
Second, the distinctness of the Andean and Mesoamerican domesticated gene pools is maintained in Brazil, in spite of the close geographic interspersion of the two gene pools (M. Burle and P. Gepts, unpublished data). In the center of origin of common bean, which stretches from northern Mexico to northwestern Argentina, two major gene pools are generally recognized, corresponding to two geographically separate domestications, in the southern Andes and Mexico, respectively. The two domesticated gene pools resulting from these domestications are generally geographically isolated, although exceptions exist such as in Colombia, where the two gene pools meet (Gepts and Bliss 1986). Thus, under these circumstances, there are limited possibilities for reciprocal introgression between these gene pools. Furthermore, this isolation could be reinforced by biological reproductive isolation, namely F1 hybrid lethality conditioned by two complementary semi-dominant genes in the F1 and recessive genes in later generations (Shii et al. 1980; Gepts and Bliss 1985; Koinange and Gepts 1992; Singh and Molina 1996).
In Brazil, geographic isolation cannot be invoked to account for the continued distinctness of the two gene pools. An alternative cause may be a high frequency of inter-gene pool reproductive isolation (Gepts and Bliss 1985; Singh and Molina 1996). Such isolation has been documented in wild (Koinange and Gepts 1992) as domesticated (Gepts and Bliss 1985) accessions. In the domesticated gene pool, the lethality genes may be more widespread in races Nueva Granada in the Andean gene pool and Mesoamerica in the Mesoamerican gene pool, which are precisely the main races represented mainly in Brazil. A similar distinctiveness between Andean and Mesoamerican has been observed in Kenya and Ethiopia (Asfaw et al. 2009).
Third, the division between Andean and Mesoamerican cultivars leads to significant MAs as measured by LD, irrespective of whether loci are linked or not. This confirms observations made earlier by Kwak and Gepts (2009) in a sample of 349 accessions, including 100 wild and 249 domesticated accessions representing the primary center of origin in the Americas. In their results, 96% of tested locus pairs showed a departure from random association, compared to 80% in the present study, when the entire sample was considered. Conducting the same analysis on the Andean and Mesoamerican subsamples lead to a reduction of the proportion of locus pairs in LD to 68 and 75% in the Andean and Mesoamerican subsamples, respectively (Kwak and Gepts 2009). In the current study, LD decreased more strongly, to 8 and 23%, respectively. Differences in the levels of LD may be due to differences in the sample analyzed. The current sample did not contain wild P. vulgaris as wild beans have never been reported from the Brazilian territory (Freytag and Debouck 2002). Wild bean populations show a higher level of population differentiation, as shown by measures of both spatial autocorrelation (Papa and Gepts 2003) and inter-population genetic diversity (G ST; Zizumbo-Villarreal et al. 2005). Clearly, any association analysis will have to be conducted within the two major gene pools, instead of across the entire P. vulgaris species. An MA analysis based solely on mapped markers revealed that significant MAs do not occur only or even predominantly within linkage groups, but occurs in similar proportion both within and across linkage groups. The prevalence of MAs has been observed before by Kwak and Gepts (2009) and Rossi et al. (2009). It has important consequences for bean breeding as it suggests that epistatic interactions may play an important role in the expression of agronomic traits. Johnson and Gepts (2002) observed that digenic QTLs had magnitudes similar to independently acting QTLs in the control of seed yield, biological and seed yield per day, and harvest index. Thus, our observations are consistent with these earlier results. The lack of relationship between the magnitude of MA within linkage group in contrast to LD measurements could be attributed to differences in scale. In this study, the average genetic distance between locus pairs within chromosome was about 55 cM, well above the usual distances used in LD studies around specific genes; the latter range from several hundreds to thousands of base pairs. Fourth, the overall level of genetic diversity observed in this sample of Brazilian bean landraces—mean gene diversity of 0.48—is intermediate compared to other estimates of microsatellite diversity in common bean. Kwak and Gepts (2009) observed a gene diversity of 0.63 for domesticated entries in their sample, compared to 0.47 in the current study. Blair et al. (2006) observed a similar value (0.64) in their sample, which included only domesticated accessions. Thus, dissemination from the center of origins and domestication has, as expected, led to a reduction in genetic diversity.
In the present study, the differences in diversity between the different types of microsatellite markers—genomic markers being more diverse, whereas gene-based markers being less diverse—were not as strong as the differences observed by Blair et al. (2006) in common bean. In our study, the two groups of microsatellite markers with either the lowest or highest diversity included both genomic and gene-based markers. Nevertheless, genomic markers detected a slightly higher gene diversity and average allele numbers when compared to gene-based markers. The differences for averages between these types of markers in the present study were similar to those identified by Díaz and Blair (2006).
Fifth, a striking feature of the Mesoamerican gene pool in Brazilian bean landraces was the high frequency of accessions of hybrid origin. At K = 3 in the Structure analysis, the Mesoamerican gene pool consisted of two “pure” (posterior membership probability over 0.80) groups as well as a group of accessions that resulted from hybridization between these two groups (Fig. 2). At higher K values, this hybrid group was maintained or expanded 9 (data not shown). The significance of this hybrid group remains to be determined. However, the frequency of these hybrid accessions is much higher compared to that in the primary diversification center (Mesoamerican, Central America, and northern South America) (Kwak and Gepts 2009). Morphological analyses are under way to determine differences among these groups, if any. Likewise, correlations between membership in the different groups identified in the Structure and NG analyses are being determined to better understand the nature of this subdivision.
Sixth, for the SCAR markers linked to disease-resistance genes tested in our sample, the frequency of accessions that presented the molecular marker (or amplification product) ranged from 34 to 77% of the accessions. SCAR markers have been used routinely in different common bean breeding programs for marker-assisted selection, aimed at disease resistance (Young et al. 1998; Broughton et al. 2003; Ragagnin et al. 2005). However, the presence of the marker does not guarantee the presence of the corresponding tagged genes. Recombination may have separated the gene and the marker. Johnson et al. (1997) reported a distance of 0.0 ± 7.5 cM between the ROC11 marker and bc-3 gene. Haley et al. (1994) reported distances between the SW13 SCAR and the I gene of 1.0 ± 0.7, 1.3 ± 0.8, and 5.0 ± 2.2 cM, in different genetic backgrounds. SCAR marker SB12 is located at a distance of 2.9 cM (Mendez de Vigo et al. 2002). According to Corrêa et al. (2000), SCAR marker SF12 is located at 6.0 ± 1.3 cM of the corresponding resistance gene. Thus, with the exception of the ROC11 marker, there is a possibility of recombination. Furthermore, even when the resistance gene is present, it is also possible that the gene will not be active against local strains. A more comprehensive field evaluation is therefore needed to assess the presence of actual resistance genes.
As for the PvTFL1y locus, 86% of the accessions in the Brazilian sample showed the 1.3 kbp haplotype associated with an indeterminate growth habit (Kwak et al. 2008; M. Kwak and P. Gepts, unpublished results). The 4.1 kbp insertion, correlated with a determinate growth habit, was more frequent in the Andean group (31% of accessions) than in the Mesoamerican group (9%). These results are in agreement with Koinange et al. (1996), who argued that determinate common bean genotypes would have been favored in the Andean domestication region, because in this region the crop may have been domesticated without maize as a physical support.
This study provides a first comprehensive picture of the diversity and structure in a geographically broadly representative collection of common bean landraces from Brazil. The assessment of genetic diversity and structure obtained in the present study are, probably, at least of medium robustness, considering the relatively high number of markers used for this estimation, with markers spread over all linkage groups of the species. As a basis of comparison, other recent studies assessing genetic diversity of crop plants (with or without their wild relatives) with microsatellites used the following samples: Semon et al. (2005) in Oryza glaberrima: 198 accessions, 93 SSRs; Vigouroux et al. (2008) in Zea mays subsp. Mays: 964 plants, 96 SSRs; and Orabi et al. (2009) in Hordeum vulgare: 185 accessions, 36 SSRs. This collection of common bean landraces presented intermediate diversity, when compared to the complete gene pool of common bean or other common bean collections. However, the importance of this collection should not be neglected. The high frequencies of SCARs molecular phenotypes related to disease resistance observed in this study sample suggest further research into the rusticity of the accessions of this collection. Our study also confirmed the very high degree of structure in the domesticated common bean gene pool in Brazil. Andean and Mesoamerican groups could be clearly distinguished; they showed low levels of admixture. The high degree of genome-wide MA among the molecular markers identified in this study confirmed the high levels of structure, and emphasizes the importance of recognizing these distinct gene pools for upcoming studies, such as association mapping. The high frequency of MA is also consistent with the high frequency of epistatic interactions observed by Johnson and Gepts (2002). The further subdivision of the Brazilian sample in higher number of sub-populations deserves more investigation, by integrating other kinds of data, such as morphological and agronomic information as well as environmental information on local climate, vegetation, and soils.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
A CAPES (Brazil) fellowship to MLB is gratefully acknowledged. USDA/FAS/ICD/RSED provided funds for laboratory and field work. We are grateful to Professor Daniel Potter (UC Davis) for his suggestions on the statistical analyses of this study. We are also thankful to Hanh Le, Lily Pham and James Tanedo, undergraduate students at UC Davis, who helped with the molecular analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Asfaw A, Blair M, Almekinders C (2009) Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African highlands. Theor Appl Genet. http://www.springerlink.com/content/6mx7242tr7060r7608/fulltext.pdf [DOI] [PubMed]
- Blair MW, Pedraza F, Buendia HF, Gaitán-Solís E, Beebe SE, Gepts P, Tohme J. Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2003;107:1362–1374. doi: 10.1007/s00122-003-1398-6. [DOI] [PubMed] [Google Scholar]
- Blair MW, Giraldo MC, Buendia HF, Tovar E, Duque MC, Beebe SE. Microsatellite marker diversity in common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2006;113:100–109. doi: 10.1007/s00122-006-0276-4. [DOI] [PubMed] [Google Scholar]
- Borém A, Carneiro JES, et al. A cultura. In: Vieira C, et al., editors. Feijão: aspectos gerais e cultura no Estado de Minas. Viçosa: Universidade Federal de Viçosa; 1998. [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Hernández G, Blair M, Beebe S, Gepts P, Vanderleyden J. Beans (Phaseolus spp.)—model food legumes. Plant Soil. 2003;252:55–128. doi: 10.1023/A:1024146710611. [DOI] [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF. Phylogenetics analysis models and estimation procedures. Am J Hum Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Chiorato AF, Carbonell SAM, Dias LAS, Moura RR, Chiavegato MB, Colombo CA. Identification of common bean (Phaseolus vulgaris) duplicates using agromorphological and molecular data. Genet Mol Biol. 2006;29:105–111. doi: 10.1590/S1415-47572006000100020. [DOI] [Google Scholar]
- Corrêa RX, Costa MR, Good-God PI, Ragagnin VA, Faleiro FG, Moreira MA, Barros EG. Sequence characterized amplified regions linked to rust resistance genes in the common bean. Crop Sci. 2000;40:804–807. doi: 10.2135/cropsci2000.403804x. [DOI] [Google Scholar]
- Debouck DG. Primary diversification of Phaseolus in the Americas: three centers? Plant Genet Res Newsl. 1986;67:2–8. [Google Scholar]
- Díaz LM, Blair MW. Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers. Theor Appl Genet. 2006;114:143–154. doi: 10.1007/s00122-006-0417-9. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Emydgio BM, Antunes IF, Choer E, Nedel JL. Eficiência de coeficientes de similaridade em genótipos de feijão mediante marcadores RAPD. Pesq Agrop Brasil. 2003;38:243–250. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software SRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Farnir F, Coppieters W, Arranz JJ, et al. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 2000;10:220–227. doi: 10.1101/gr.10.2.220. [DOI] [PubMed] [Google Scholar]
- Flint-Garcia S, Thornsberry JF, ES Bucker., IV Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- Fonseca JR, da Silva HT. Emprego da análise multivariada na caracterização de acessos de feijão (Phaseolus vulgaris L.) Rev Bras Sem. 1977;19:335–341. [Google Scholar]
- Freitas FO. Evidências genético-arqueológicas sobre a origem do feijão comum no Brasil. Pesq Agropec Bras. 2006;41:1199–1203. [Google Scholar]
- Freyre R, Skroch P, et al. Towards an integrated linkage map of common bean. 4. Development of a core map andalignment of RFLP maps. Theor Appl Genet. 1998;97:847–856. doi: 10.1007/s001220050964. [DOI] [Google Scholar]
- Freytag GF, Debouck DG. Taxonomy, distribution, and ecology of the genus Phaseolus (Leguminosae–Papilionoideae) in North America. Forth Worth: Mexico and Central America. Botanical Research Institute of Texas; 2002. [Google Scholar]
- Gaitán-Solís E, Duque MC, Edwards KJ, Tohme J. Microsatellite repeats in bean (Phaseolus vulgaris): isolation, characterization and cross-species amplification in Phaseolus spp. Crop Sci. 2002;42:2128–2136. doi: 10.2135/cropsci2002.2128. [DOI] [Google Scholar]
- Gepts P. A Middle American and an Andean common bean gene pool. In: Gepts P, editor. Genetic resources of Phaseolus beans. Dordrecht: Kluwer; 1988. pp. 375–407. [Google Scholar]
- Gepts P. Origin and evolution of common bean: past events and recent trends. HortScience. 1998;33:1124–1130. [Google Scholar]
- Gepts P, Aragão F, et al. Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the tropics. In: Moore P, Ming R, et al., editors. Genomics of Tropical Crop Plants. Berlin: Springer; 2008. pp. 113–143. [Google Scholar]
- Gepts P, Bliss FA. F1 hybrid weakness in the common bean: differential geographic origin suggests two gene pools in cultivated bean germplasm. J Hered. 1985;76:447–450. [Google Scholar]
- Gepts P, Bliss FA. Phaseolin variability among wild and cultivated common beans (Phaseolus vulgaris) from Colombia. Econ Bot. 1986;40:469–478. [Google Scholar]
- Gepts P, Kmiecik K, Pereira P, Bliss FA. Dissemination pathways of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoretic variability. I. The Americas. Econ Bot. 1988;42:73–85. [Google Scholar]
- Grisi MCM, Blair MW, Gepts P, Brondani C, Pereira PAA, Brondani RPV. Genetic mapping of a new set of microsatellite markers in a reference common bean (Phaseolus vulgaris) population BAT93 × Jalo EEP558. Genet Mol Res. 2007;6:691–706. [PubMed] [Google Scholar]
- Haley SD, Afanador L, Kelly JD. Identification and application of a random amplified polymorphic DNA marker for the I gene (Potyvirus resistance) in common bean. Phytopathology. 1994;84:157–160. doi: 10.1094/Phyto-84-157. [DOI] [Google Scholar]
- Hoehne FC. Botânica e agricultura no Brasil no seculo XVI. São Paulo: Cia. Editora Nacional; 1937. [Google Scholar]
- Ibarra-Pérez F, Ehdaie B, Waines G. Estimation of outcrossing rate in common bean. Crop Sci. 1997;37:60–65. doi: 10.2135/cropsci1997.0011183X003700010009x. [DOI] [Google Scholar]
- Johnson WC, Gepts P. The role of epistasis in controlling seed yield and other agronomic traits in an Andean x Mesoamerican cross of common bean (Phaseolus vulgaris L.) Euphytica. 2002;125:69–79. doi: 10.1023/A:1015775822132. [DOI] [Google Scholar]
- Johnson WC, Guzman P, Mandala D, Mkandawire ABC, Temple S, Gilbertson RL, Gepts P. Molecular tagging of the bc-3 gene for introgression into Andean common bean. Crop Sci. 1997;37:248–254. doi: 10.2135/cropsci1997.0011183X003700010044x. [DOI] [Google Scholar]
- Kami J, Velásquez VB, Debouck DG, Gepts P. Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proc Natl Acad Sci. 1995;92:1101–1104. doi: 10.1073/pnas.92.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami J, Poncet V, Geffroy V, Gepts P. Development of four phylogenetically-arrayed BAC libraries and sequence of the APA locus in Phaseolus vulgaris. Theor Appl Genet. 2006;112:987–998. doi: 10.1007/s00122-005-0201-2. [DOI] [PubMed] [Google Scholar]
- Koenig R, Gepts P. Allozyme diversity in wild Phaseolus vulgaris: further evidence for two major centers of diversity. Theor Appl Genet. 1989;78:809–817. doi: 10.1007/BF00266663. [DOI] [PubMed] [Google Scholar]
- Koinange EMK, Gepts P. Hybrid weakness in wild Phaseolus vulgaris L. J Hered. 1992;83:135–139. [Google Scholar]
- Koinange EMK, Singh SP, Gepts P. Genetic control of the domestication syndrome in common-bean. Crop Sci. 1996;36:1037–1045. doi: 10.2135/cropsci1996.0011183X003600040037x. [DOI] [Google Scholar]
- Kwak M, Gepts P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae) Theor Appl Genet. 2009;118:979–992. doi: 10.1007/s00122-008-0955-4. [DOI] [PubMed] [Google Scholar]
- Kwak M, Velasco D, Gepts P. Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris) J Hered. 2008;99:283–291. doi: 10.1093/jhered/esn005. [DOI] [PubMed] [Google Scholar]
- Kwak M, Kami J, Gepts P. The putative Mesoamerican domestication center of Phaseolus vulgaris is located in the Lerma-Santiago Basin of Mexico. Crop Sci. 2009;49:554–563. doi: 10.2135/cropsci2008.07.0421. [DOI] [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Maciel FL, Gerald LTS, Echeverrigaray S. Random amplified polymorphic DNA (RAPD) markers variability among cultivars of common beans (Phaseolus vulgaris L.) of south-Brazil. Euphytica. 2001;120:257–263. doi: 10.1023/A:1017579014961. [DOI] [Google Scholar]
- Maciel FL, Echeverrigaray S, Gerald LTS, Grazziotin FB. Genetic relationships and diversity among Brazilian cultivars and landraces of common beans (Phaseolus vulgaris L.) revealed by AFLP markers. Genet Res Crop Evol. 2003;50:887–893. doi: 10.1023/A:1025994513043. [DOI] [Google Scholar]
- McClean PE, Lavin M, Gepts P, Jackson SA. Phaseolus vulgaris: a diploid model for soybean. In: Stacey G, editor. Genetics and genomics of soybean. New York: Springer; 2008. pp. 55–76. [Google Scholar]
- Melotto M, Afanador L, Kelly JD. Development of a SCAR marker linked to the I gene in common bean. Genome. 1996;39:1216–1219. doi: 10.1139/g96-155. [DOI] [PubMed] [Google Scholar]
- Mendez de Vigo B, Rodriguez C, Paneda A, Giraldez R, Ferreira JJ. Development of a SCAR marker linked to Co-9 in common bean. Annu Rep Bean Improv Coop. 2002;45:116–117. [Google Scholar]
- Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3(7):e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orabi J, Jahoor A, Backes G. Genetic diversity and population structure of wild and cultivated barley from West Asia and North Africa. Plant Breed. 2009;128:606–614. doi: 10.1111/j.1439-0523.2009.01637.x. [DOI] [Google Scholar]
- Pallottini L, Garcia E, Kami J, Barcaccia G, Gepts P. The genetic anatomy of a patented yellow bean. Crop Sci. 2004;44:968–977. doi: 10.2135/cropsci2004.0968. [DOI] [Google Scholar]
- Papa R, Gepts P. Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theor Appl Genet. 2003;106:239250. doi: 10.1007/s00122-002-1085-z. [DOI] [PubMed] [Google Scholar]
- Pereira PA, Souza CRB. Tipos de faseolina em raças “crioulas” de feijão no Brasil. Pesq Agrop Bras. 1992;27:1219–1221. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragagnin VA, Alzate-Marin AL, Souza TLPO, Sanglard DA, Moreira MA, Barros EG. Use of molecular markers to pyramiding multiple genes for resistance to rust, anthracnose and angular leaf spot in the common bean. Annu Rep Bean Improv Coop. 2005;48:94–95. [Google Scholar]
- Ramalho MAP, Abreu AFB, et al. Cultivares. In: Vieira C, et al., editors. Feijão. 2. Viçosa: UFV; 2006. pp. 415–436. [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- Rossi M, Bitocchi E, Bellucci E, Nanni L, Rau D, Attene G, Papa R. Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evol Appl. 2009;2:504–522. doi: 10.1111/j.1752-4571.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Semon M, Nielsen R, Jones MP, McCouch SR. The population structure of African cultivated rice Oryza glaberrima (Steud.): Evidence for elevated levels of linkage disequilibrium caused by admixture with O-sativa and ecological adaptation. Genetics. 2005;169:1639–1647. doi: 10.1534/genetics.104.033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shii CT, Mok MC, Temple SR, Mok DWS. Expression of developmental abnormalities in hybrids of Phaseolus vulgaris L. J Hered. 1980;71:218–222. [Google Scholar]
- Singh SP, Molina A. Inheritance of crippled trifoliate leaves occuring in interracial crosses of common bean and its relationship with hybrid dwarfism. J Hered. 1996;87:464–469. [Google Scholar]
- Singh SP, Gepts P, Debouck DG. Races of common bean (Phaseolus vulgaris L., Fabaceae) Econ Bot. 1991;45:379–396. [Google Scholar]
- Vieira C. Resistência horizontal às doenças e diversidade genética no melhoramento do feijoeiro no Brasil. Rev Ceres. 1972;19:261–279. [Google Scholar]
- Vieira C. Phaseolus genetic resources and breeding in Brazil. In: Gepts P, editor. Genetic resources of Phaseolus beans. Dordrecht: Kluwer; 1988. pp. 467–483. [Google Scholar]
- Vigouroux Y, Glaubitz JC, Matsuoka Y, Goodman MM, Sánchez González J, Doebley J. Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am J Bot. 2008;95:1240–1253. doi: 10.3732/ajb.0800097. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II. Sunderland: Sinauer Associates Inc; 1996. [Google Scholar]
- Yokoyama LP, Stone LF. Cultura do feijoeiro no Brasil: características da produção. Santo Antonio de Goiás: Embrapa Arroz e Feijão; 2000. [Google Scholar]
- Young RA, Melotto M, Nodari RO, Kelly JD. Marker-assisted dissection of the oligogenic anthracnose resistance in the common bean cultivar, ‘G2333’. Theor Appl Genet. 1998;96:87–94. doi: 10.1007/s001220050713. [DOI] [Google Scholar]
- Yu K, Park J, Poysa V, Gepts P. Integration of Simple Sequence Repeats (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris) J Hered. 2000;91:429–434. doi: 10.1093/jhered/91.6.429. [DOI] [PubMed] [Google Scholar]
- Zizumbo-Villarreal D, Colunga-GarcíaMarín P, de la Payró Cruz E, Delgado-Valerio P, Gepts P. Population structure and evolutionary dynamics of wild-weedy-domesticated complexes of common bean in a Mesoamerican region. Crop Sci. 2005;35:1073–1083. doi: 10.2135/cropsci2004.0340. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.