Abstract
Glucocorticoid hormones control diverse physiological processes, including metabolism and immunity, by activating the major glucocorticoid receptor (GR) isoform, GRα. However, humans express an alternative isoform, human (h)GRβ, that acts as an inhibitor of hGRα to produce a state of glucocorticoid resistance. Indeed, evidence exists that hGRβ contributes to many diseases and resistance to glucocorticoid hormone therapy. However, rigorous testing of the GRβ contribution has not been possible, because rodents, especially mice, are not thought to express the β-isoform. Here, we report expression of GRβ mRNA and protein in the mouse. The mGRβ isoform arises from a distinct alternative splicing mechanism utilizing intron 8, rather than exon 9 as in humans. The splicing event produces a form of β that is similar in structure and functionality to hGRβ. Mouse (m)GRβ has a degenerate C-terminal region that is the same size as hGRβ. Using a variety of newly developed tools, such as a mGRβ-specific antibody and constructs for overexpression and short hairpin RNA knockdown, we demonstrate that mGRβ cannot bind dexamethasone agonist, is inhibitory of mGRα, and is up-regulated by inflammatory signals. These properties are the same as reported for hGRβ. Additionally, novel data is presented that mGRβ is involved in metabolism. When murine tissue culture cells are treated with insulin, no effect on mGRα expression was observed, but GRβ was elevated. In mice subjected to fasting-refeeding, a large increase of GRβ was seen in the liver, whereas mGRα was unchanged. This work uncovers the much-needed rodent model of GRβ for investigations of physiology and disease.
This study demonstrates the existence of glucocorticoid receptor-β in mice, its actions as an inhibitor of GRα, and its potential role in metabolism.
Human glucocorticoid receptor (hGR) is expressed as two major isoforms: hGRα and hGRβ (1,2). Glucocorticoid hormones (GCs) control diverse physiological processes (3,4), such as metabolism, immunity/inflammation, development, and behavior. These responses are a direct result of GRα activity as a hormone-activated transcription factor (5,6). In contrast, the role of GRβ in GC control of physiology is still poorly understood. Most recent studies suggest that GRβ acts as an inhibitor of GRα (7,8,9,10) to produce a state of glucocorticoid resistance (1,2). Indeed, there is indirect evidence that elevated expression of GRβ may be responsible for a variety of immunological diseases. Severe asthma, leukemia, ulcerative colitis, chronic sinusitis, systemic lupus erythematosus, and possibly cigarette smoking all correlate with overexpression of GRβ (2,11,12,13). Many patients suffering from these diseases are refractory to GC treatment. Not surprisingly, increased activation of proinflammatory transcription factors and cytokines has also been noted in cases of GC resistance with elevated GRβ expression. These observations suggest an important role for GRβ as a homeostatic mechanism in the normal attenuation of GC responses and as a possible culprit in hormone-resistant disease states.
The hGR gene was cloned and sequenced in 1985, revealing the expression of hGRα and hGRβ (14). Additional studies showed that the isoforms result from alternative splicing to yield GRs identical through amino acid 727, but which differ in their C-terminal regions. The hGRα C terminus is composed of 50 amino acids containing important sites for hormone binding, as well as helix 12, which provides critical transcriptional activation activity as a site for coregulator interaction (15). In contrast, the unique and nonhomologous C terminus of hGRβ is a disordered 15-amino acid region of no known function. Not surprisingly, hGRβ cannot bind GC agonists (7,16). However, binding by RU486 antagonist, although disputed (17), has been shown by one laboratory (18). Although hGRβ contains activation function-1 and DNA-binding domains identical to those in hGRα, no transcriptional activation or repression activities in response to hormone have yet been found for this isoform. Instead, most data point to hGRβ as an inhibitor of hGRα activity, either through competition for coregulators or through formation of inactive α/β heterodimers. Consistent with this mechanism is the predominant presence of hGRβ in the nucleus of most cells, whereas hGRα resides in the cytoplasm, undergoing nuclear translocation in response to ligand (19). Thus, hGRβ can be viewed as a dominant-negative inhibitor of hGRα, a mechanism of action which may underlie the potential role of GRβ in GC resistance. However, two recent studies using gene array analyses have revealed that hGRβ can constitutively regulate genes not controlled by hGRα (17,18). Therefore, hormone-free hGRβ, in addition to its dominant-negative activity, appears to have an intrinsic gene regulatory function important to physiological responses distinct from hGRα.
The only observation of GRβ outside humans has been in zebrafish (20). However, when the mouse GR (mGR) was originally cloned and sequenced, one active GR was discovered that responded to GCs (21), but two different mRNAs were found with distinct poly-A tails (22). Moreover, an intact mGR protein was identified that was unable to bind hormone (23). Curiously, the alternative isoform of mGR was not pursued, and it is now generally accepted that rodents do not express GRβ. This conventional wisdom owes its existence to studies designed to discover mGRβ based on the hGRβ process. In humans, GRα and GRβ share exons 1–8 but diverge to contain exons 9α and 9β, respectively, based on alternative usage of splice acceptor sites in exon 9 (24). Efforts to discover GRβ based on similar splicing events in rodents and sheep have been unsuccessful (25,26). The recent discovery of GRβ in zebrafish has shown that splicing can also occur, not in exon 9, but through alternative donor sites in intron 8, to yield a zebrafish GRβ (zGRβ) with properties similar to hGRβ. In this work, we demonstrate the existence of GRβ mRNA and protein in the mouse. Like zGRβ, mGRβ results from alternative usage of donor sites, in this case, in a region previously assigned as intron 8 of the mGR gene. Although the splicing mechanism differs from that which generates hGRβ, mGRβ contains a 15-amino acid C terminus that is identical in length and highly similar in sequence to hGRβ. In contrast, the C terminus of zGRβ is much larger (50 amino acids). Mouse GRβ shares similar properties with hGRβ, including ubiquitous expression in mouse organs, lack of responsiveness to the GC agonist dexamethasone (Dex), and an ability to inhibit mGRα activity. This study has uncovered the much-needed rodent model of GRβ, providing a new tool for future in vivo investigations of glucocorticoid resistance and sensitivity in physiology and disease.
Results
The mGRα and mGRβ isoforms
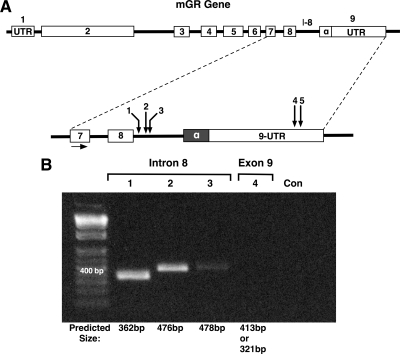
To identify mGRβ, the National Center for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov) was used for the nuclear receptor subfamily 3, group C, member 1 (Mus musculus), and information was downloaded for the complete GR genomic DNA sequence. Information regarding introns and exons in the mGR gene (ENSMUSG00000024431) was downloaded from Ensembl Mouse GeneView (www.ensembl.org). A single mGR genomic sequence was identified. The mGR gene is located on chromosome 18 and consists of at least nine exons (Fig. 1). The Drosophila website for splice site predictions (http://www.fruitfly.org/seq_tools/splice.html) was used to characterize possible acceptor and donor alternative splice sites within mGR (see Supplemental Figs. 1 and 2, published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). In silico mapping revealed that intron 8 was the most likely target of alternative splicing, yielding several donor and acceptor sites with high splice-site prediction scores. Interestingly, no splice-site predictions were found in exon 8. Predicted splice sites were also found in exon 9, in the distal region of the exon where alternative splicing is known to occur in the hGR gene (7,16). For initial screening of mGRβ, primers were designed to test for utilization of the intron 8 and exon 9 sites, followed by RT-PCR analysis of total RNA from mouse embryonic fibroblast (MEF) cells (Fig. 1A). Consistent with prior published efforts (25), no PCR product was observed using a primer specific to the distal region of exon 9 (Fig. 1B). However, all three primers in the intron 8 region yielded PCR products, suggesting the existence of mGR transcripts derived from intron 8.
Figure 1.
Detection of intron 8 transcripts of the mGR gene. A, Genomic organization (not to scale) of the mGR gene, showing location of forward primer (exon 7) and reverse primers (1,2,3,4,5) used for RT-PCR analysis. B, Predicted PCR product sizes for reverse primers 1–4 are indicated. Only primers in the intron 8 (I-8) region yielded products. In the case of primer 4, two products were predicted based on alternate utilization of a donor or acceptor site (Supplemental Fig. 1). Primers 4 and 5 (data not shown) yielded no products on several attempts, consistent with published results (25). UTR, Untranslated region; Con, amplification without template.
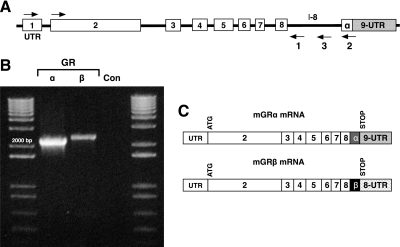
To determine the presence of full-length transcripts, RT-PCR was performed on MEF cell total RNA using a common forward primer at the 5′ end of exon 1 and reverse primers to sequences within intron 8 (primer no. 1) or to the proximal region of exon 9 (primer no. 2) (Fig. 2A). As expected, a full-length transcript was seen corresponding to mGRα (Fig. 2B). More importantly, a full-length transcript containing sequences derived from intron 8 was also found. The intron 8-containing mRNA had reduced abundance relative to mGRα, consistent with the known relationship of hGRβ and hGRα (16). To sequence the novel full-length product, RT-PCR was repeated using a forward primer from the ATG start site in exon 2 and a reverse primer (no. 3) specific to a more distal portion of intron 8, and the product was cloned and sequenced (GenBank accession no. HM236293). The results (Fig. 2C) showed presence of coding sequences that extend beyond exon 8 into intron 8, with a stop codon at position 46–48 base pairs within the intron (for complete mRNA sequence, please see Supplemental Fig. 3). Additional noncoding sequences derived from the distal portion of intron 8 were present. These results suggest that the β-specific amino acids of mGR are located in the proximal portion of intron 8, in contrast to humans, where GRβ sequences are found in the distal portion of exon 9. It is likely, therefore, that mGRβ arises through use of alternative donor sites located within intron 8, as opposed to the hGR mechanism of alternative acceptor sites in exon 9.
Figure 2.
Isolation and sequencing of full-length mGRβ mRNA. A, Genomic organization (not to scale) of the mGR gene, showing location of forward primer (exon 1) and reverse primers (1 and 2) used for RT-PCR analysis. B, Primer 3 in conjunction with the forward primer in exon 1 was used to generate a second full-length mGRβ product that was cloned and sequenced to yield the organization of mGRβ mRNA is seen in C. Primer 3 in conjunction with the forward primer in exon 2 was used to generate the cDNA cloned into pcDNA3.1+ (see Fig. 5). Con, Amplification without template.
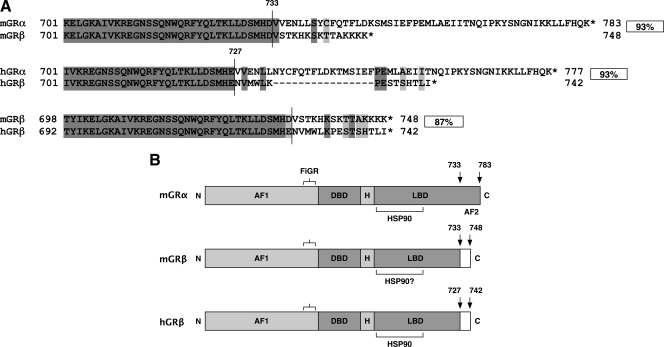
The translation product of mGRβ spans 748 amino acids (Fig. 3A). Alignment of mGRβ with the mGRα peptide sequence revealed 93% homology. The encoded mGRβ protein diverges from mGRα at amino acid 733, beyond which is a disordered C terminus of 15 amino acids. In comparison, hGRα and hGRβ are also 93% identical. Interestingly, both hGRβ and mGRβ add an additional 15 amino acids to exon 8, even though the means of alternative splicing are different. The two β-isoforms are an overall 87% match and share a functional domain organization that appears identical (Fig. 3B). In both hGR and mGR, the point of divergence between α and β isoforms is located in the transition region between the 10th and 11th helix of the ligand-binding domain (7,16). This results in a degenerate helix 12 in the ligand-binding domains of hGRβ and mGRβ. As a consequence, mGRβ, like hGRβ, is unresponsive to the glucocorticoid agonist Dex (see below).
Figure 3.
Sequence and functional domain comparisons of mGRβ and hGRβ. A, C-terminal regions of GRα and GRβ were aligned for each species. Overall percent homologies for entire proteins are indicated. Vertical lines indicate borders between exon 8 and distal domains. Light gray boxes indicate conservative substitutions. B, The mGRβ isoform exhibits a functional domain structure that is nearly identical to hGRβ. Compared with mGRα, the β-isoforms of both species have reduced and distinct C-terminal regions that lack the activation function-2 (AF-2) domain (helix 12). These features account for their reduced ability to bind hormone and activate transcription. DBD, DNA-binding domain; H, hinge region; LBD, ligand-binding domain; FiGR, epitope recognized by FiGR monoclonal antibody. HSP90 binding regions of mGRα and hGRβ are shown, along with putative site in mGRβ.
Expression of mGRβ mRNA in vivo
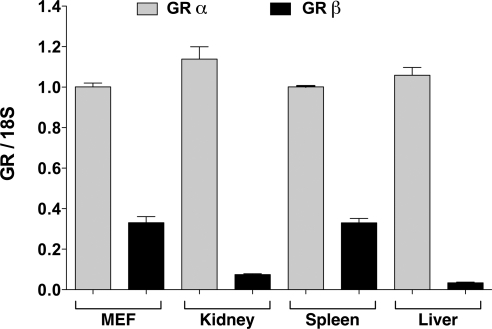
As a first test of significance, expression of mGRβ in mouse organs and tissues was assayed by quantitative real-time PCR (qPCR). Total RNA was extracted from mouse tissues and analyzed with forward primers to exon 8 and reverse primers for exon 9 (mGRα) and intron 8 (mGRβ) indicated in Fig. 2. The qPCR results are from two different C57/Bl6 mice. The data are expressed relative to mGRα expression. In all tissues studied, mGRα and mGRβ mRNA appear to be coexpressed, with mGRα mRNA levels being significantly higher than mGRβ (Fig. 4). The highest level of mGRβ was observed in the spleen, with lesser amounts in kidney and liver. These results are in good agreement with expression profiling of GRβ mRNA in human (16) and zebrafish (20) tissues. Moreover, the high levels of GRβ in mouse spleen are consistent with the important role played by hGRβ as an dominant-negative inhibitor of GRα in lymphoid cells (10).
Figure 4.
Tissue expression profile of mGR mRNA isoforms. Real-time PCR analysis of tissues and MEF cells was performed with primers to intron 8 (mGRβ) or exon 9 (mGRα). All values were normalized to MEF cell mGRα and represent means ± sem for two independent samples/tissues. Adult, male C57/BL6 mice fed normal chow ad libitum were used as donors.
Characterization of mGRβ protein
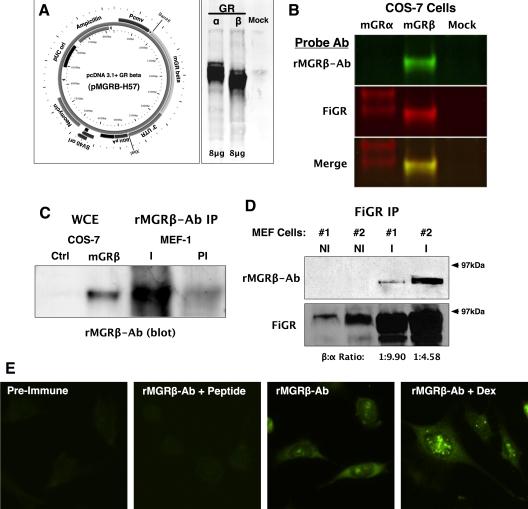
A cDNA expression vector was made by cloning the full-length mGRβ PCR product (Fig. 2) into pcDNA3.1 to generate the pMGRβ-H57 vector (Fig. 5A). Receptor-less COS-7 cells were transfected with equal amounts of pMGRβ-H57 and pSV2Wrec encoding mGRα, followed by Western blot analysis using FiGR monoclonal antibody against mGR that recognizes a shared epitope in the N-terminal domain (see Fig. 3) (27). The results (Fig. 5A) show expression of mGRβ protein with a molecular mass smaller (81.7 kDa) than mGRα (86.0 kDa). Importantly, this shows that the endogenously produced mGRβ mRNA can be translated into protein. To detect endogenous mGRβ, a rabbit polyclonal antibody (rMGRB-Ab) was made. Specificity of rMGRB-Ab was determined by transfecting COS-7 cells with pSV2Wrec, pMGRβ-H57, or empty vector, followed by probing of Western blottings with rMGRB-Ab and FiGR antibodies. Results were analyzed with the Odyssey infrared system utilizing red- and green-emitting counter antibodies to detect FiGR and rMGRB-Ab, respectively. As predicted, rMGRB-Ab detected mGRβ but showed no reactivity to mGRα (Fig. 5B). To demonstrate existence of endogenous mGRβ protein in cells, untransfected MEF cell lysates were immunoadsorbed with rMGRB-Ab or FiGR followed by Western blot analysis (Fig. 5, C and D). The results show immune-specific pull-down of mGRβ by the rMGRB-Ab and FiGR antibodies. As expected, the FiGR purification results showed expression of mGRβ protein to be lower than mGRα. As a last test, indirect immunofluorescence with rMGRB-Ab was performed in MEF cells (Fig. 5E), showing localization of mGRβ protein in the cytoplasm and nuclear foci. Interestingly, Dex treatment elevated the mGRβ signal in both compartments. This result is consistent with the ability of Dex to increase mGRβ mRNA expression (please see figure 8 below).
Figure 5.
Cloning, Western blot, and indirect immunofluorescence analyses of mGRβ. A, Primers spanning the ATG start site in exon 2 and the distal region of intron 8 (see Fig. 2) were used to isolate the full-length mGRβ cDNA, followed by cloning into pcDNA3.1 to yield the pMGRβ-H57 vector. COS-7 cells were transfected with 8 μg each of pSV2Wrec (mGRα), pMGRβ-H57 (mGRβ), or empty vector (mock), followed by Western blot analysis with FiGR mouse monoclonal antibody that recognizes a common epitope on both mGR isoforms. B, A rabbit polyclonal antibody specific to mGRβ (rMGRβ-Ab) was generated using the unique 15-amino acid terminal sequence. Whole-cell extracts from COS-7 cells transfected with pSV2Wrec, pMGRβ-H57, or empty vector (mock) were simultaneously probed with FiGR and rMGRβ-Ab antibodies. The Odyssey infrared detection system utilizing 680 nm (red) and 800 nm (green) emitting counter antibodies was used to detect FiGR (mGRα) and rMGRβ-Ab, respectively. Results show that the rMGRβ-Ab antibody reacts only with mGRβ not with mGRα. C, MEF cell lysates were immunoadsorbed with rMGRβ-Ab (I) or preimmune (PI) serum, followed by blotting with rMGRβ-Ab, using enhanced chemiluminescence. Whole-cell extracts from COS-7 cells transfected with pMGRβ-H57 were used for comparison. D, Two separate MEF cell lysates were immunoadsorbed with FiGR (I) or nonimmune (NI) IgG, followed by sequential blotting with rMGRβ-Ab and FiGR, using enhanced chemiluminescence. Relative densitometric ratios of mGRβ to mGRα are shown. E, Indirect immunofluorescence using preimmune serum, rMGRβ-Ab, or rMGRβ-Ab blocked with mGRβ peptide was performed on MEF cells treated or untreated with Dex (100 nm, 2 h).
Figure 8.
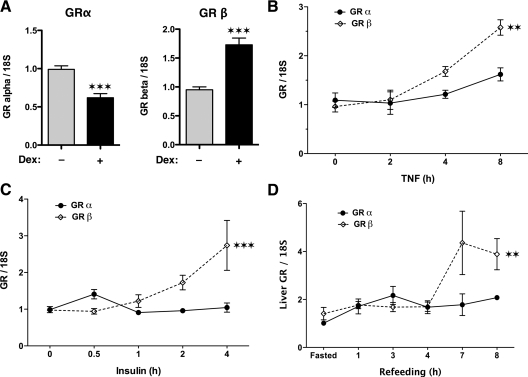
Hormonal and dietary control of mGRβ expression. Real-time PCR analysis of mGR isoforms in (A) MEF cells treated with 100 nm Dex (***, P < 0.001), (B) RAW 264.7 monocytic macrophage cells treated with 10 nm TNFα (**, P < 0.01), (C) MEF cells treated with 100 nm insulin (***, P < 0.001), and (D) livers of adult male C57/BL6 mice subjected to fasting refeeding (**, P = 0.01). All values were normalized to 18S RNA and represent means ± sem of three independent treatments assayed in triplicate.
mGRβ is a hormone-insensitive dominant-negative inhibitor of mGRα
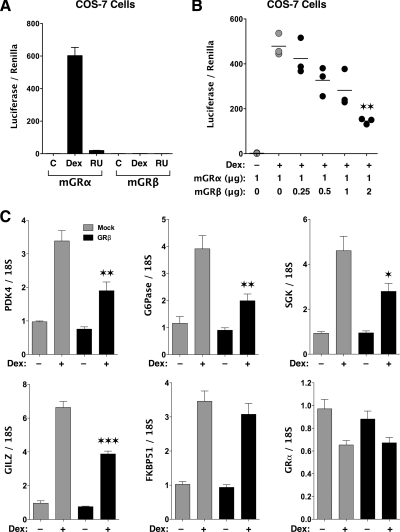
Because mGRβ and hGRβ express C-terminal domains that are almost identical, we reasoned that mGRβ would be unresponsive to activation by glucocorticoid ligands. In addition, Cidlowski and co-workers (9) have shown that the ability of hGRβ to inhibit hGRα activity on gene expression is encoded by its C-terminal 15-amino acid domain. Thus, the mGRβ discovered in this work seemed likely to act as a hormone-insensitive, dominant-negative inhibitor of mGRα. To test both of these functions, COS-7 cells were transfected with pGRE2E1B-Luc reporter and various combinations of pMGRβ-H57 and pSV2Wrec. The results of Fig. 6A showed that mGRβ cannot increase pGRE2E1B-Luc activity in response to Dex or RU486. RU486 antagonist was chosen because of one report suggesting reactivity to this ligand by hGRβ (18). Figure 6B demonstrates dose-dependent inhibition of mGRα activity by mGRβ that begins at a β:α molar ratio of 0.5:1. To determine the dominant-negative activity of mGRβ at endogenous genes, real-time PCR analysis was performed in mGRα-expressing MEF cells after transfection of pMGRβ-H57 (Fig. 6C). The results showed that Dex-induced expression of glucocorticoid-inducible leucine zipper (GILZ), serum- and glucocorticoid-inducible kinase, pyruvate dehydrogenase kinase-4 (PDK4), and glucose-6-phosphatase (G6Pase) were effectively inhibited by mGRβ. In contrast, mGRβ had no effect on expression of FK506 binding protein 51 (FKBP51) and on the ability of Dex to down-regulate mGRα expression. The latter results suggest that mGRβ is not a global inhibitor of mGRα actions, and that the inhibitory effect of mGRβ is not due to decreased mGRα expression.
Figure 6.
Hormone sensitivity and dominant-negative activity of mGRβ. A, COS-7 cells transfected with pSV2Wrec (mGRα) or pMGRβ-H57 (mGRβ) were assayed for luciferase activity at the pGRE2E1B-Luc reporter after treatment with Dex (1 μm), RU486 (1 μm), or vehicle control. Values were normalized to transfection efficiency (renilla) and represent means ± sem for three independent treatments. B, Luciferase (pGRE2E1B-Luc) activity in COS-7 cells was measured after transfection with mGRα and increasing amounts of mGRβ and treatment with 100 nm Dex (sem, n =3; **, P < 0.01 vs. Dex, mGRα alone). C, To assess dominant-negative activity of mGRβ at endogenous genes, MEF cells expressing mGRα were transfected with pMGRβ-H57 or control vector and treated with or without Dex (100 nm), followed by real-time PCR analysis. Values represent means ± sem of three independent treatments assayed in triplicate. ***, P < 0.001; **, P < 0.01; *, P < 0.05 vs. Dex, mGRα alone.
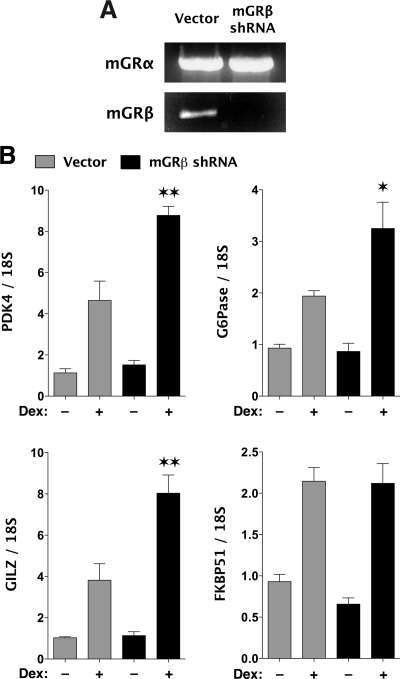
To directly test the inhibitory actions of endogenous mGRβ, short hairpin RNA (shRNA) specific to mGRβ was designed based on its unique intron 8 sequence. MEF cells were infected with lentiviral constructs expressing mGRβ shRNA or empty vector. Figure 7A demonstrates that the mGRβ shRNA effectively blocks expression of the mGRβ full-length mRNA, whereas leaving mGRα mRNA untouched. More importantly, Dex-induced expression of three endogenous genes (PDK4, G6Pase, and GILZ) was increased in response to mGRβ knockdown (Fig. 7B). These results are the first direct evidence that endogenously expressed GRβ (mouse or human) plays a functional role in mGRα actions. Consistent with the results of Fig. 6C, mGRβ knockdown had no effect on FKBP51 expression.
Figure 7.
Gene silencing of mGRβ. Lentiviral delivery of mGRβ shRNA was use to make a MEF cell line with stable down-regulation of mGRβ. A control cell line was infected with lentivirus expressing empty vector. A, Complete mRNA constructs for mGRα or mGRβ were amplified via PCR to show mGRβ knockdown. B, mGRβ shRNA and vector MEF cells were treated with 100 nm Dex for 2 h or vehicle, followed by real-time PCR analysis. Values represent means ± sem of three independent treatments assayed in triplicate. **, P < 0.01; *, P < 0.05 vs. Dex, vector control.
Hormonal and dietary control of mGRβ expression
As an agonist to mGRα, Dex not only causes activation or repression of GR-regulated genes, it also causes autologous down-regulation of GRα expression in many species, as a means by which to attenuate overstimulation by GC ligands (28,29). Because GRβ can be viewed as an alternative mechanism of GRα attenuation, and because GRβ cannot bind Dex, we determined what effect Dex had on mGRβ expression. MEF cells were treated with 100 nm Dex for 2 h, followed by measurement mGRα and mGRβ mRNA expression via qPCR (Fig. 8A). As expected, a significant decrease in mGRα mRNA was seen in response to hormone treatment. Interestingly, mGRβ mRNA was increased. This suggests the existence of a negative feedback loop, most likely mediated by GRα, that up-regulates GRβ expression to control sensitivity of cells to glucocorticoids.
GCs acting through hGRα are potent antiinflammatory drugs that inhibit the ability of nuclear factor κB (NF-κB) to activate expression of proinflammatory cytokine genes (30). In turn, activation of NF-κB by TNF-α leads to reciprocal attenuation of hGRα transcriptional activity (31). Although NF-κB can inhibit hGRα through direct protein-protein binding (30), an additional mechanism was recently discovered, in which NF-κB causes selective up-regulation of hGRβ expression (10). To test whether this mechanism applies to mGRβ, we measured mGRα and mGRβ expression in RAW 264.7 monocytic macrophage cells subjected to TNF-α treatment (Fig. 8B). The results show a modest approximately 1.5-fold induction of mGRα at the 8-h time point but a greater approximately 2.5-fold induction of mGRβ. These data are in good agreement with those of Webster et al. (10) and suggest that mGRβ, like its human cognate, serves to induce a state of GC resistance to maximize inflammatory responses.
As a last test of relevance, we asked whether mGRβ might play a role in GC control of metabolism. Glucocorticoids are well-known antagonists to insulin that promote gluconeogenesis over glycogenesis, especially in the liver (4,32). Conversely, insulin acts to inhibit gluconeogenesis, in part, by blocking GR activity at the hepatic phosphoenolpyruvate carboxykinase promoter (33,34). Because mGRβ can similarly block mGRα activity at the PDK4 and G6Pase genes (Fig. 6C), we tested whether insulin might achieve this effect on gluconeogenic genes by up-regulating expression of mGRβ (Fig. 8C). The results show a short-lived increase of mGRα immediately after insulin treatment of MEF cells, but a more dramatic increase of mGRβ with longer treatment. As a further test, we assayed for mGR isoform expression in the livers of mice subjected to a fasting-refeeding regimen (Fig. 8D). During refeeding, there is a well-documented and robust stimulation of hepatic metabolism by insulin (35,36). The results show a small increase in mGRα during the early stages of refeeding but a much larger increase in mGRβ at the later stages. Taken as a whole, these results represent a potential new role for GRβ (mouse or human) in the GC-insulin axis, in which up-regulation of GRβ may serve to maintain insulin sensitivity.
Discussion
Due to underlying pathology or drug treatment, glucocorticoid resistance can develop and is now a major concern in many disease states. Thus, the need for a mammalian GRβ model is imperative. In humans, GC resistance can occur by two major mechanisms: loss-of-function mutations in GRα (37) or by increased expression of GRβ, which acts as a dominant-negative inhibitor of GRα. Although GRα mutations can result in a type of GC resistance that is both systemic and severe, these mutations are rare. In contrast, the evolving evidence suggests that GC resistance based on GRβ is much more common and likely to be tissue specific in nature. To date, GC resistance based on GRβ has been principally characterized in immunological diseases and drug-resistant states. Immune system homeostasis is balanced by glucocorticoids, which regulate immune cell turnover by suppressing cytokine production and promoting apoptosis. GC insensitivity due to elevated hGRβ expression increases proinflammatory cytokines, leading to escalated cell growth and reduced cell death (38). Superantigens, such as staphylococcal enterotoxin B and toxic shock syndrome toxin, have been demonstrated to cause increased GRβ expression and GC resistance (39). Also, proinflammatory cytokines, such as TNFα and IL-1, increase expression of GRβ via the NF-κB pathway (10). Although cytokine production is increased in all asthma patients, some subjects do not benefit from GC therapy because of elevated GRβ (40,41). Indeed, fatal asthma has been linked to extremely high levels of GRβ in the airways (42) and a complete loss of GC drug response (43). Other inflammatory diseases linked to high levels of GRβ include: ulcerative colitis, ankylosing spondylitis (44), cigarette smoking (45), leukemia (12), and systemic lupus erythematosus (46).
GC resistance via GRβ is almost unknown in diseases of metabolism. Only one study on this topic has been published. It showed that exposure of skeletal muscle to GCs leads to a decline in GRα expression and a concomitant increase of GRβ (47). Because GC hormones are potent regulators of glucose and lipid metabolism, and because GCs are broad and chronic antagonists to the actions of insulin (48), we reasoned that GRβ may play a role as a modulator of GRα actions, similar to insulin antagonism. In this work, we provide evidence that mGRβ is indeed involved in metabolic processes. GC induction of the gluconeogenic enzymes PDK4 and G6Pase was inhibited by mGRβ, suggesting that one function of the GRβ isoform, like insulin, is to block glucose production. Not surprisingly, in cells treated with insulin, mGRα expression was unchanged, but mGRβ went up. In cells treated with Dex, mGRβ was also increased, suggesting that both GCs and insulin share up-regulation of mGRβ as a common mechanism for antagonism of mGRα. Similarly, in mice subjected to fasting refeeding, a large increase of GRβ was seen in the liver, whereas mGRα was once again unchanged. The fasting-refeeding regimen employed is known to produce a robust stimulation of hepatic metabolism by insulin (35,36). Taken as a whole, these data are the first evidence that GRβ up-regulation may be an important mechanism for maintaining organ sensitivity to insulin.
In this work, we present data that mice express a GRβ isoform that derives from alternative splicing of intron 8, similar to the mechanism in zebrafish (20). The sequence encoding the GRβ-specific amino acids is located in the middle portion of exon 9 in the human gene but is found in intron 8 in the zebrafish gene. In zGR, exon 9 is skipped or silenced as a result of alternative splicing, and intron 8 is retained. Therefore, hGRα and hGRβ mRNA are produced through alternative usage of a splice acceptor site in exon 9, whereas alternative use of a splice donor site in intron 8 appears to be the underlying mechanism in both mice and zebrafish GR. This mechanism is often referred to as intron retention and is not unique to mGRβ. Indeed, C-terminal isoform variants of vitamin D, peroxisome proliferator-activated-α and peroxisome proliferator-activated-γ receptors can be generated by intron retention (24). It has long been thought that GRβ does not exist in rodents, in large part because one high-profile study concluded that the alternative splicing event does not occur in mice (25). The study assumed that the splicing event in mice must be similar to humans and used primers that focused on the distal portion of exon 9. It is now clear that mice express a GRβ isoform derived from intron 8. In contrast to zGRβ, which has a 50-amino acid C-terminal region, mGRβ has a protein structure in which the C-terminal region is the same size (15 amino acids) as the human β-isoform. Moreover, for the properties so far tested, mGRβ is highly similar to hGRβ.
The mechanism controlling alternative splicing of the GR gene is poorly understood but is generally thought to involve the generation of a spliceosome composed of ribonucleoproteins and serine-arginine (SR)-rich proteins, among others, that bind structures found in both introns and exons, such as branch point sequences (BPS) and polypyrimidine tracts (ror review, see Ref. 49). Intron 8 in mice is nearly twice as large as intron 8 in humans (1061 vs. 526 bp, respectively). Because intron size correlates to the likelihood of alternative splicing, this reason alone may account for why murine species utilize intron 8 for isoform control. Interestingly, we have identified two BPS sites within murine intron 8, as well as a single polypyrimidine tract (Supplemental Fig. 1). We speculate that mGRα and mGRβ are generated through a mechanism that uses the separate, but distinct, BPS sites to initiate splicing. Targeting of these sites or the spliceosome proteins that bind them may eventually form the basis by which to inhibit mGRβ expression, resulting in cells and tissues that are more sensitive to glucocoritocoids. Recent advances lend credence to this hypothesis. Bombesin, a ligand for G protein-coupled receptors, is known to up-regulate expression of SR protein p30c, causing elevated expression of GRβ in prostate cancer cells (50). A related protein, SRp40, regulates splicing of GRβ in HeLa and 293T cells (51). As can be seen by these examples, major factors that regulate spliceosome action on the GR gene are only now being discovered. Our newly discovered mouse model of GRβ can now be used to establish feasibility of these targets for eventual alteration of hGRβ expression.
Our data show that mGRβ expressed in receptor-less COS cells cannot respond to Dex or RU486 by activating expression of a pGRE-Luc reporter. This result is in good agreement with most studies of hGRβ showing that it cannot bind GC agonists nor activate reporter or endogenous gene expression in response to hormone (7,8,9,16). However, there is one report demonstrating hGRβ binding by RU486 with consequent induction of nuclear translocation (18). In contrast to this unresolved issue, it is now clear that hGRβ can exert constitutive control on gene expression (17,18). These studies used gene array approaches after overexpression of hGRβ and showed that, in addition to its ability to suppress hGRα-regulated genes, hGRβ exerted both positive and negative control over a unique set of genes not regulated by hGRα. In addition, a constitutive ability of hGRβ to induce histone deacetylation has been found (52,53), providing a possible mechanism for hGRβ-mediated repression of gene expression. Taken as a whole, these results show that hormone-free GRβ has an unexpected constitutive and intrinsic gene expression function that may regulate cellular and physiological responses distinct from GRα. Although it remains to be seen whether mGRβ can replicate the latter property, the mGRβ mouse model described here is likely to foster study of glucocorticoid resistance and sensitivity in diverse disease states, such as inflammation, hematological cancers, diabetes, and obesity.
Materials and Methods
In silico prediction of mGRβ
To identify mGRβ, the NCBI website (www.ncbi.nlm.nih.gov) was used for the nuclear receptor subfamily 3, group C, member 1 (M. musculus), and gene information was downloaded for the complete GR genomic DNA sequence. Information regarding introns and exons (ENSMUSG00000024431) was downloaded from Ensembl Mouse GeneView (www.ensembl.org). A single mGR genomic sequence was identified. The NCBI website was used to search for expressed sequence tag and cDNA sequences derived from mGR transcripts. Text searches and Basic Local Alignment Search Tool (BLAST) searches were performed. BLAST searches of the mouse genomic sequence were carried out using the BLAST nucleotide tool at the Sanger Institute Ensembl server.
Characterization of alternative splice sites within mGR gene
Recognition of alternative splice sites in exons 7–9 and intron 8 of the mGR gene were investigated using the Drosophila website for splice site predictions (http://www.fruitfly.org/seq_ tools/splice.html). Sequences with the highest scores in exon 8, exon 9, and intron 8 were identified as potential targets for alternative splice sites that could lead to production of mGRβ (Supplemental Fig. 1). Primers were developed for these prospective splice sites (see below), and an initial screen for mGRβ was performed via RT-PCR.
Initial screening of mGRβ
Total RNA was isolated from MEF cells using 5-Prime PerfectPure RNA Cell kit (Fisher Scientific Co., LLC, Auburn, AL) according to the manufacturer’s instructions. Total RNA concentration and purity was determined by measuring absorbance at 260/280 nm and confirmed on an RNA denaturing formaldehyde gel. Purified RNA (1 μg) was used to produce complementary strands of DNA (cDNA) using a 1st strand synthesis kit (Roche Applied Science, Indianapolis, IN). Newly synthesized DNA (3 μl) was amplified by RT-PCR using forward primers containing sequences from exon 7 (GCAGAGAATGACTCTACCCTGCA) and reverse primers based on prospective splice sites ratings from the Drosophila website. Three different reverse primers for intron 8 (TAAAGGCATCTGCCACCACC, CTGTCTTTGGGCTTTTGAGATAGG, and CTTTGGGCTTTTGAGATAGGATC) and two different reverse primers for the latter part of exon 9 (TCCCAGCTCCCTCTCCCTAG and TCCCTCTCCCTAGCTTAGAG) were used to identify the location of mGRβ (see Fig. 1). 18S RNA was amplified as an internal control. PCR conditions used were: 95 C for 5 min, 95 C for 1 min, 60 C for 1 min, 72 C for 40 sec, and 72 C for 10 min. PCR products were electrophoresed in a 1% agarose gel and visualized with ethidium bromide. The 1 Kb Plus DNA Ladder (Invitrogen, Carlsbad, CA) was used as a size standard.
Generation of complete mGRβ transcript
Primers for exon 1 (GTAGAGACGAAGTCCCCAGCA) and reverse primers were based on sequences from intron 8 (GRβ) (TAAAGGCATCTGCCACCACC) and exon 9 (GRα) (AGCTAAGGAGATTTTCAACCACA) of mGR and used to demonstrate the complete mGRα and mGRβ mRNA constructs. The expected mGRα and mGRβ products were 2251 and 2361 bp, respectively. PCR conditions used were: 95 C for 5 min, 95 C for 1 min, 60 C for 1 min, 72 C for 3.5 min, and 72 C for 10 min.
Cloning and sequencing of mGRβ
Cloning and sequencing of mGRβ from MEF cells was performed as follows. After total RNA isolation, cDNA synthesis was achieved using KOD Xtreme Hot Start DNA Polymerase (Novagen, Madison, WI) and a forward primer for the ATG start site of the open reading frame in exon 2 (CGGGATCCATGGGACTGTATATGGGAGAG) and a reverse primer to the distal portion of intron 8 (GCTCTAGAGTAATGTATCTTGATTGTGGC). The expected product was 2852 bp. GRβ PCR products were ligated to pcDNA 3.1+ vector using BamHI and XbaI and transformed into One Shot INV F cells (Invitrogen). Plasmid DNA from positive clones was determined by RT-PCR and further extracted using the QIAGEN Spin Miniprep Kit (QIAGEN, Crawley, UK). BamHI and XbaI sites flank the position on the vector at which the mGRβ gene is inserted. The presence of GRβ in mice was confirmed by restriction digestion with BamHI and XbaI to determine the size of the insert (Boehringer Mannheim, Mannheim, Germany) and further digested with HindIII to determine sequence specificity. Sequencing was performed by the University of Iowa DNA Facility (Iowa City, IA) using T7 forward and BGH reverse primers that flank the gene insertion site of the plasmid. The sequence confirmed plasmid was named pMGRβ-H57. The mGRβ sequence has been deposited to GenBank under accession no. HM236293.
Quantitative real-time PCR analysis
Total RNA was extracted from mouse tissues using 5-Prime PerfectPure RNA Tissue kit (Fisher Scientific Co., LLC). Total RNA from MEF cells was extracted as described above. cDNA was synthesized using iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). PCR amplification of the cDNA was performed by qPCR using qPCR Core kit for SYBR Green I (Applied Biosystems, Foster City, CA). The thermocycling protocol consisted of 10 min at 95 C, 40 cycles of 15 sec at 95 C, 30 sec at 61 C, and 20 sec at 72 C and finished with a melting curve ranging from 60 to 95 C to allow distinction of specific products. Primers were designed using Primer Express 3.0 software (Applied Biosystems) to amplify a region in intron 8 that was revealed in the initial screening for mGRβ. A common forward primer in exon 8 (AAAGAGCTAGGAAAAGCCATTGTC) was used in conjunction with a reverse primer in intron 8 for mGRβ (CTGTCTTTGGGCTTTTGAGATAGG) or a reverse primer in exon 9 for mGRα (TCAGCTAACATCTCTGGGAATTCA). Normalization was performed in separate reactions with primers to 18S mRNA (TTCGAACGTCTGCCCTATCAA and ATGGTAGGCACGGCGACTA). To study whether genomic sequences were amplified, a control sample was used, in which no reverse transcriptase was added (non-RT control). The following primers were used for endogenously expressed genes: serum- and glucocorticoid-inducible kinase, forward GAGAAGGATGGGCCTGAACGAT and reverse CGGACCCAGGTTGATTTGTTGA; GILZ, forward AATGCGGCCACGGATG and reverse GGACTTCACGTTTCAGTGGACA; PDK4, forward TTTCTCGTCTCTACGCCAAG and reverse GATACACCAGTCATCAGCTTCG; G6Pase, forward TGCAAGGGAGAACTCAGCAA and reverse GGACCAAGGAAGCCACAATG; and FKBP51, forward GCTGGCAAACAACACGAGAG and reverse GAGGAGGGCCGAGTTCATT. All primer sequences were uploaded to the PrimerFinder database at http://www.primerfinder.com/.
Generation of mGRβ antibody
A rabbit polyclonal antibody to mGRβ via the method used to produce antibody to hGRβ (33). A peptide corresponding to amino acids 733–748 (VSTKHKSKTTAKKKK) at the C terminus of the mGRβ protein was synthesized and purified by Pacific Immunology (Ramona, CA). An N-terminal cysteine was added to the peptide as a linker, followed by conjugation to a peptide carrier protein keyhole limpet hemocyanin and adjuvant-based immunization in a female New Zealand White rabbit. Preimmune serum was collected before injecting the rabbits with mGRβ conjugate peptide. The rabbits were boosted 2 wk after injection with the mGRβ peptide with Complete Freund’s Adjuvant and subsequently boosted two more times with Incomplete Freund’s Adjuvant every 2 wk. Serum was collected at 2 months and analyzed via ELISA for mGRβ specific antibodies. Serum of high titer was obtained and subjected to one round of affinity purification using the mGRβ peptide.
Generation of mGRβ shRNA lentiviral construct
To identify an small interfering RNA to knockdown mGRβ, a free Web-based tool (http://www.genelink.com/sirna/shRNAi. asp) was used to design a putative small interfering RNA against the mGRβ and to design oligonucleotides that encode a corresponding shRNA. The resulting shRNA recognized a sequence beginning at exon 8 within the mGR mRNA and extended into intron 8. XbaI and XhoI restriction sites were added to flanking regions of the sequence. Oligonucleotides were: GGACTCCATGCATGATGTAAGTACCAAACATCAAGAGTGTTTGGTACTTACATCATGCATGGAGTCTTTTTT and the homologous sequence. Synthetic oligonucleotides were annealed, digested with restriction enzymes, and then ligated into the XbaI/XhoI sites of the FG12 vector that has an independent green fluorescent protein (GFP) marker and transformed in DH5α cells (Invitrogen). Clones were selected and tested by transient transfection to determine knockdown of mGRβ. After confirmation of knockdown, the construct was cotransfected together with vectors expressing gag-pol, REV, and VSV-G into 293FT cells (Invitrogen) to generate a third generation lentiviral construct. Transfection was achieved using Lipofectamine 2000 (Invitrogen) using 100 ng total DNA per cm2 of the growth plate or well. The supernatants were harvested, and the cell debris was removed by centrifugation at 2000 × g. The supernatant was used to infect MEF cells after addition of polybrene (5 ng/ml; Sigma Chemical Co., St. Louis, MO) to establish cell lines with stable down-regulation of mGRβ mRNA or expressing empty vector. After 72 h the cells were sorted by flow cytometry for GFP by the Flow Cytometry Core Facility at the University of Toledo Health Science Campus. GFP positive cells were used for all experiments.
Transfection and reporter assays
Expression vector for mGRβ (pMGRβ-H57) was constructed as described above. The Ringold laboratory had already developed a plasmid for mGRα, pSV2Wrec (21). Both plasmids were transiently transfected into COS-7 cells (African green monkey kidney cells lacking an endogenous GR), and protein expression was measured via Western blot analysis. Dominant-negative activity was measured by luciferase assay using the GR-responsive minimal reporter pGRE2EIB-Luc (54) and pRL-CMV Renilla reporter for normalization to transfection efficiency. Transient transfection was achieved using Lipofectamine 2000. Twenty-four-hour posttransfected cells were treated with vehicle or 1 μm Dex or 1 μm RU486 for an additional 24 h until harvest. Cell lysates and assay were performed using the Promega luciferase assay system (Promega, Madison, WI). Statistical analyses employed the Student’s t test or ANOVA using GraphPad Prism version 5.0a for Mac (GraphPad Software, San Diego, CA).
Gel electrophoresis and Western blot analysis
Whole-cell extracts were prepared from COS-7 cells that were transiently transfected for 48 h with either pMGRβ-H57 or pSV2Wrec using Lipofectamine 2000. Control cells were untransfected COS-7 cells that do not express GR. Protein content was determined by BCA method of Pierce. Protein samples were resolved by SDS-PAGE and electrophoretically transferred to Immobilon-FL membranes. Membranes were blocked at room temperature for 1 h in Tris-buffered saline (TBS) [10 mm Tris-HCl (pH 7.4) and 150 mm NaCl] containing 3% BSA. Subsequently, the membrane was incubated overnight at 4 C with FiGR antibody for total GR or rMGRβ antibody for mGRβ at a dilution of 1:1000 in TBS. After three washes in TBS with 0.1% Tween 20, the membrane was incubated with an infrared antirabbit (IRDye 800, green) or antimouse (IRDye 680, red) secondary antibody labeled with IRDye infrared dye (LI-COR Biosciences, Lincoln, NE) (1:15,000 dilution in TBS) for 2 h at 4 C. Immunoreactivity was visualized and quantified by infrared scanning in the Odyssey system (LI-COR Biosciences).
Immunoadsorption of GR complexes
Cells were harvested in HEMG [10 mm HEPES, 3 mm EDTA, 20 mm sodium molybdate, and 10% glycerol (pH 7.4)] plus protease inhibitor cocktail and set on ice for 10 min followed by Dounce homogenization. Supernatants (cytosol) were collected proceeding a 10 min 4 C centrifugation at 20,800 × g, then precleared with protein A or G-Sepharose nutating for 1 h at 4 C. Samples were spun down, split into equal aliquots of cytosol, and immunoadsorbed overnight with FiGR antibodies against total GR, rMGRβ antibody for mGRβ, and appropriate controls (nonimmune mouse IgG or preimmune rabbit serum) at 4 C under constant rotation. Pellets were washed five to seven times with TEG [10 mm Tris, 3 mm EDTA, 10% glycerol, 50 mm NaCl, and 20 mm sodium molybdate (pH 7.4)], and complexes were eluted with 6× sodium dodecyl sulfate sample buffer.
Animals
Adult, male C57/BL6 mice maintained on a normal diet ad libitum or subjected to a fasting-refeeding regimen were used as tissue donors. Fasting encompassed 16 h (including the overnight 12-h dark cycle), followed by 8 h of refeeding ad libitum with normal chow at the start of the light cycle. All procedures were approved by the Institutional Animal Care and Utilization Committee of The University of Toledo.
Supplementary Material
Acknowledgments
We thank Rudel Saunders and Qiong Wu for advice on plasmid construction.
Footnotes
This work was supported by National Institutes of Health Grants DK70127 (to E.R.S.), DK54254, and DK83850 (to S.M.N.) and by the United States Department of Agriculture Grant 38903-02315 (to S.M.N.). T.D.H. was supported by the Predoctoral National Institutes of Health National Research Service Award F31DK84958.
Disclosure Summary: The authors have nothing to disclose.
First Published Online July 21, 2010
Abbreviations: BPS, Branch point sequences; BLAST, Basic Local Alignment Search Tool; Dex, dexamethasone; FKBP51, FK506 binding protein 51; GC, glucocorticoid hormone; GFP, green fluorescent protein; GILZ, glucocorticoid-inducible leucine zipper; G6Pase, glucose-6-phosphatase; GR, glucocorticoid receptor; hGR, human GR; MEF, mouse embryonic fibroblast; mGR, mouse GR; NCBI, National Center for Biotechnology Information; NF-κB, nuclear factor κB; PDK4, pyruvate dehydrogenase kinase-4; qPCR, quantitative real-time PCR; rMGRB-Ab, rabbit polyclonal antibody to mouse GRβ; shRNA, short hairpin RNA; SR, serine-arginine; TBS, Tris-buffered saline; zGR, zebrafish GR.
References
- Chrousos GP, Kino T 2005 Intracellular glucocorticoid signaling: a formerly simple system turns stochastic. Sci STKE 2005:pe48 [DOI] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, Cidlowski JA 2006 The physiology of human glucocorticoid receptor β (hGRβ) and glucocorticoid resistance. Ann NY Acad Sci 1069:1–9 [DOI] [PubMed] [Google Scholar]
- Sapolsky RM, Romero LM, Munck AU 2000 How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr Rev 21:55–89 [DOI] [PubMed] [Google Scholar]
- Vegiopoulos A, Herzig S 2007 Glucocorticoids, metabolism and metabolic diseases. Mol Cell Endocrinol 275:43–61 [DOI] [PubMed] [Google Scholar]
- Tata JR 2002 Signalling through nuclear receptors. Nat Rev Mol Cell Biol 3:702–710 [DOI] [PubMed] [Google Scholar]
- Kumar R, Johnson BH, Thompson EB 2004 Overview of the structural basis for transcription regulation by nuclear hormone receptors. Essays Biochem 40:27–39 [DOI] [PubMed] [Google Scholar]
- Bamberger CM, Bamberger AM, de Castro M, Chrousos GP 1995 Glucocorticoid receptor β, a potential endogenous inhibitor of glucocorticoid action in humans. J Clin Invest 95:2435–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro M, Elliot S, Kino T, Bamberger C, Karl M, Webster E, Chrousos GP 1996 The non-ligand binding β-isoform of the human glucocorticoid receptor (hGRβ): tissue levels, mechanism of action, and potential physiologic role. Mol Med 2:597–607 [PMC free article] [PubMed] [Google Scholar]
- Oakley RH, Jewell CM, Yudt MR, Bofetiado DM, Cidlowski JA 1999 The dominant negative activity of the human glucocorticoid receptor β isoform. Specificity and mechanisms of action. J Biol Chem 274:27857–27866 [DOI] [PubMed] [Google Scholar]
- Webster JC, Oakley RH, Jewell CM, Cidlowski JA 2001 Proinflammatory cytokines regulate human glucocorticoid receptor gene expression and lead to the accumulation of the dominant negative β isoform: a mechanism for the generation of glucocorticoid resistance. Proc Natl Acad Sci USA 98:6865–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DY, de Castro M, Szefler SJ, Chrousos GP 1998 Mechanisms of glucocorticoid-resistant asthma. Ann NY Acad Sci 840:735–746 [DOI] [PubMed] [Google Scholar]
- Longui CA, Vottero A, Adamson PC, Cole DE, Kino T, Monte O, Chrousos GP 2000 Low glucocorticoid receptor α/β ratio in T-cell lymphoblastic leukemia. Horm Metab Res 32:401–406 [DOI] [PubMed] [Google Scholar]
- Zhang X, Ognibene CM, Clark AF, Yorio T 2007 Dexamethasone inhibition of trabecular meshwork cell phagocytosis and its modulation by glucocorticoid receptor β. Exp Eye Res 84:275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM 1985 Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature 318:635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG 1997 A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–776 [DOI] [PubMed] [Google Scholar]
- Oakley RH, Sar M, Cidlowski JA 1996 The human glucocorticoid receptor β isoform. Expression, biochemical properties, and putative function. J Biol Chem 271:9550–9559 [DOI] [PubMed] [Google Scholar]
- Kino T, Manoli I, Kelkar S, Wang Y, Su YA, Chrousos GP 2009 Glucocorticoid receptor (GR)β has intrinsic, GRα-independent transcriptional activity. Biochem Biophys Res Commun 381:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, Jewell CM, Bienstock RJ, Collins JB, Cidlowski JA 2007 Human glucocorticoid receptor β binds RU-486 and is transcriptionally active. Mol Cell Biol 27:2266–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley RH, Webster JC, Sar M, Parker Jr CR, Cidlowski JA 1997 Expression and subcellular distribution of the β-isoform of the human glucocorticoid receptor. Endocrinology 138:5028–5038 [DOI] [PubMed] [Google Scholar]
- Schaaf MJ, Champagne D, van Laanen IH, van Wijk DC, Meijer AH, Meijer OC, Spaink HP, Richardson MK 2008 Discovery of a functional glucocorticoid receptor β-isoform in zebrafish. Endocrinology 149:1591–1599 [DOI] [PubMed] [Google Scholar]
- Danielsen M, Northrop JP, Ringold GM 1986 The mouse glucocorticoid receptor: mapping of functional domains by cloning, sequencing and expression of wild-type and mutant receptor proteins. EMBO J 5:2513–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop JP, Danielsen M, Ringold GM 1986 Analysis of glucocorticoid unresponsive cell variants using a mouse glucocorticoid receptor complementary DNA clone. J Biol Chem 261:11064–11070 [PubMed] [Google Scholar]
- Northrop JP, Gametchu B, Harrison RW, Ringold GM 1985 Characterization of wild type and mutant glucocorticoid receptors from rat hepatoma and mouse lymphoma cells. J Biol Chem 260:6398–6403 [PubMed] [Google Scholar]
- van der Vaart M, Schaaf MJ 2009 Naturally occurring C-terminal splice variants of nuclear receptors. Nucl Recept Signal 7:e007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto C, Reichardt HM, Schütz G 1997 Absence of glucocorticoid receptor-β in mice. J Biol Chem 272:26665–26668 [DOI] [PubMed] [Google Scholar]
- Root B, Abrassart J, Myers DA, Monau T, Ducsay CA 2008 Expression and distribution of glucocorticoid receptors in the ovine fetal adrenal cortex: effect of long-term hypoxia. Reprod Sci 15:517–528 [DOI] [PubMed] [Google Scholar]
- Bodwell JE, Ortí E, Coull JM, Pappin DJ, Smith LI, Swift F 1991 Identification of phosphorylated sites in the mouse glucocorticoid receptor. J Biol Chem 266:7549–7555 [PubMed] [Google Scholar]
- Burnstein KL, Jewell CM, Sar M, Cidlowski JA 1994 Intragenic sequences of the human glucocorticoid receptor complementary DNA mediate hormone-inducible receptor messenger RNA down-regulation through multiple mechanisms. Mol Endocrinol 8:1764–1773 [DOI] [PubMed] [Google Scholar]
- Schaaf MJ, Cidlowski JA 2002 Molecular mechanisms of glucocorticoid action and resistance. J Steroid Biochem Mol Biol 83:37–48 [DOI] [PubMed] [Google Scholar]
- Scheinman RI, Gualberto A, Jewell CM, Cidlowski JA, Baldwin Jr AS 1995 Characterization of mechanisms involved in transrepression of NF-κB by activated glucocorticoid receptors. Mol Cell Biol 15:943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldenhoven E, Liden J, Wissink S, Van de Stolpe A, Raaijmakers J, Koenderman L, Okret S, Gustafsson JA, Van der Saag PT 1995 Negative cross-talk between Rel A and the glucocorticoid receptor: a possible mechanism for the anti-inflammatory action of glucocorticoids. Mol Endocrinol 9:401–412 [DOI] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR 2001 Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414:799–806 [DOI] [PubMed] [Google Scholar]
- Hall RK, Wang XL, George L, Koch SR, Granner DK 2007 Insulin represses phosphoenolpyruvate carboxykinase gene transcription by causing the rapid disruption of an active transcription complex: a potential epigenetic effect. Mol Endocrinol 21:550–563 [DOI] [PubMed] [Google Scholar]
- Pierreux CE, Ursø B, De Meyts P, Rousseau GG, Lemaigre FP 1998 Inhibition by insulin of glucocorticoid-induced gene transcription: involvement of the ligand-binding domain of the glucocorticoid receptor and independence from the phosphatidylinositol 3-kinase and mitogen-activated protein kinase pathways. Mol Endocrinol 12:1343–1354 [DOI] [PubMed] [Google Scholar]
- Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM 1998 Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest 101:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar SM, Yang Y, Fernström MA, Lee SJ, Deangelis AM, Rjaily GA, Al-Share QY, Dai T, Miller TA, Ratnam S, Ruch RJ, Smith S, Lin SH, Beauchemin N, Oyarce AM 2005 Insulin acutely decreases hepatic fatty acid synthase activity. Cell Metab 2:43–53 [DOI] [PubMed] [Google Scholar]
- Kino T, Chrousos GP 2001 Glucocorticoid and mineralocorticoid resistance/hypersensitivity syndromes. J Endocrinol 169:437–445 [DOI] [PubMed] [Google Scholar]
- Strickland I, Kisich K, Hauk PJ, Vottero A, Chrousos GP, Klemm DJ, Leung DY 2001 High constitutive glucocorticoid receptor β in human neutrophils enables them to reduce their spontaneous rate of cell death in response to corticosteroids. J Exp Med 193:585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk PJ, Goleva E, Strickland I, Vottero A, Chrousos GP, Kisich KO, Leung DY 2002 Increased glucocorticoid receptor β expression converts mouse hybridoma cells to a corticosteroid-insensitive phenotype. Am J Respir Cell Mol Biol 27:361–367 [DOI] [PubMed] [Google Scholar]
- Sousa AR, Lane SJ, Cidlowski JA, Staynov DZ, Lee TH 2000 Glucocorticoid resistance in asthma is associated with elevated in vivo expression of the glucocorticoid receptor β-isoform. J Allergy Clin Immunol 105:943–950 [DOI] [PubMed] [Google Scholar]
- Goleva E, Li LB, Eves PT, Strand MJ, Martin RJ, Leung DY 2006 Increased glucocorticoid receptor β alters steroid response in glucocorticoid-insensitive asthma. Am J Respir Crit Care Med 173:607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulopoulos P, Leung DY, Elliott MW, Hogg JC, Muro S, Toda M, Laberge S, Hamid QA 2000 Increased number of glucocorticoid receptor-β-expressing cells in the airways in fatal asthma. J Allergy Clin Immunol 106:479–484 [DOI] [PubMed] [Google Scholar]
- Leung DY, Hamid Q, Vottero A, Szefler SJ, Surs W, Minshall E, Chrousos GP, Klemm DJ 1997 Association of glucocorticoid insensitivity with increased expression of glucocorticoid receptor β. J Exp Med 186:1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Lee EY, Cho YS, Moon KA, Yoo B, Moon HB 2005 Increased expression of glucocorticoid receptor β messenger RNA in patients with ankylosing spondylitis. Korean J Intern Med 20:146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston E, Darroch CE, Chaudhuri R, McPhee I, McMahon AD, Mackenzie SJ, Thomson NC 2004 Glucocorticoid receptor α:β ratio in blood mononuclear cells is reduced in cigarette smokers. J Allergy Clin Immunol 114:1475–1478 [DOI] [PubMed] [Google Scholar]
- Piotrowski P, Burzynski M, Lianeri M, Mostowska M, Wudarski M, Chwalinska-Sadowska H, Jagodzinski PP 2007 Glucocorticoid receptor β splice variant expression in patients with high and low activity of systemic lupus erythematosus. Folia Histochem Cytobiol 45:339–342 [PubMed] [Google Scholar]
- Whorwood CB, Donovan SJ, Wood PJ, Phillips DI 2001 Regulation of glucocorticoid receptor α and β isoforms and type I 11β-hydroxysteroid dehydrogenase expression in human skeletal muscle cells: a key role in the pathogenesis of insulin resistance? J Clin Endocrinol Metab 86:2296–2308 [DOI] [PubMed] [Google Scholar]
- Weinstein SP, Wilson CM, Pritsker A, Cushman SW 1998 Dexamethasone inhibits insulin-stimulated recruitment of GLUT4 to the cell surface in rat skeletal muscle. Metabolism 47:3–6 [DOI] [PubMed] [Google Scholar]
- Black DL 2003 Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336 [DOI] [PubMed] [Google Scholar]
- Zhu J, Gong JY, Goodman Jr OB, Cartegni L, Nanus DM, Shen R 2007 Bombesin attenuates pre-mRNA splicing of glucocorticoid receptor by regulating the expression of serine-arginine protein p30c (SRp30c) in prostate cancer cells. Biochim Biophys Acta 1773:1087–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XB, Tang CH, Huang Y, Fang H, Yu ZQ, Wu LM, Liu RY 2010 Alternative splicing in exon 9 of glucocorticoid receptor pre-mRNA is regulated by SRp40. Mol Biol Rep 37:1427–1433 [DOI] [PubMed] [Google Scholar]
- Kelly A, Bowen H, Jee YK, Mahfiche N, Soh C, Lee T, Hawrylowicz C, Lavender P 2008 The glucocorticoid receptor β isoform can mediate transcriptional repression by recruiting histone deacetylases. J Allergy Clin Immunol 121:203–208.e1 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim DH, Lavender P, Seo JH, Kim YS, Park JS, Kwak SJ, Jee YK 2009 Repression of TNF-α-induced IL-8 expression by the glucocorticoid receptor-β involves inhibition of histone H4 acetylation. Exp Mol Med 41:297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allgood VE, Oakley RH, Cidlowski JA 1993 Modulation by vitamin B6 of glucocorticoid receptor-mediated gene expression requires transcription factors in addition to the glucocorticoid receptor. J Biol Chem 268:20870–20876 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.