Abstract
Retroviral DNA integration into the host cell genome is an essential feature of the retroviral life cycle. The ability to integrate their DNA into the DNA of infected cells also makes retroviruses attractive vectors for delivery of therapeutic genes into the genome of cells carrying adverse mutations in their cellular DNA. Sequencing of the entire human genome has enabled identification of integration site preferences of both replication-competent retroviruses and retroviral vectors. These results, together with the unfortunate outcome of a gene therapy trial, in which integration of a retroviral vector in the vicinity of a protooncogene was associated with the development of leukemia, have stimulated efforts to elucidate the molecular mechanism underlying integration site selection by retroviral vectors, as well as the development of methods to direct integration to specific DNA sequences and chromosomal regions. This review outlines our current knowledge of the mechanism of integration site selection by retroviruses in vitro, in cultured cells, and in vivo; the outcome of several of the more recent gene therapy trials, which employed these vectors; and the efforts of several laboratories to develop vectors that integrate at predetermined sites in the human genome.
Introduction
Early steps of the retroviral life cycle include entry, reverse transcription, import of viral DNA and certain viral proteins into the cell nucleus, and finally, insertion of viral DNA into the host cell genome (Fig. 1; Flint et al., 2004). The joining of viral to host cell DNA is termed integration, and like the other early steps, is shared by both retroviruses and retroviral vectors. The ability to catalyze integration makes retroviral vectors a valuable tool for gene therapy. The inserted DNA becomes a part of the cellular genome and is thus stably maintained in the infected cells.
FIG. 1.
Early steps of the retroviral life cycle. After entry, the RNA genome of the retrovirus or retroviral vector undergoes reverse transcription in the cytoplasm of infected cells. Viral DNA, integrase (IN), and certain viral and cellular proteins constitute the preintegration complex, which is imported into the nucleus (Flint et al., 2004). IN then catalyzes the integration of viral DNA into the host cell DNA. As indicated by the asterisk (*), this process applies only to some retroviruses, such as human immunodeficiency virus (HIV)-1 and avian sarcoma virus (ASV; Flint et al., 2004). Murine leukemia virus (MLV) and MLV-based vectors cannot pass readily through the nuclear pore and access the host cell DNA unless the nuclear envelope is dissolved during mitosis (Roe et al., 1993). IN(n), integrase multimer.
The mechanism of integration (how viral DNA is joined to host cell DNA) has been thoroughly studied in vitro and the role of viral proteins in the process has been revealed to a large extent. In addition to viral proteins, integration also involves host cell proteins. Some of these have been identified. Their roles, as well as those of viral proteins, are discussed in the first section of this review. In contrast to the mechanism of integration, integration site selection, that is, where in the host cell genome integration occurs, and the process of selection of integration site(s), has been until recently less well understood. The integration site selection process has been studied in vitro, as well as in cultured cells, and in vivo, in both human and murine models. Studies taking advantage of the human genome project have identified integration site preferences for several species of retroviruses and retroviral vectors. These results, as well as our current understanding of the underlying molecular mechanism of integration site selection, are summarized in the second part of this review. The next section discusses the role integration site selection plays in gene therapy in light of results that point to highly significant clinical consequences of the process in several gene therapy trials. The final section of this review describes attempts to control the process by genetic manipulation of the proteins that are involved in integration.
Integration
The retroviral enzyme integrase (IN) plays an essential role in integration. This enzyme is contained in the virion and later the preintegration complex (see Fig. 1). IN works as a multimer (probably a tetramer) and can catalyze integration reactions in vitro, even in the absence of other viral or cellular proteins (Coffin et al., 1997; Flint et al., 2004). The integration reaction itself can be divided into two distinct steps. In the first step, IN removes two nucleotides from the 3′ ends of the viral DNA, the synthesis of which was completed by the viral enzyme reverse transcriptase (Fig. 2; Coffin et al., 1997; Flint et al., 2004). This step of integration is termed processing (Katz and Skalka, 1994; Coffin et al., 1997; Flint et al., 2004). The second step of integration, called joining, occurs when the viral preintegration complex reaches the host cell chromatin (see Figs. 1 and 2) (Coffin et al., 1997; Flint et al., 2004). During joining, IN catalyzes a coupled cleavage-joining reaction, in which the 3′ ends of viral DNA are joined to host cell DNA (Fig. 2). The product of the integration reaction is an intermediate, in which viral DNA is flanked by short single-stranded gaps in host DNA. The two steps of integration are then followed by postintegration repair, which includes trimming of the 5′ ends of viral DNA, filling of the single-stranded gaps in host DNA, ligation of the host cell DNA sequences to the 5′ ends of viral DNA, and, finally, reconstitution of appropriate chromatin structure at the integration site. Postintegration repair is not performed by IN, but requires host cell DNA repair proteins (Daniel et al., 1999; Lau et al., 2005; Skalka and Katz, 2005; Daniel, 2006; Smith and Daniel, 2006).
FIG. 2.
Retroviral DNA integration. For details see text. IN(n), integrase multimer.
Integration can be reconstituted in vitro with purified IN and model DNA substrates. Whereas most of these assays employ oligonucleotides, and only one end of a DNA substrate that represents viral DNA is joined to target DNA, some assays use miniviral DNA substrates and demonstrate concerted processing and joining of two viral ends (Flint et al., 2004). IN is thus sufficient to perform both steps of the integration reaction in vitro. In vivo, however, integration is likely to depend on host cell proteins that may facilitate the reaction. Several cellular proteins have attracted considerable interest as potential cofactors of integration. A yeast two-hybrid screen identified a human immunodeficiency virus (HIV)-1 IN-binding protein termed INI1 (integrase interactor 1; Kalpana et al., 1994). At first INI1 was shown to increase integration efficiency when added to the integration reaction in vitro (Kalpana et al., 1994). Experiments with small interfering RNA (siRNA) targeting INI1 (Ariumi et al., 2006) showed that HIV-1 replication was significantly reduced in cells with knocked-down INI1. However, integration was reported to occur in INI1-deficient cultured cells just as efficiently as when INI1 is present (Boese et al., 2004). Thus, it is now believed that INI1 does not seem to affect integration per se. However, INI1 has been demonstrated to be involved in other steps of the retroviral life cycle (Ariumi et al., 2006; Mahmoudi et al., 2006; Treand et al., 2006). Similar to INI1, a cellular high-mobility group protein-1 (HMG-1) was found to enhance integration in vitro (Aiyar et al., 1996). HMG proteins are nonhistone chromatin proteins and such enhancement could be due to their DNA-bending ability (Aiyar et al., 1996; Flint et al., 2004). A closely related protein, HMG-I(Y), was found in HIV-1 preintegration complexes isolated from infected cultured cells (Farnet and Bushman, 1997). As with HMG-1, HMG-I(Y), as well as HMG-2, enhance integration in vitro (Aiyar et al., 1996; Farnet and Bushman, 1997; Hindmarsh et al., 1999). However, experiments with cells lacking HMG-I(Y) failed to demonstrate a role for this protein in integration (Beitzel and Bushman, 2003).
The role of HMG proteins in integration thus remains unclear. The proper target for integration is host cell DNA, and integration into viral DNA itself, called autointegration, is expected to abort the retroviral life cycle. A biochemical analysis of murine leukemia virus (MLV) preintegration complexes identified the presence of a small protein (89 amino acids) that prevents autointegration of viral DNA, and was thus called the barrier-to-autointegration factor (BAF) (Lee and Craigie, 1998). BAF was also found in HIV-1 preintegration complexes, also blocking autointegration (Lin and Engelman, 2003). Finally, in 2003, use of the yeast two-hybrid system led to the isolation of a new HIV-1 IN-binding protein, a previously identified cellular protein termed LEDGF/p75 (lens epithelium-derived growth factor; Cherepanov et al., 2003). Ironically, knockout experiments demonstrated that LEDGF/p75 is not a lens growth factor (Sutherland et al., 2006). Instead, animals lacking the mouse LEDGF/p75 homolog, PSIP1 (PC4 and SFRS1-interacting protein-1), have skeletal abnormalities, indicating that this protein is involved in bone development (Sutherland et al., 2006). However, suppression of LEDGF/p75 with siRNA, as well as experiments with primary LEDGF/p75 null cells from the LEDGF/p75 null transgenic animals, showed that integration of HIV-1-based vectors is reduced 89–96% in the absence of LEDGF/p75 (Llano et al., 2006a; Shun et al., 2007). Therefore, LEDGF/p75 is required for efficient integration of HIV-1 DNA. Interestingly, LEDGF/p75 does not bind to MLV IN, nor is it required for MLV integration (Llano et al., 2004b; Busschots et al., 2005; Shun et al., 2007). LEDGF/p75 enhances integration in vitro and, in addition to this effect, plays a major role in integration site selection of HIV-1 and HIV-1-based vectors, as discussed below (Ciuffi et al., 2005, 2006; Llano et al., 2006b; Shun et al., 2007).
In summary, retroviral DNA integration is catalyzed by the viral protein integrase, but host cell proteins appear to play a significant role in enhancing the efficiency of the reaction, and in preventing autointegration.
Integration Site Preferences of Retroviruses and Retroviral Vectors
Integration can occur anywhere throughout the host cell genome and there are no strict host sequence requirements for site selection. However, integration is not random (Schroder et al., 2002; Wu et al., 2003; Mitchell et al., 2004). In vitro systems, which employ purified IN and naked target DNA, demonstrated that certain DNA-binding proteins can prevent access of IN to target DNA and thus block integration at their binding sites (Pryciak and Varmus, 1992; Bushman, 1994). In contrast, bending or distortion of target DNA appears to stimulate integration (Pryciak and Varmus, 1992; Pryciak et al., 1992a; Pruss et al., 1994a,b; Katz et al., 1998). When naked target DNA is replaced by DNA that is partly wrapped around nucleosomes, distortion of DNA promotes integration within the nucleosome-bound DNA (Pryciak and Varmus, 1992; Pryciak et al., 1992b; Pruss et al., 1994a). These systems thus reveal certain integration site preferences within model DNA substrates. However, as host DNA exists in a higher order chromatin structure, the results of these in vitro studies may not be relevant to events that take place in infected cells. To remedy this deficiency of in vitro systems, one system employed an extended 13-nucleosome array, which could be compacted into a higher order chromatin structure by addition of the histone H1 (Taganov et al., 2004). It was observed that the chromatin structure affects integration site selection of HIV-1 and avian sarcoma virus (ASV) IN proteins in opposite ways. Specifically, HIV-1 IN-mediated integration was decreased after compaction of the target chromatin, whereas ASV IN-mediated integration was more efficient after compaction. These data suggested that a higher order chromatin structure may indeed affect site selection, and different retroviral species may exhibit different integration preferences.
There are approximately 25.000 genes in the human genome (International Human Genome Sequencing Consortium, 2004). Early studies suggested that retroviruses may integrate in or in the vicinity of transcription units (Mooslehner et al., 1990; Scherdin et al., 1990). However, these studies were hampered by a relatively low number of the identified transcription sites (Bushman et al., 2005). Moreover, it was not clear what percentage of the genome contains these “favored” integration sites. The situation changed after the sequencing of the human genome was completed, which enabled a true statistical analysis of integration sites. Large-scale analyses of integration site selection of HIV-1 in human T cell lines demonstrated that approximately 70% of integration events occurred in genes (Schroder et al., 2002; Bushman et al., 2005). As about 30% should be expected if integration were random, this preference for genes is highly significant. In addition, some integration “hotspots” were found (11q13 chromosomal region). No preferences were detected within transcription units. Similar data were obtained with pseudotyped HIV-1-based vectors (Schroder et al., 2002). Integration preferences of the related simian immunodeficiency virus (SIV), an SIV-based vector, and HIV-2 closely resemble that of HIV-1 (Hematti et al., 2004; Crise et al., 2005; MacNeil et al., 2006). Integration preferences of the feline immunodeficiency virus (FIV) also resemble HIV-1 preferences (Kang et al., 2006).
MLV shows different integration preferences compared with HIV-1. Approximately 20% of integration events occur in the vicinity of the 5′ ends of transcription units (Wu et al., 2003). The remaining integration sites are distributed in a somewhat random fashion. In addition, approximately 17% of MLV integration events occur in the vicinity of CpG islands (Mitchell et al., 2004), and an increased frequency of integration was also noted in the vicinity of DNase I-hyper-sensitive sites (11%; Lewinski et al., 2006). Similar data were obtained with MLV-based vectors (Wu et al., 2003; Mitchell et al., 2004). Finally, a preference, albeit a weak one, for integration near transcription start sites was shown by foamy viruses (FVs; Trobridge et al., 2006; Beard et al., 2007). FV also shows a preference for CpG islands that is similar to that of MLV (Trobridge et al., 2006).
Avian retroviruses and vectors exhibit yet another integration preference: only a weak preference for genes was detected (about 40%) and no MLV-like preference for 5′ ends of transcription units (Mitchell et al., 2004; Narezkina et al., 2004). Interestingly, high levels of transcription may even inhibit ASV integration in genes (Weidhaas et al., 2000; Maxfield et al., 2005). These preferences are consistent with the above-described data from the in vitro system, which used nucleosomal arrays (Taganov et al., 2004). Interestingly, the human T-leukemia virus type 1 (HTLV-1) and mouse mammary tumor virus (MMTV), like avian retroviruses, do not specifically target genes and transcription start sites (Derse et al., 2007; Faschinger et al., 2008).
Finally, careful examination of a large number of integration sites from HIV-1, SIV, MLV, and avian sarcoma-leukosis viruses revealed symmetric base preferences surrounding integration sites (Holman and Coffin, 2005; Wu et al., 2005). These weak consensus sequences are virus specific and possibly reflect the influence of IN on integration site selection (Holman and Coffin, 2005). This hypothesis is also supported by the symmetry of the target site sequence, because IN likely functions as a tetramer (Coffin et al., 1997; Flint et al., 2004; Wu et al., 2005; and see above).
In summary, integration preferences described in this section are distinct for different groups of retroviruses. The first group, exemplified by HIV-1 and including HIV-2, SIV, and FIV, preferentially integrates into genes. The second group consists of MLV and FV. These retroviruses show preferences for 5′ ends of transcription units and CpG islands. Finally, the last group includes ASV, HTLV-1, and MMTV. Members of this group show weak or no preferences for genes or transcription start sites. In addition to these preferences, the specific DNA sequence appears to play a role in integration site selection, but the primary mechanism seems to be independent of the DNA sequence, and likely includes cellular cofactors and other cellular structures. The next section summarizes our current understanding of virus–cell interactions that partake in integration site selection.
Mechanism of Integration Site Selection
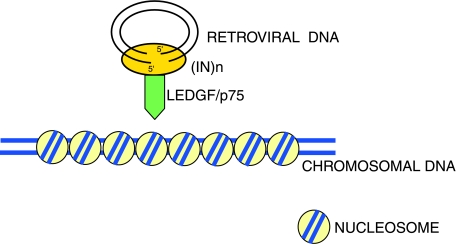
As noted previously, IN shows low specificity for binding to host cell DNA. Therefore, it would seem natural for host cell proteins to participate in the integration process. In 2003, Debyser and coworkers used the yeast two-hybrid system to identify a new HIV-1 IN-binding protein, termed LEDGF/p75 (Cherepanov et al., 2003). As noted above, LEDGF/p75 is required for efficient integration of HIV-1 DNA. A molecular analysis showed that LEDGF/p75 is a transcription factor and has a C-terminal IN-binding domain and N-terminal chromatin-binding domain (Maertens et al., 2003; Cherepanov et al., 2004; Vanegas et al., 2005; Llano et al., 2006b; Turlure et al., 2006). Chromatin binding is mediated by PWWP and AT-hook motifs in the N-terminal domain of LEDGF/p75 (Llano et al., 2006b; Turlure et al., 2006). In cultured cells, LEDGF/p75 was found associated with preintegration complexes of HIV-1 and FIV (Llano et al., 2004b). In addition to stimulating IN activity in vitro, LEDGF/p75 appears to prevent degradation of ectopically expressed HIV-1 IN by the proteasome, and therefore might contribute to the stability of preintegration complexes during infection (Maertens et al., 2003; Llano et al., 2004a). In addition to these potential LEDGF/p75 effects on IN and integration, an analysis of the integration sites in LEDGF/p75 null cells showed that the residual integration in these cells no longer occurs preferentially in active genes (Shun et al., 2007). Instead, integration occurred preferentially in the vicinity of promoters and CpG islands (Shun et al., 2007). The symmetric base preferences surrounding the integration site remained preserved (Holman and Coffin, 2005; Shun et al., 2007). Thus, in the absence of LEDGF/p75, HIV-1 integration site preferences resemble those of MLV (Shun et al., 2007). Taken together, these results strongly support the hypothesis that LEDGF/p75 targets HIV-1 (and other lentiviral) integration into active genes by tethering the IN protein to chromatin (Fig. 3).
FIG. 3.
LEDGF/p75 role in integration site selection. LEDGF/p75 binds to specific sites, thereby mediating contact between IN and host cell chromatin. This association tethers the HIV-1 preintegration complex near or in genes.
Although LEDGF/p75 appears to be a major HIV-1 IN-binding cellular protein, other factors are likely involved in integration site selection by HIV-1 and HIV-1-based vectors. Analysis of a large number of integration sites showed that favored integration sites occur in the vicinity of certain computer-predicted epigenetic marks, such as histone H3 K4 methylation, H4 acetylation, or H3 acetylation (Wang et al., 2007). Taken together, these data may suggest that the chromatin structure, including the histone code, may also affect integration site selection (Fig. 4). However, final proof that these marks play a role in integration site selection has yet to be shown. Additional factors that may affect integration site selection have been identified. Knockdown of the T-cell lineage-specific chromatin organizer, SATB1 (special AT-rich sequence-binding protein-1), reduces HIV-1 integration near SATB1-binding sites (Kumar et al., 2007). SATB1 thus appears to be involved in integration site selection, by an unknown mechanism. Finally, it has been suggested that the cellular protein Ku80, which is present in the preintegration complex, targets integration toward chromatin domains prone to silencing (Li et al., 2001; Masson et al., 2007).
FIG. 4.
A possible role for chromatin structure in HIV-1 DNA integration. HIV-1 DNA integration occurs preferentially in the vicinity of certain epigenetic modifications, such as H3 K4 methylation (H3K4Me) and H3 and H4 acetylation (H3-Ac and H4-Ac, respectively). Integration seems to be disfavored in the vicinity of H3K27 trimethylation (H3K27Me3) and CpG islands in the DNA (see text and Wang et al., 2007).
In contrast to HIV-1, integration of MLV-based and ASV-based vectors does not seem to be influenced by LEDGF/p75 (see above; and see Mitchell et al., 2004; Narezkina et al., 2004). What determines ASV integration site selection is not known. In the case of MLV, a study of HIV chimeras with MLV genes showed that MLV IN seems to be the principal determinant of integration site selection (Lewinski et al., 2006). Interestingly, Gag-derived proteins play an auxiliary role in the process, as an HIV chimera containing MLV Gag displayed targeting preferences different from those of both HIV and MLV (Lewinski et al., 2006). These results thus support a different mechanism of integration site selection for MLV versus HIV. Finally, in addition to Gag proteins, other viral proteins could play a role in integration site selection. However, examination of auxiliary proteins in SIV integration showed that Vif, Vpr, Vpx, Nef, Env, and promoter or enhancer regions are not required for preferential SIV integration into genes (Monse et al., 2006).
In summary, more recent results have increased our understanding of the mechanism of retroviral integration site selection and show a major role of host cell proteins in the process. However, the process has yet to be completely understood and it is likely that new players in retroviral integration site selection will be revealed in future studies.
Integration Site Selection and Gene Therapy
Gene therapy trials, as well as gene therapy experiments in animal systems, commonly employ vectors that are based on either MLV or HIV-1. What could be the significance of the integration preferences of these vectors in gene therapy trials?
The tendency of MLV-based vectors to integrate at the 5′ end of transcription units, and HIV-1 preferences for genes, could suggest increased danger of an adverse event during gene therapy, due to either activation or disruption of a cellular gene which could potentially stimulate tumorigenesis. Alternatively, it is possible that integration at an undesirable site occurs at such a low frequency that it may not affect a therapeutic outcome. In this context, it is important to note that carcinogenesis is a multistep process and even if integration occurs in a “wrong spot,” it may not lead to tumor development (Hahn and Weinberg, 2002; Baum et al., 2006).
Supporting evidence for the hypothesis that integration may have adverse consequences was provided by a human gene therapy trial involving children with X-linked severe combined immunodeficiency (SCID-X1) (Gunzburg, 2003; Hacein-Bey-Abina et al., 2003; Alexander et al., 2007; Bushman, 2007; Deichmann et al., 2007). In this trial, which used an MLV-based vector, 4 of 11 patients developed T cell leukemia. In addition, it has been reported that a single patient (of 10 enrolled) developed leukemia in another SCID-X1 gene therapy trial (Alexander et al., 2007; Schwarzwaelder et al., 2007; Thrasher and Gaspar, 2007). A sequencing analysis of the T cells from two of the patients in the first trial, who developed leukemia first, demonstrated clonal expansion of T cell clones that contained an insertion of the vector in the vicinity of (and subsequent activation of) the Lin-1, Isl-1, Mec-3 (LIM) domain only-2 (LMO2) protooncogene by the long terminal repeat (LTR) enhancer of the vector (Hacein-Bey-Abina et al., 2003). In addition, it appears that insertion in the vicinity of the LMO2 protooncogene also occurred in the patient from the second trial (Thrasher and Gaspar, 2007). These results strongly suggest that vector integration at a “dangerous” location in the human genome contributed to the development of leukemia in these patients. There could be other factors that played a role in the development of leukemia and have not yet been fully delineated. These may include expression of the transgene and a chromosomal rearrangement (Hacein-Bey-Abina et al., 2003; Pike-Overzet et al., 2006; Thrasher et al., 2006; Woods et al., 2006). Nevertheless, the association of leukemia with integration in the vicinity of a protooncogene suggests a high significance of integration site selection for the success or failure of gene therapy approaches using retroviral vectors.
A follow-up analysis of patients in these trials showed nonrandom distribution of integration sites in vivo (Deichmann et al., 2007; Schwarzwaelder et al., 2007). Integrations were found to occur preferentially near the 5′ ends of genes and associated CpG islands (Bushman, 2007; Deichmann et al., 2007; Schwarzwaelder et al., 2007). This is consistent with results obtained with MLV in cultured cells (see above). Comparison of integration sites in vector-transduced cells before infusion into patients, with integration sites in cells that were recovered from patients after infusion, indicated that vector integration likely influences cell engraftment, survival, and proliferation in vivo (Deichmann et al., 2007; Schwarzwaelder et al., 2007). Likewise, clonal evolution was observed in an ADA-SCID gene therapy trial (Aiuti et al., 2007). However, in this case, no adverse effects were associated with vector integration (Aiuti et al., 2007). In a similar vein, integration was observed to deregulate gene expression, but did not lead to development of leukemias in other gene therapy trials (Ott et al., 2006; Recchia et al., 2006). In addition, the effect of integration sites on the outcome of gene therapy experiments in animal models generally resembled those observed in human gene therapy trials (Li et al., 2002; Calmels et al., 2005; Kustikova et al., 2005; Modlich et al., 2005; Montini et al., 2006).
In summary, insertion of retroviral DNA into certain chromosomal locations was associated with the development of leukemias in both animal models and human gene therapy trials. However, these insertions may not be sufficient to induce malignant transformation and other events may contribute to the development of malignancies in these cases (Hacein-Bey-Abina et al., 2003; Dave et al., 2004). Nevertheless, these cases highlight a necessity for further improvements in the design of retroviral vectors so that they are less likely to integrate at undesirable sites in the human genome, thereby having an increased safety margin.
Efforts to Direct Integration to Predetermined DNA Sequences and Chromosomal Regions
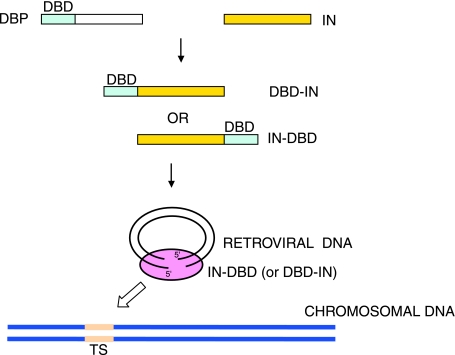
Given the potential of retroviral vectors to integrate into undesirable sites in the human genome, it is not surprising that efforts in several laboratories have been directed toward the development of a vector that would integrate in a predetermined DNA sequence or chromosomal region. Because integration is catalyzed by IN, this protein became the focus of early efforts. Three laboratories (Bushman, 1994; Goulaouic and Chow, 1996; Katz et al., 1996) initially used a strikingly similar approach to the problem. All three groups constructed fusion proteins, which consisted of IN protein either from HIV-1 or ASV, and a DNA-binding sequence from a cellular or bacterial protein. In two cases, the DNA-binding domain (DBD) was from the Escherichia coli LexA repressor (Goulaouic and Chow, 1996; Katz et al., 1996), and in one case it was from the phage λ repressor (Bushman, 1994), fused to the N or C terminus of IN. The resulting fusion proteins directed integration to their target sites in DNA in vitro (Bushman, 1994; Goulaouic and Chow, 1996; Katz et al., 1996). Targeting integration in cultured cells, however, proved to be more difficult. The ASV IN–LexA fusion proved to be a target for the viral protease protein, which deleted a majority of the heterologous DBD (Katz et al., 1996). We then attempted to mutate the protease recognition site between the DBD and the rest of the protein. The resulting mutant protein indeed proved to be resistant to viral protease in vitro and stable when the mutant gene was transfected together with the rest of viral DNA into chicken DF-1 cells; however, we did not obtain any viral particles containing the mutant protein, possibly because of a failure to incorporate into the virion (R. Daniel and A.M. Skalka, unpublished data). Similar problems were encountered when the LexA DBD was replaced with the DBD of the cellular GATA-6 protein (R. Daniel and A.M. Skalka, unpublished data). In another report, HIV-1 IN was fused to the zinc finger protein zif268 (Bushman and Miller, 1997). This fusion protein was incorporated into HIV-1 virions, but the virus lost its infectivity. However, viruses containing mixtures of wild-type IN and the fusion protein IN–zif268 were infective. Preintegration complexes of these viruses, when purified from cells, were able to target integration near zif268 recognition sites in vitro (Bushman and Miller, 1997). It is not clear whether these viruses exhibited the same properties in cultured cells.
Chow and coworkers demonstrated that an HIV-1 IN–LexA fusion protein can target integration to LexA-binding sequences in vitro (Goulaouic and Chow, 1996). Direct cloning in the HIV-1 IN gene region is difficult because of an overlap with the open reading frame of vif and the presence of a splice acceptor site (Purcell and Martin, 1993). Thus, these investigators took advantage of the packaging properties of the HIV-1 Vpr protein, which enable other proteins to be incorporated into HIV-1 particles in trans, when they are fused to Vpr (Wu et al., 1995). Vpr is incorporated into the particle through an interaction with the C terminus of the p6 protein in Gag (for review and summary articles on particle formation see Lu et al., 1995; Kondo and Gottlinger, 1996; Flint et al., 2004; Hill et al., 2005). It should be noted that there are about 100–200 copies of Vpr and approximately 50–100 copies of IN per virion (Flint et al., 2004). The wild-type IN protein in these viruses was inactivated by the D64V mutation in the IN catalytic site and the IN–LexA protein was then fused to Vpr so it would be incorporated into the virion. The viral particle carrying IN–LexA could infect and integrate its DNA in host cells, at an efficiency of 17 to 24% of the wild-type virus (Holmes-Son and Chow, 2000). To understand the mechanism of complementation, virus–host DNA junctions from cells infected with these viruses were then cloned and sequenced. Correct integration hallmarks were observed, including the duplication of host DNA sequence flanking the provirus, as well as 5′-TG/CA-3′ ends of viral DNA (Holmes-Son and Chow, 2000). LexA binds to its operator DNA sequence with a relatively low specificity (Lewis et al., 1994). To increase specificity, IN was then fused to a designed polydactyl zinc finger protein E2C, which binds to a unique 18-bp sequence in the human genome (Beerli et al., 1998, 2000; Tan et al., 2004). These fusion proteins again targeted integration preferentially to the E2C-binding site in vitro. These proteins were then incorporated into virions in trans, as with IN–LexA fusion proteins (Tan et al., 2006). Viruses containing E2C fused to the C terminus of IN had 7-fold higher preference for integrating near the E2C-binding site than viruses containing wild-type IN. Viruses containing E2C fused to the N terminus of IN had a 10-fold higher preference for the E2C-binding site when compared with wild-type IN. Therefore, these fusion proteins can affect integration site selection in cultured cells. However, it should be noted that the overall efficiency of retroviral DNA integration in this case was 4- to 100-fold lower than with wild-type IN, which is an undesirable side effect (Tan et al., 2006). Moreover, integration in the vicinity of the E2C site is still relatively rare even when compared with the overall number of integration events in these cells (up to 1.5% of the total integration sites when the N-terminal fusion protein is used; Tan et al., 2006). However, these experiments provide proof-of-principle evidence that it is possible to target integration in cultured cells by manipulation of the IN protein (Fig. 5).
FIG. 5.
A model for targeting integration by fusion proteins of integrase and a heterologous DNA-binding domain. A DNA-binding domain (DBD) of a cellular DNA-binding protein (DBP) can be fused to either the N terminus or C terminus of IN. The resulting fusion protein can be incorporated into the virion and the preintegration complex, and targets integration toward the DNA sequences recognized by the DBD, the target site (TS).
As described above, LEDGF/p75 is a major cellular cofactor for integration of HIV-1 DNA. Therefore, hypothetically it should be possible to target integration by manipulation of this cellular protein. One approach is to use fusion proteins that contain the LEDGF/p75 IN-binding domain and a DNA-binding domain of a transcription factor of known specificity. Indeed, it had been shown that a fusion of LEDGF/p75 with the DNA-binding domain of phage λ repressor directs integration to λR-binding sites in vitro (Ciuffi et al., 2006). However, it remains to be seen whether these proteins can target integration in cells.
Finally, a new and innovative approach has been described that uses engineered zinc finger nucleases (ZFNs) and integrase-defective lentiviral vectors (IDLVs) to target gene addition to specific regions (Lombardo et al., 2007). ZFNs are designed to induce a double-stranded DNA break in the chromosome of a target cell, at a predetermined location (Kim et al., 1996). Two different ZFNs are required for a break to occur, because of a requirement for dimer formation (Bitinaite et al., 1998). This break is then repaired by the cellular DNA repair system, which involves either nonhomologous end joining or homologous recombination. The latter repair system copies a homologous DNA sequence during the repair process (Cahill et al., 2006). ZFNs were shown to mediate integration of extrachromosomal DNA into a specified location when expressed in the target cell. The extrachromosomal DNA, which was delivered by transfection, carried locus-specific homology arms, in addition to a gene of interest (Moehle et al., 2007). To improve efficiency of the ZFN gene delivery system, the Naldini laboratory used the above-mentioned IDLVs, which carry an inactivating mutation in the IN gene (Lombardo et al., 2007). IDLVs cannot integrate; however, they can still perform reverse transcription and a gene of interest can be expressed, albeit transiently, from unintegrated DNA. Two different ZFNs and a gene of interest were subcloned into two IDLVs, which were then used to cotransduce a variety of cell types. Between 5 and 50% of infected cells were stably transduced by this method, with gene addition occurring at the ZFN target site (Lombardo et al., 2007). The specificity of this ZFN method thus compares favorably with the above-described method, which uses IN fusion proteins.
In summary, a persistent effort by several laboratories has led to the development of a variety of systems for potential targeting of integration to predetermined regions of the human genome. These encouraging results may lead to safer methods used to employ retroviral vectors in gene therapy applications. New therapeutic approaches that take advantage of the novel properties of these systems are promising, and are being explored.
Summary and Future Directions
Results from a variety of experiments have increased our understanding of the integration preferences of retroviruses and retroviral vectors, as well as of the molecular mechanism underlying integration site selection. Clinical evidence shows that integration at an undesirable site of the patient's genome can have negative consequences for the outcome of the gene therapy-based treatment. Therefore, new approaches are needed to increase the safety of retroviral vectors. Among the possibilities outlined in this review are a switch to retroviral vectors that are based on avian, foamy, and possibly HIV-1 retroviruses rather than MLV; genetic manipulation of the vector integrases and/or cellular factors that are involved in integration; and use of IDLV vectors that carry designed zinc finger nucleases in order to target integration to predetermined chromosomal regions. It is hoped that these research directions will result in new vectors, which may minimize the risk of adverse events in gene therapy applications.
Acknowledgments
The authors thank Drs. Richard Katz and Anna Marie Skalka for reading the manuscript and providing helpful comments. This work has been supported by NCI grants CA98090 and CA125272 and by a W.W. Smith Foundation AIDS Research Award to R.D.
Author Disclosure Statement
No competing financial interests exist.
References
- Aiuti A. Cassani B. Andolfi G. Mirolo M. Biasco L. Recchia A. Urbinati F. Valacca C. Scaramuzza S. Aker M. Slavin S. Cazzola M. Sartori D. Ambrosi A. Di Serio C. Roncarolo M.G. Mavilio F. Bordignon C. Multilineage hematopoietic reconstitution without clonal selection in ADASCID patients treated with stem cell gene therapy [see comment] J. Clin. Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar A. Hindmarsh P. Skalka A.M. Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: Dependence on both long terminal repeat termini. J. Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander B.L. Ali R.R. Alton E.W. Bainbridge J.W. Braun S. Cheng S.H. Flotte T.R. Gaspar H.B. Grez M. Griesenbach U. Kaplitt M.G. Ott M.G. Seger R. Simons M. Thrasher A.J. Thrasher A.Z. Ylä-Herttuala S. Severe combined immunodeficiency, progress and prospects: Gene therapy clinical trials (part 1) Gene Ther. 2007;14:1439–1447. doi: 10.1038/sj.gt.3303001. [Erratum in Gene Ther. 2007;14:1754.]. [DOI] [PubMed] [Google Scholar]
- Ariumi Y. Serhan F. Turelli P. Telenti A. Trono D. The integrase interactor 1 (INI1) proteins facilitate Tat-mediated human immunodeficiency virus type 1 transcription. Retrovirology. 2006;3:47. doi: 10.1186/1742-4690-3-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum C. Kustikova O. Modlich U. Li Z. Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum. Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- Beard B.C. Keyser K.A. Trobridge G.D. Peterson L.J. Miller D.G. Jacobs M. Kaul R. Kiem H.P. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, or foamy virus. Hum. Gene Ther. 2007;18:423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- Beerli R.R. Segal D.J. Dreier B. Barbas C.F., III Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli R.R. Schopfer U. Dreier B. Barbas C.F., III Chemically regulated zinc finger transcription factors. J. Biol. Chem. 2000;275:32617–32627. doi: 10.1074/jbc.M005108200. [DOI] [PubMed] [Google Scholar]
- Beitzel B. Bushman F. Construction and analysis of cells lacking the HMGA gene family. Nucleic Acids Res. 2003;31:5025–5032. doi: 10.1093/nar/gkg684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaite J. Wah D.A. Aggarwal A.K. Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boese A. Sommer P. Gaussin A. Reimann A. Nehrbass U. Ini1/hSNF5 is dispensable for retrovirus-induced cytoplasmic accumulation of PML and does not interfere with integration. FEBS Lett. 2004;578:291–296. doi: 10.1016/j.febslet.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Bushman F. Lewinski M. Ciuffi A. Barr S. Leipzig J. Hannenhalli S. Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- Bushman F.D. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F.D. Retroviral integration and human gene therapy [see comment] J. Clin. Invest. 2007;117:2083–2086. doi: 10.1172/JCI32949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F.D. Miller M.D. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J. Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busschots K. Vercammen J. Emiliani S. Benarous R. Engelborghs Y. Christ F. Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- Cahill D. Connor B. Carney J.P. Mechanisms of eukaryotic DNA double strand break repair. Front. Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- Calmels B. Ferguson C. Laukkanen M.O. Adler R. Faulhaber M. Kim H.J. Sellers S. Hematti P. Schmidt M. Von Kalle C. Akagi K. Donahue R.E. Dunbar C.E. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. Maertens G. Proost P. Devreese B. Van Beeumen J. Engelborghs Y. De Clercq E. Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P. Devroe E. Silver P.A. Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- Ciuffi A. Llano M. Poeschla E. Hoffmann C. Leipzig J. Shinn P. Ecker J.R. Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Ciuffi A. Diamond T.L. Hwang Y. Marshall H.M. Bushman F.D. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum. Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- Coffin J.M. Hughes S.H. Varmus H.E. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. [PubMed] [Google Scholar]
- Crise B. Li Y. Yuan C. Morcock D.R. Whitby D. Munroe D.J. Arthur L.O. Wu X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 2005;79:12199–12204. doi: 10.1128/JVI.79.19.12199-12204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R. DNA repair in HIV-1 infection: A case for inhibitors of cellular co-factors? Curr. HIV Res. 2006;4:411–421. doi: 10.2174/157016206778560027. [DOI] [PubMed] [Google Scholar]
- Daniel R. Katz R.A. Skalka A.M. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- Dave U.P. Jenkins N.A. Copeland N.G. Gene therapy insertional mutagenesis insights. Science. 2004;303:333. doi: 10.1126/science.1091667. [DOI] [PubMed] [Google Scholar]
- Deichmann A. Hacein-Bey-Abina S. Schmidt M. Garrigue A. Brugman M.H. Hu J. Glimm H. Gyapay G. Prum B. Fraser C.C. Fischer N. Schwarzwaelder K. Siegler M.L. De Ridder D. Pike-Overzet K. Howe S.J. Thrasher A.J. Wagemaker G. Abel U. Staal F.J. Delabesse E. Villeval J.L. Aronow B. Hue C. Prinz C. Wissler M. Klanke C. Weissenbach J. Alexander I. Fischer A. Von Kalle C. Cavazzana-Calvo M. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy [see comment] J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D. Crise B. Li Y. Princler G. Lum N. Stewart C. McGrath C.F. Hughes S.H. Munroe D.J. Wu X. Human T-cell leukemia virus type 1 integration target sites in the human genome: Comparison with those of other retroviruses. J. Virol. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnet C.M. Bushman F.D. HIV-1 cDNA integration: Requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- Faschinger A. Rouault F. Sollner J. Lukas A. Salmons B. Gunzburg W.H. Indik S. Mouse mammary tumor virus integration site selection in human and mouse genomes. J. Virol. 2008;82:1360–1367. doi: 10.1128/JVI.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint S.E. Racaniello V.R. Skalka A.M. Principles of Virology. ASM Press; Washington, D.C: 2004. [Google Scholar]
- Goulaouic H. Chow S.A. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J. Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzburg W.H. Retroviral gene therapy: Where now? Trends Mol. Med. 2003;9:277–278. doi: 10.1016/s1471-4914(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M. McCormack M.P. Wulffraat N. Leboulch P. Lim A. Osborne C.S. Pawliuk R. Morillon E. Sorensen R. Forster A. Fraser P. Cohen J.I. De Saint Basile G. Alexander I. Wintergerst U. Frebourg T. Aurias A. Stoppa-Lyonnet D. Romana S. Radford-Weiss I. Gross F. Valensi F. Delabesse E. Macintyre E. Sigaux F. Soulier J. Leiva L.E. Wissler M. Prinz C. Rabbitts T.H. Le Deist F. Fischer A. Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCIDX1 [see comment] Science. 2003;302:415–419. doi: 10.1126/science.1088547. Erratum appears in Science 2003;302:568. [DOI] [PubMed] [Google Scholar]
- Hahn W.C. Weinberg R.A. Rules for making human tumor cells. N. Engl. J. Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. Erratum appears in N. Engl. J. Med. 2003;348:674. [DOI] [PubMed] [Google Scholar]
- Hematti P. Hong B.K. Ferguson C. Adler R. Hanawa H. Sellers S. Holt I.E. Eckfeldt C.E. Sharma Y. Schmidt M. Von Kalle C. Persons D.A. Billings E.M. Verfaillie C.M. Nienhuis A.W. Wolfsberg T.G. Dunbar C.E. Calmels B. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M. Tachedjian G. Mak J. The packaging and maturation of the HIV-1 Pol proteins. Curr. HIV Res. 2005;3:73–85. doi: 10.2174/1570162052772942. [DOI] [PubMed] [Google Scholar]
- Hindmarsh P. Ridky T. Reeves R. Andrake M. Skalka A.M. Leis J. HMG-protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman A.G. Coffin J.M. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [Erratum appears in Proc. Natl. Acad. Sci. U.S.A. 2005;102:6238] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes-Son M.L. Chow S.A. Integrase–LexA fusion proteins incorporated into human immunodeficiency virus type 1 that contains a catalytically inactive integrase gene are functional to mediate integration. J. Virol. 2000;74:11548–11556. doi: 10.1128/jvi.74.24.11548-11556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome [see comment] Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kalpana G.V. Marmon S. Wang W. Crabtree G.R. Goff S.P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5 [see comment] Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- Kang Y. Moressi C.J. Scheetz T.E. Xie L. Tran D.T. Casavant T.L. Ak P. Benham C.J. Davidson B.L. McCray P.B., Jr. Integration site choice of a feline immunodeficiency virus vector. J. Virol. 2006;80:8820–8823. doi: 10.1128/JVI.00719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R.A. Skalka A.M. The retroviral enzymes. Annu. Rev. Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- Katz R.A. Merkel G. Skalka A.M. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: In vitro activities and incorporation of a fusion protein into viral particles. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- Katz R.A. Gravuer K. Skalka A.M. A preferred target DNA structure for retroviral integrase in vitro. J. Biol. Chem. 1998;273:24190–24195. doi: 10.1074/jbc.273.37.24190. [DOI] [PubMed] [Google Scholar]
- Kim Y.G. Cha J. Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo E. Gottlinger H.G. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J. Virol. 1996;70:159–164. doi: 10.1128/jvi.70.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.P. Mehta S. Purbey P.K. Notani D. Jayani R.S. Purohit H.J. Raje D.V. Ravi D.S. Bhonde R.R. Mitra D. Galande S. SATB1-binding sequences and Alulike motifs define a unique chromatin context in the vicinity of human immunodeficiency virus type 1 integration sites. J. Virol. 2007;81:5617–5627. doi: 10.1128/JVI.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova O. Fehse B. Modlich U. Yang M. Dullmann J. Kamino K. Von Neuhoff N. Schlegelberger B. Li Z. Baum C. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Lau A. Swinbank K.M. Ahmed P.S. Taylor D.L. Jackson S.P. Smith G.C. O'Connor M.J. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat. Cell Biol. 2005;7:493–500. doi: 10.1038/ncb1250. [DOI] [PubMed] [Google Scholar]
- Lee M.S. Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski M.K. Yamashita M. Emerman M. Ciuffi A. Marshall H. Crawford G. Collins F. Shinn P. Leipzig J. Hannenhalli S. Berry C.C. Ecker J.R. Bushman F.D. Retroviral DNA integration: Viral and cellular determinants of target-site selection. PLoS Pathogens. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L.K. Harlow G.R. Gregg-Jolly L.A. Mount D.W. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- Li L. Olvera J.M. Yoder K.E. Mitchell R.S. Butler S.L. Lieber M. Martin S.L. Bushman F.D. Role of the nonhomologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Dullmann J. Schiedlmeier B. Schmidt M. Von Kalle C. Meyer J. Forster M. Stocking C. Wahlers A. Frank O. Ostertag W. Kuhlcke K. Eckert H.G. Fehse B. Baum C. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- Lin C.W. Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 2003;77:5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M. Delgado S. Vanegas M. Poeschla E.M. Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 2004a;279:55570–55577. doi: 10.1074/jbc.M408508200. [DOI] [PubMed] [Google Scholar]
- Llano M. Vanegas M. Fregoso O. Saenz D. Chung S. Peretz M. Poeschla E.M. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 2004b;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M. Saenz D.T. Meehan A. Wongthida P. Peretz M. Walker W.H. Teo W. Poeschla E.M. An essential role for LEDGF/p75 in HIV integration. Science. 2006a;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Llano M. Vanegas M. Hutchins N. Thompson D. Delgado S. Poeschla E.M. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006b;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- Lombardo A. Genovese P. Beausejour C.M. Colleoni S. Lee Y.L. Kim K.A. Ando D. Urnov F.D. Galli C. Gregory P.D. Holmes M.C. Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Lu Y.L. Bennett R.P. Wills J.W. Gorelick R. Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J. Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil A. Sankale J.L. Meloni S.T. Sarr A.D. Mboup S. Kanki P. Genomic sites of human immunodeficiency virus type 2 (HIV-2) integration: Similarities to HIV-1 in vitro and possible differences in vivo. J. Virol. 2006;80:7316–7321. doi: 10.1128/JVI.00604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens G. Cherepanov P. Pluymers W. Busschots K. De Clercq E. Debyser Z. Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T. Parra M. Vries R.G. Kauder S.E. Verrijzer C.P. Ott M. Verdin E. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J. Biol. Chem. 2006;281:19960–19968. doi: 10.1074/jbc.M603336200. [Erratum appears in J Biol. Chem. 2006;281:26768.] [DOI] [PubMed] [Google Scholar]
- Masson C. Bury-Mone S. Guiot E. Saez-Cirion A. Schoevaertbrossault D. Brachet-Ducos C. Delelis O. Subra F. Jeanson-Leh L. Mouscadet J.F. Ku80 participates in the targeting of retroviral transgenes to the chromatin of CHO cells. J. Virol. 2007;81:7924–7932. doi: 10.1128/JVI.02015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield L.F. Fraize C.D. Coffin J.M. Relationship between retroviral DNA-integration-site selection and host cell transcription [see comment] Proc. Natl. Acad. Sci. U.S.A. 2005;102:1436–1441. doi: 10.1073/pnas.0409204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R.S. Beitzel B.F. Schroder A.R. Shinn P. Chen H. Berry C.C. Ecker J.R. Bushman F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PloS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U. Kustikova O.S. Schmidt M. Rudolph C. Meyer J. Li Z. Kamino K. Von Neuhoff N. Schlegelberger B. Kuehlcke K. Bunting K.D. Schmidt S. Deichmann A. Von Kalle C. Fehse B. Baum C. Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood. 2005;105:4235–4246. doi: 10.1182/blood-2004-11-4535. [DOI] [PubMed] [Google Scholar]
- Moehle E.A. Rock J.M. Lee Y.L. Jouvenot Y. Dekelver R.C. Gregory P.D. Urnov F.D. Holmes M.C. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. Erratum appears in Proc. Natl. Acad. Sci. U.S.A. 2007;104:6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monse H. Laufs S. Kuate S. Zeller W.J. Fruehauf S. Uberla K. Viral determinants of integration site preferences of simian immunodeficiency virus-based vectors. J. Virol. 2006;80:8145–8150. doi: 10.1128/JVI.00373-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E. Cesana D. Schmidt M. Sanvito F. Ponzoni M. Bartholomae C. Sergi Sergi L. Benedicenti F. Ambrosi A. Di Serio C. Doglioni C. Von Kalle C. Naldini L. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Mooslehner K. Karls U. Harbers K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J. Virol. 1990;64:3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narezkina A. Taganov K.D. Litwin S. Stoyanova R. Hayashi J. Seeger C. Skalka A.M. Katz R.A. Genomewide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M.G. Schmidt M. Schwarzwaelder K. Stein S. Siler U. Koehl U. Glimm H. Kuhlcke K. Schilz A. Kunkel H. Naundorf S. Brinkmann A. Deichmann A. Fischer M. Ball C. Pilz I. Dunbar C. Du Y. Jenkins N.A. Copeland N.G. Luthi U. Hassan M. Thrasher A.J. Hoelzer D. Von Kalle C. Seger R. Grez M. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1 [see comment] Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Pike-Overzet K. De Ridder D. Weerkamp F. Baert M.R. Verstegen M.M. Brugman M.H. Howe S.J. Reinders M.J. Thrasher A.J. Wagemaker G. Van Dongen J.J. Staal F.J. Gene therapy: Is IL2RG oncogenic in T-cell development? [see comment] Nature. 2006;443 doi: 10.1038/nature05218. discussion E6–E7. [DOI] [PubMed] [Google Scholar]
- Pruss D. Bushman F.D. Wolffe A.P. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. U.S.A. 1994a;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss D. Reeves R. Bushman F.D. Wolffe A.P. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 1994b;269:25031–25041. [PubMed] [Google Scholar]
- Pryciak P.M. Varmus H.E. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- Pryciak P.M. Muller H.P. Varmus H.E. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc. Natl. Acad. Sci. U.S.A. 1992a;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak P.M. Sil A. Varmus H.E. Retroviral integration into minichromosomes in vitro. EMBO J. 1992b;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell D.F. Martin M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia A. Bonini C. Magnani Z. Urbinati F. Sartori D. Muraro S. Tagliafico E. Bondanza A. Stanghellini M.T. Bernardi M. Pescarollo A. Ciceri F. Bordignon C. Mavilio F. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe T. Reynolds T.C. Yu G. Brown P.O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherdin U. Rhodes K. Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J. Virol. 1990;64:907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder A.R. Shinn P. Chen H. Berry C. Ecker J.R. Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Schwarzwaelder K. Howe S.J. Schmidt M. Brugman M.H. Deichmann A. Glimm H. Schmidt S. Prinz C. Wissler M. King D.J. Zhang F. Parsley K.L. Gilmour K.C. Sinclair J. Bayford J. Peraj R. Pike-Overzet K. Staal F.J. De Ridder D. Kinnon C. Abel U. Wagemaker G. Gaspar H.B. Thrasher A.J. Von Kalle C. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo [see comment] J. Clin. Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun M.C. Raghavendra N.K. Vandegraaff N. Daigle J.E. Hughes S. Kellam P. Cherepanov P. Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka A.M. Katz R.A. Retroviral DNA integration and the DNA damage response. Cell Death Differentiation. 2005;12(Suppl. 1):971–978. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- Smith J.A. Daniel R. Following the path of the virus: The exploitation of host DNA repair mechanisms by retroviruses. ACS Chemical Biology [Electronic Journal] 2006;1:217–226. doi: 10.1021/cb600131q. [DOI] [PubMed] [Google Scholar]
- Sutherland H.G. Newton K. Brownstein D.G. Holmes M.C. Kress C. Semple C.A. Bickmore W.A. Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations. Mol. Cell. Biol. 2006;26:7201–7210. doi: 10.1128/MCB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K.D. Cuesta I. Daniel R. Cirillo L.A. Katz R.A. Zaret K.S. Skalka A.M. Integrase-specific enhancement and suppression of retroviral DNA integration by compacted chromatin structure in vitro. J. Virol. 2004;78:5848–5855. doi: 10.1128/JVI.78.11.5848-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W. Zhu K. Segal D.J. Barbas C.F., III Chow S.A. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J. Virol. 2004;78:1301–1313. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W. Dong Z. Wilkinson T.A. Barbas C.F., III Chow S.A. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrasher A.J. Gaspar H.B. Severe adverse event in clinical trial of gene therapy for X-SCID. Institute of Child Health; London, UK: 2007. [Apr;2008 ]. [Google Scholar]
- Thrasher A.J. Gaspar H.B. Baum C. Modlich U. Schambach A. Candotti F. Otsu M. Sorrentino B. Scobie L. Cameron E. Blyth K. Neil J. Abina S.H. Cavazzana-Calvo M. Fischer A. Gene therapy: X-SCID transgene leukaemogenicity [see comment] Nature. 2006;443:E5–E6. doi: 10.1038/nature05219. discussion E6-E7. [DOI] [PubMed] [Google Scholar]
- Treand C. Du Chene I. Bres V. Kiernan R. Benarous R. Benkirane M. Emiliani S. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 2006;25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobridge G.D. Miller D.G. Jacobs M.A. Allen J.M. Kiem H.P. Kaul R. Russell D.W. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlure F. Maertens G. Rahman S. Cherepanov P. Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1653–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas M. Llano M. Delgado S. Thompson D. Peretz M. Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- Wang G.P. Ciuffi A. Leipzig J. Berry CC. Bushman F.D. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J.B. Angelichio E.L. Fenner S. Coffin J.M. Relationship between retroviral DNA integration and gene expression. J. Virol. 2000;74:8382–8389. doi: 10.1128/jvi.74.18.8382-8389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods N.B. Bottero V. Schmidt M. Von Kalle C. Verma I.M. Gene therapy: Therapeutic gene causing lymphoma [see comment] Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- Wu X. Liu H. Xiao H. Kim J. Seshaiah P. Natsoulis G. Boeke J.D. Hahn B.H. Kappes J.C. Targeting foreign proteins to human immunodeficiency virus particles via fusion with Vpr and Vpx. J. Virol. 1995;69:3389–3398. doi: 10.1128/jvi.69.6.3389-3398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Li Y. Crise B. Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration [see comment] Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Wu X. Li Y. Crise B. Burgess S.M. Munroe D.J. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J. Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]