Abstract
Old astrocyte specifically induced substance (OASIS) has previously been shown to be a putative endoplasmic reticulum (ER) stress sensor in astrocytes with a mechanism of activation that is similar to ATF6. In this study we investigated the expression and activation of endogenous and overexpressed OASIS in pancreatic β-cells. OASIS mRNA expression was detected in pancreatic β-cell lines and rodent islets, and the expression level was up-regulated by ER stress-inducing compounds. Endogenous OASIS protein, however, is expressed at low levels in pancreatic β-cell lines and rodent islets, possibly due to abundant levels of the micro-RNA miR-140 present in these cells. In contrast, expression of both full-length and cleaved (active) OASIS was readily detectable in the developing mouse pancreas (embryonic d 15.5). Microarray analysis after expression of an active nuclear-localized version of OASIS in an inducible INS-1 β-cell line resulted in the up-regulation of many genes implicated in extracellular matrix production and protein transport but not classical ER stress response genes. Consistent with this, expression of active OASIS failed to induce glucose-regulated protein 78 kDa promoter activity in pancreatic β-cells. These results suggest that the repertoire of genes induced by OASIS is cell type-dependent and that the OASIS protein may have a role in pancreas development.
The endoplasmic reticulum stress sensor, OASIS, induces expression of extracellular matrix genes and may be important in pancreas development.
The endoplasmic reticulum (ER) serves as a site for the biosynthesis of membrane and secretory proteins and maintains an environment that is conducive for efficient synthesis of such proteins. Perturbations in the ER luminal environment such as alteration in calcium concentration, ER redox potential, or ineffective posttranslational modification of secretory proteins may result in an accumulation of unfolded and/or misfolded proteins creating a state of ER stress. Cells sense the levels of unfolded proteins in the ER and elicit the unfolded protein response (UPR). Activation of the UPR leads to a transient translational inhibition, followed by activation of a transcriptional response to enhance ER chaperone capacity and ER-associated degradation (1,2). If these effects are not sufficient to relieve the protein folding demands and unfolded/misfolded proteins continue to accumulate, the cell initiates apoptotic pathways (3).
Mammalian cell ER stress sensing and signaling involves three well-established and ubiquitous ER stress sensors: RNA-dependent protein kinase-like ER kinase (PERK), inositol-requiring enzyme-1α (IRE1α) and activating transcription factor 6 (ATF6) (2). These proteins are activated by either sensing misfolded proteins directly or disassociation of the chaperone glucose-regulated protein 78 (GRP78) that binds the luminal domains of these proteins under basal conditions. PERK and IRE1α activation involves autophosphorylation to activate subsequent signaling, whereas ATF6 activation involves its transport from the ER and proteolysis in the Golgi. The cleaved ATF6 cytosolic domain then translocates to the nucleus to mediate gene expression changes (1,4,5). In addition to these classic ER stress sensors, other putative ER stress sensors related to ATF6 have also been reported including cAMP responsive element binding protein (CREB)-4, CREB-H, luman, BBF2H7, and old astrocyte specifically induced substance (OASIS)/CREB 3-like 1 (CREB3L1) (6,7,8,9).
OASIS (also referred to as CREB3L1) has been reported to be a potential ER stress sensor in astrocytes (10). OASIS was initially identified as a highly induced gene in long-term cultured mouse astrocytes (11). Subsequent studies have shown that OASIS may have a role in the differentiation and development of odontoblasts and osteoblasts (12,13) and regulation of expression of cell-specific transcription factors (14). However, an early study indicated that OASIS contains a transmembrane domain, was localized to the ER, and could potentially be activated by regulated intramembrane proteolysis (15). It was subsequently shown that OASIS translocates from the ER to the Golgi in response to ER stress in astrocytes, where it is cleaved by membrane proteases (site-1 and -2 proteases) to release the cytosolic domain that translocates to the nucleus (7,16). OASIS can potentially bind to both cAMP response elements and ER stress responsive elements and has been demonstrated to induce the GRP78 promoter, suggesting that this protein is a bona fide ER stress transducer (7). More recently studies in OASIS-deficient mice have uncovered that the protein provides protection to astrocytes from kainic acid toxicity, which is known to induce ER stress (17) and is involved in bone formation by osteoblasts (18). Whether OASIS has a role in the ER stress response in tissues other than osteoblasts and astrocytes has not been examined.
ER stress has been implicated in pancreatic β-cell dysfunction in the context of obesity and type 2 diabetes (19,20). Pancreatic β-cells are particularly sensitive to ER stress due to their function in synthesizing and secreting insulin to meet metabolic demands. Indeed expression of ER stress sensors is particularly high in this cell type. Given that OASIS mRNA expression has been detected in pancreas tissue (15), we investigated the expression and activation of OASIS in pancreatic β-cell lines and rodent islets.
Materials and Methods
Cell culture
Rat INS-1 (21) (obtained from Dr. C. Wollheim, University of Geneva, Geneva, Switzerland) and rat INS-1 832/13 (22) (obtained from Dr. C. Newgard, Duke University, Durham, NC), pancreatic β-cell lines were cultured and maintained as described in the above references. MIN6 cells were obtained from Dr. Michael Wheeler (University of Toronto, Toronto, Canada) and cultured as described previously (23). Rat C6 glioma (American Type Culture Collection, Manassas, VA; CCL-107), human HeLa, and HT1080 cells were propagated in DMEM medium (25 mm glucose, 2 mm l-glutamine, 10% fetal bovine serum).
Preparation of islets from rodent pancreas and mouse embryonic pancreas isolation
Islets were isolated from male Wistar rats or C57BL/6 mice by collagenase digestion followed by Ficoll gradient separation as reported previously (24). Animals were housed and anesthetized following protocols approved by the Animal Use Committee at the University Health Network (Toronto, Canada).
Pancreas tissue from mouse embryos at d 15.5 of gestation was dissected and immediately frozen on dry ice. All mouse studies were performed in accordance with the Vanderbilt University Institutional Animal Care and Use Committee guidelines. For embryonic analyses, the morning of the vaginal plug was considered embryonic day (E) 0.5. The tissue was thawed on ice and lysed directly in 40 μl 2 × NuPAGE sample buffer containing β-mercaptoethanol or TRIzol reagent (Invitrogen, Carlsbad, CA) was added for total RNA isolation.
Plasmid generation and cell transfection
Full-length rat OASIS cDNA was amplified from rat islet total RNA and subcloned into pCR II Topo vector (Invitrogen, Carlsbad, CA). It was then subcloned into the expression vector pcDNA 3.1(−) to generate pCMV-rOASIS-FL (rOASIS-FL). Human OASIS cDNA (NM_052854) was amplified by PCR from the pCMV6-XL5-hCREB3L1 plasmid (OriGene, Rockville, MD) and subcloned in pCR II Topo vector. The human OASIS cDNA was cut from pCR II-hOASIS-FL with EcoRI and ligated downstream of the myc epitope tag in the pCMV 3C vector (Stratagene, La Jolla, CA) to generate pCMV 3C-hOASIS-FL (h-mycOASIS-FL). This vector (h-mycOASIS-FL) was subjected to site-targeted mutagenesis using the QuickChange kit (Stratagene) to replace a TGC to TGA (stop codon) at amino acid 374 to generate pCMV3C-h-mycOASIS-374 (h-mycOASIS-374). This region comprises the cytosolic domain of OASIS because the stop codon occurs just before the predicted transmembrane domain (7). The region comprising the Kozak sequence, myc tag, and human mycOASIS cDNA (without transmembrane and luminal domains) (∼1.1 kb) was amplified from h-mycOASIS-374 plasmid with KpnI and NheI site overhangs and ligated into pTRE-Tight vector (CLONTECH, Palo Alto, CA). The resulting pTRE-Tight-h-mycOASIS-374 expression vector was used for the generation of INS-1 h-mycOASIS-374 doxycyline-inducible stable cell line.
Rat ATF6 cytosolic domain (amino acids1-377) was amplified from the vector p3×FLAG-CMV-7.1-ATF6 containing full-length ATF6 (amino acids 1-670) obtained from Dr. R. Prywes (Columbia University, New York, NY). The amplified ATF6 cytosolic domain containing the N-terminal 3×FLAG tag was subcloned into the pCR II Topo vector. The FLAG-ATF6-377 region was cloned into pcDNA 3.1(−) vector generating pcDNA 3.1(−)-FLAG-ATF6-377 (FLAG-ATF6-377). Cloning steps were monitored by the restriction digestion and confirmed by DNA sequencing. All primers sequences are available upon request.
Transfection of the various plasmids into the different cell lines was performed with Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol provided.
Generation of an inducible OASIS-expressing INS-1 cell line
To generate an inducible pancreatic INS-1 cell line expressing nuclear-localized OASIS, a stable pTet-ON INS-1 cell line (INS-1 pTet-ON no. 46) was first generated by transfection with pTet-ON vector followed by selection with G418, according to the instructions provided (CLONTECH). The pTRE-Tight-h-mycOASIS-374 expression vector was cotransfected with a hygromycin resistance plasmid into INS-1 pTet-ON no. 46 and double-stable clones resistant to G418 and hygromycin were isolated. Several clones were tested for human mycOASIS-374 expression by immunoblotting and immunofluorescence staining.
Total RNA isolation, RT-PCR, and real-time PCR analysis
Total RNA was isolated from rodent islets, embryonic pancreas, or cultured cell lines using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Isolated RNA was then purified using the RNeasy RNA isolation kit (QIAGEN, Valencia, CA). The RNA was used in a reverse transcription reaction using the One Step RT-PCR kit (QIAGEN) according to the protocol provided. Reactions were run in a PTC-200 (MJ Research, Watertown, MA) thermal cycler for 35 cycles. Primer sequences used to amplify OASIS cDNA are available upon request.
For real-time PCR analysis, total RNA was isolated from cells treated as described in the figure legends. Total RNA was reverse transcribed to single-stranded cDNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). The resulting cDNA was used for real-time PCR analysis using the TaqMan gene expression system (Applied Biosystems) as described elsewhere (25). The following TaqMan MGB probes were used: rat GRP78 (Rn01435771_g1), rat β-actin (Rn00667869_m1), rat OASIS (Rn00496405_m1), mouse OASIS (Mm01331535_m1), rat Matn1 (Rn01519782_g1), and rat Papss2 (Rn01479533_m1).
X-box binding protein (XBP)-1 splicing assay
Total mRNA from mouse embryonic pancreas tissue was extracted using TRIzol reagent (Invitrogen) and RNeasy minikit (QIAGEN). Mouse XBP-1 cDNA was amplified by RT-PCR (QIAGEN One Step RT-PCR kit) using the following primers: XBP-1 sense primer, 5′-GAA CCA GGA GTT AAG AAC ACG-3′; XBP-1 antisense primer, 5′-AGG CAA CAG TGT CAG AGT CC-3′. The protocol used for RT-PCR was 50 C (30 min); 95 C (15 min); 35 cycles [94 C (1 min), 55 C (1 min), 72 C (1 min)]; 72 C (10 min). PCR products were resolved on a 3% agarose gel and visualized using ethidium bromide.
Luciferase reporter assays
INS-1 832/13 and C6 cells were cotransfected with OASIS or ATF6 expression vectors and reporter plasmids. Human mycOASIS-374 or FLAG-ATF6-377 cytosolic/nuclear constructs or control vectors (pCMV3C or pCDNA 3.1 vectors) (0.5 μg) were cotransfected with GRP78 reporter plasmid (0.5 μg). The GRP78 reporter plasmid contained the human GRP78 promoter (−304) from the transcriptional start site upstream of the luciferase gene in the pGL2-plasmid. After 24 h transfection, the cells were lysed using the buffer provided with the luciferase assay kit (Stratagene). Firefly luciferase activity was measured using a LUMINOSKAN ascent luminometer (Thermo Fisher Scientific, Waltham, PA). Relative activity was normalized for total cellular protein in each condition.
Micro-RNA detection
Total RNA was isolated from C6, HeLa, HT1080, INS-1 832/13, and MIN6 cells using miRCURY RNA isolation kit (Exiqon, Inc., Vedbaek, Denmark). Twenty micrograms of total RNA in Novex Tris-borate EDTA (TBE) urea sample buffer (2 times) were resolved using 15% Novex TBE-urea-acrylamide gels. RNA was transferred to BrightStar-Plus nylon membrane (Ambion, Austin, TX) using semidry transblot apparatus (Bio-Rad Laboratories, Hercules, CA) in 0.5× TBE buffer. Membrane, filter papers, and gel were rinsed for 30 min in 0.5× TBE buffer. The wet blot was UV cross-linked and baked at 70 C for 30 min. Prehybridization was done with ULTRAhyb-oligo hybridization buffer at 42 C for 30 min. Digoxigenin (DIG)-5′-labeled locked nucleic acid detection probe for miR-140 was purchased from Exiqon. A scrambled detection probe was used to assess the specificity of detection. The detection probe (25 pm) was added to hybridization buffer (same buffer used in prehybridization) and incubated at 42 C for 16–18 h. After hybridization, the blot was subjected to washings (23 C/5min, 42 C/15 min (times 2) with NorthernMax low-stringency wash buffer (Ambion). The blot was rinsed, blocked with solutions from DIG wash/block buffer set (Roche Diagnostics, Indianapolis, IN) and subsequently incubated in blocking solution containing anti-DIG- alkaline phosphotase-Fab fragment (DIG label detection kit; Roche) for 2 h at 23 C. Micro-RNA was detected by a colorimetric assay (DIG label detection kit; Roche).
Microarray analysis
Microarray expression profiling was used to assess the global transcriptional changes in response to expression of active nuclear localized myc-tagged OASIS in an inducible INS-1 clone. Cells were induced or not with doxycycline for 24 and 48 h, and total RNA was isolated using TRIzol reagent (Invitrogen) followed by isolation using an RNeasy mini kit (QIAGEN). Three independent experiments were performed. Assessment of RNA quality and microarray analysis was performed at the University Health Network Microarray Centre. Briefly, after RNA quality assessment with an Agilent BioAnalyzer (Palo Alto, CA), samples were reverse transcribed to cDNA. cDNA was purified with cDNA purification module from Affymetrix (Santa Clara, CA). Biotin was incorporated during in vitro transcription and purified cRNA was then fragmented. Fifteen micrograms of labeled and fragmented cRNA were hybridized to Rat Genome 230 2.0 arrays (Affymetrix Genechip) for 17 h at 45 C at 60 rpm speed. The arrays were stained and washed using fluidic stations with antibody and streptavidin-phycoerythrin conjugate solutions. All arrays were scanned using the Affymetrix 3000 7G laser scanner. The image data were quantified to yield gene expression values. For statistical analysis CEL files were analyzed using GeneSpring software (version 7.1; Agilent Technologies). Unpaired t test with a cutoff of P < 0.05 was performed between groups (doxycycline 24 h vs. control, etc.), and the Benjamini and Hochberg method was used for multiple testing correction. Well-substantiated genes showing a minimum 2-fold change between control and doxycycline-treated cells in at least two of three independent experiments at 24 and 48 h h are listed in Table 1 and Supplemental Table 1, published on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org. The complete list of gene changes in each experiment is provided in the supplemental data.
Table 1.
Affymetrix (rat genome array) expression profiling comparing uninduced cells to cells expressing active fragment of human OASIS for 24 h
| Gene ID | Fold change (mean) | Description/putative function |
|---|---|---|
| 294103 | 23.54 | Papss2, sulfate conjugation |
| 25107 | 20.09a | Avrp1A, inositol-calcium signaling |
| 360959 | 8.32 | HtrA3, cytoskeletal remodeling, ERAD, inhibitor of TGF-β signaling |
| 312607 | 6.77 | Pdzrf3, ubiquitin protein ligase activity |
| 287877 | 5.34 | P5cr1, arginine and proline metabolism |
| 294048 | 5.2 | Pi3kap, signaling |
| 297894 | 4.3a | Matn1 cartilage matrix protein |
| 498609 | 4.28 | Lpar2, signal transduction |
| 360611 | 4.22 | Copζ2, protein transport |
| 25027 | 3.4 | Slc16a1, transporter |
| 29241 | 3.18a | Adcy8, adenylyl cyclase 8 |
| 282712 | 3.03 | Slc6a15, transporter |
| 315131 | 3.03 | Kdelr3, protein transport |
| 79247 | 2.85a | Htr5b, serotonin receptor |
| 304322 | 2.66a | Chst12, carbohydrate sulfotransferase |
| 29643 | 2.54 | Sv2c, protein secretion/transport |
| 64313 | 2.39 | Oat, proline metabolism |
| 690050 | 2.33 | Tpmt, thiopurine methylation |
| 680493 | 2.1 | Cd3εap, Notch signaling |
Common genes induced in three independent experiments greater than 2-fold are shown. ERAD, ER-associated degradation.
Genes induced in two of three experiments.
Immunofluorescence microscopy
Cells were cultured on 18-mm glass coverslips and the following day treated as described in the figure legends (see Figs. 3–5). Cells were fixed in 4% paraformaldehyde/PBS for 20 min, incubated with 100 mm glycine/PBS buffer for 15 min, and then permeabilized with 0.1% Triton X-100/0.5% BSA in PBS for 15 min. After blocking with 2% BSA/2% nonfat dry milk for 1 h, primary antibodies were added for 1 h. Primary antibodies used were mouse anti-myc tag (9E10; 1:500) (see Figs. 3 and 4) or mouse antichondroitin sulfate proteoglycan (Abcam, Cambridge, MA; ab78689, 1:2500) (see Fig. 5). Cells were washed three times with PBS (5 min each), and secondary antibodies (Oregon green 488 antimouse IgG, 1:1000; or Alexa Fluor 594 antimouse IgG, 1:1000; Molecular Probes, Eugene, OR) were added and incubated for 1 h. After washing the unbound secondary antibody with PBS (3 × 5 min), the nuclear stain 4′, 6′-diamidino-2-phenylindole was added for 5 min before the slides were mounted using Fluoromount-G (EM Sciences, Hatfield, PA). Fluorescence was visualized with an Olympus inverted fluorescence microscope (IX71; Tokyo, Japan), and images were captured and processed using Q-Capture Pro software (Q Imaging, Surrey, Canada).
Figure 3.
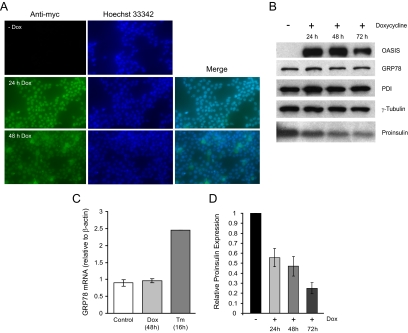
Active (nuclear localized) OASIS expression does not affect the expression of ER chaperones but reduces proinsulin expression in an inducible INS-1 β-cell line. A, INS-1 pTetON-h-mycOASIS-374 stable clone was induced or not with doxycycline (Dox; 2 μg/ml) for the times indicated, fixed, and immunostained with anti-myc tag antibody and the nuclear dye Hoechst 33342. B, Cells were treated as in A, lysed, and 10 μg total proteins resolved by SDS-PAGE and immunoblotted with anti-myc antibody (to detect human mycOASIS-374 expression) and for the other proteins indicated. C, Real-time PCR analysis of GRP78 mRNA levels in cells induced by doxycyline (Dox) to express nuclear OASIS for 48 h compared with control, uninduced cells. Means ± se of three independent experiments. As a positive control for GRP78 induction INS-1 cells were treated for 16 h with tunicamycin (Tm; 2 μg/ml) (n = 1). D, Proinsulin immunoblots were quantified by gel densitometry and expression levels relative to uninduced control is presented from three independent experiments.
Figure 4.
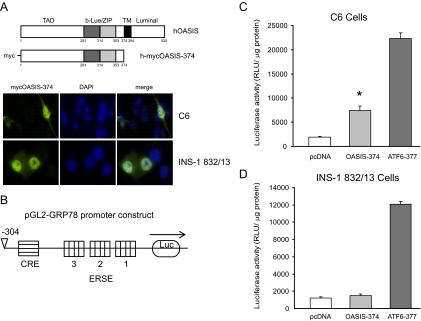
Active OASIS does not affect GRP78 promoter activity in pancreatic β-cells. A, INS-1 832/13 and C6 cells were transfected with human OASIS (hOASIS) lacking the transmembrane domain and myc tagged at the N terminus. Cells were fixed and immunostained with an anti-myc antibody. Representative transfected cells are shown. (TAD, transcriptional activation domain; b-Lue/ZIP, basic leucine zipper domain) B, Graphic representation of the GRP78 promoter luciferase construct in the pGL2-basic vector. The cAMP response element and ER stress responsive element (ERSE) sites are indicated. C and D, C6 glioma cells or INS-1 832/13 cells were cotransfected with either human OASIS-374 or human ATF6–377 and the reporter construct or with reporter construct and empty vector (pcDNA 3.1 vector) for 24 h. The cells were lysed and assayed for luciferase activity and values were normalized for protein. The mean activity ± se from three to six independent experiments is shown. *, P < 0.01 OASIS-374 vs. pcDNA control (Student t test).
Figure 5.
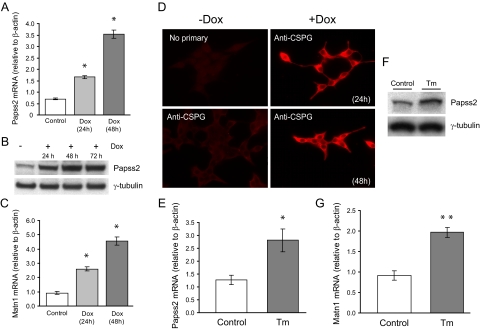
Papss2, Matn1, and chondroitin sulfate proteoglycan levels are induced by OASIS-374 expression and ER stress in pancreatic β-cells. A and C, Real-time PCR analysis of Papss2 and Matn1 mRNA levels in cells induced by doxycyline (Dox) to express nuclear OASIS for 24 or 48 h compared with control, uninduced cells (means ± se of three independent experiments). *, P < 0.05, relative to control; ANOVA and Tukey’s test. B, INS-1 pTetON-h-mycOASIS-374 stable cells were induced with doxycyline for the times indicated, and lysates were prepared and immunoblotted using antibodies to the indicated proteins. D, Control uninduced INS-1 pTetON-h-mycOASIS-374 stable cells or cells induced for the times indicated were fixed and immunostained using an antibody that detects chondroitin sulfate proteoglycan (CSPG). E and G, INS-1 832/13 β-cells were treated or not with tunicamycin (Tm; 2 μg/ml) for 16 h. Total RNA was isolated and real-time PCR analysis of Papss2 and Matn1 mRNA levels was performed. Means ± se from three independent experiments is shown. *, P < 0.05, **, P < 0.01 vs. control (Student’s t test). F. INS-1 832/13 β-cells were treated or not with tunicamycin (Tm) as in E, and lysates were prepared and immunoblotted using antibodies to the indicated proteins.
Cell lysis and Western blot analysis
Cells were treated as described in the figure legends and washed in PBS before addition of ice-cold lysis buffer (1% Triton X-100; 20 mm HEPES, pH 7.4; 100 mm KCl; 2 mm EDTA; 1 mm phenylmethylsulfonyl fluoride; 10 μg/ml leupeptin; 10 μg/ml aprotinin; 10 mm NaF; 2 mm Na3VO4; and 10 nm okadaic acid). The cells were lysed on ice for 15–20 min and centrifuged at 13,000 rpm for 10 min at 4 C. The protein concentration in the supernatant was quantified using the BCA protein assay kit (Pierce, Inc., Rockford, IL). Equal protein amounts were resolved by SDS-PAGE and transferred to Hybond nitrocellulose membrane (GE Healthcare, Piscataway, NJ). The following primary antibodies were used: rabbit anti-OASIS (Proteintech Group, Inc., Chicago, IL; 11235-2-AP, 1:1000), mouse anti-myc tag (9E10), mouse anti-Papss2 (Abnova, Walnut, CA; M07 clone 2A8), mouse antiprotein disulfide isomerase (PDI; StressGen, Victoria, Canada; SPA-891), mouse anti-KDEL (StressGen; SPA-827), guinea pig antiinsulin (Dako Inc., Carpinteria, CA; A0564), mouse anti- heat shock protein HSP90 (BD Biosciences, Mississauga, Canada; 610418), mouse anti-γ-tubulin (Sigma-Aldrich, St. Louis, MO; T6557), rabbit anti-p24 (provided by Dr. Felix Wieland, University of Heidelberg, Heidelberg. Germany). Horseradish peroxidase-conjugated secondary antibodies were used as required and the protein bands were detected by ECL (GE Healthcare). Immunoblots were scanned and analyzed using Scion Image software (Frederick, MD).
Statistical analysis
The results are expressed as means ± se. Statistical significance (Figs. 1 and 5A,C) was assessed by ANOVA followed by Tukey’s post hoc test and by Student’s t test (Figs. 4 and 5). P < 0.05 was considered significant.
Figure 1.
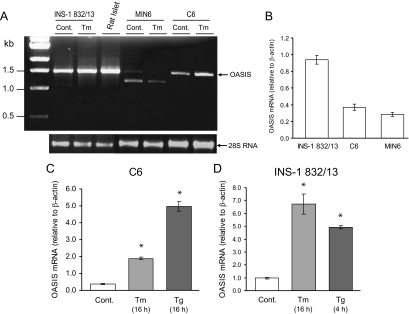
OASIS mRNA is expressed in pancreatic β-cell lines and rodent islets. A, Rat INS-1 832/13 β-cells, mouse MIN6 β-cells, or rat C6 glioma cells were treated or not (Control; Cont.) with tunicamycin (Tm; 2 μg/ml) for 16 h. Total RNA was extracted from these cells and isolated rat islets. RT-PCR was performed as described in Materials and Methods to amplify the coding region of OASIS. Ethidium bromide staining of 28S RNA is shown as a loading control. B, Real-time quantitative PCR analysis was performed for OASIS expression in the cell lines indicated and expressed relative to cellular β-actin. C and D, C6 glioma or INS-1 832/13 pancreatic β-cells were treated with tunicamycin (Tm) or thapsigargin (Tg; 1 μm) for the times indicated. Total RNA was isolated and OASIS expression analyzed by real-time PCR. *, P < 0.01, relative to control; ANOVA and Tukey’s test.
Results
OASIS is expressed in pancreatic β-cell lines and rodent islets
OASIS has been shown to be a putative ER stress sensor expressed in astrocytes (6,7,10,16). Comparative Northern blot analysis for human OASIS expression in various tissues indicated that the pancreas also showed abundant mRNA expression (15). To examine whether OASIS is expressed in pancreatic β-cell lines and islets, we first performed RT-PCR analysis using total RNA extracted from rat INS-1 832/13 and mouse MIN6 pancreatic β-cell lines and isolated rat islets. Using primers for the full-length cDNA that are conserved between rat and mouse OASIS, we amplified an approximately 1.5-kb cDNA fragment from INS-1 832/13 cells, rat islets, and C6 glioma cells (Fig. 1A). Only a weak PCR product at the expected size was observed in MIN6 cells, and no expression was detected in the rat acinar cell line AR42J (results not shown). The amount of amplified product appeared to be higher in total RNA isolated from tunicamycin-treated INS-1 832/13 and C6 cells compared with control cells. To quantify relative mRNA levels, we performed real-time PCR analysis using primers specific for rat OASIS (INS-1 832/13 and C6) or mouse OASIS (MIN6) and analyzed relative expression compared with cellular β-actin. Similar to the RT-PCR results, the highest OASIS mRNA abundance was detected in INS-1 832/13 cells, whereas MIN6 cells had the lowest relative expression (Fig. 1B). The expression of OASIS was up-regulated in cells treated with the ER stress-inducing compounds tunicamycin and thapsigargin in both C6 and INS-1 832/13 cells as revealed by real-time PCR analysis (Fig. 1, C and D). Cloning and sequencing of the full-length PCR products obtained from rat islet and INS-1 832/13 cells in Fig. 1A showed that they are identical with the rat OASIS sequence deposited in the National Center for Biotechnology Information database (Bethesda, MD).
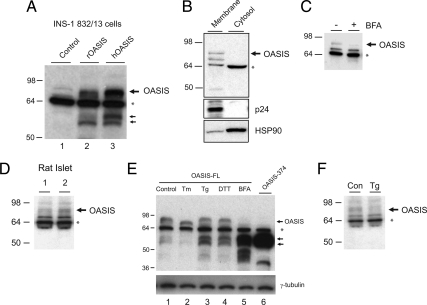
To examine whether OASIS protein is expressed in pancreatic β-cells, detergent lysates were prepared from INS-1 832/13 cells and immunoblotted with an anti-OASIS/CREB3L1 antibody (Fig. 2A). As a positive control, we transfected INS-1 832/13 cells with full-length rat and human OASIS cDNAs. A band at approximately 80 kDa was detected in cells transfected with full-length OASIS (Fig. 2, lanes 2 and 3) and in control nontransfected cells (Fig. 2, lane 1), although the endogenous signal was low. Two bands at approximately 55 kDa were also evident in transfected cells (bottom arrows in Fig. 2A). These results are similar to previous studies overexpressing full-length OASIS in astrocytes (16). The smaller bands are cleavage products of the full-length protein that escape the ER when OASIS is overexpressed and is cleaved by Site-1 and Site-2 proteases as demonstrated previously (7). With the antibody used, a prominent band at approximately 64 kDa was also evident in both control and transfected cells (Fig. 2A, asterisk). However, this unknown cross-reactive protein is primarily cytosolic and unlikely to be OASIS (Fig. 2B). Further evidence that the top band recognized by this antibody is likely to be endogenous OASIS is that it is sensitive to brefeldin A, whereas the lower cross-reactive bands are not (Fig. 2C), and it is specifically reduced by a small interfering RNA directed to rat OASIS (Supplemental Fig. 1). Endogenous OASIS was also detected in isolated rat islets, although the signal was low (Fig. 2D).
Figure 2.
Endogenous OASIS protein is detected in pancreatic β-cell lines and rodent islets and responds to ER stress. A, Control INS-1 832/13 β-cells or cells transfected with untagged full-length rat (rOASIS) or human OASIS (hOASIS) cDNAs for 24 h were lysed, and 10 μg of total protein was immunoblotted with an anti-OASIS antibody. Note that the antibody recognizes transfected OASIS at approximately 80 kDa and with what are likely cleavage products at approximately 55 kDa (indicated by double arrows). B, Control INS-1 832/13 cells were fractionated into membrane and cytosol fractions by homogenization and centrifugation. Ten micrograms of protein from each fraction were immunoblotted with the indicated antibodies. Note that the approximately 64 kDa band (asterisk) recognized by the anti-OASIS antibody is primarily cytosolic and therefore unlikely to be OASIS. HSP90, Heat shock protein 90. C, INS-1 832/13 cells were treated or not with brefeldin A (BFA; 5 μm) for 20 min before lysates were prepared and immunoblotted for OASIS. Note that only the top band is sensitive to BFA. D, Isolated rat islet lysates from two animals (20 μg total protein in each lane) were immunoblotted for OASIS. E, INS-1 832/13 β-cells were transfected with untagged full-length rat OASIS cDNA (lanes 1–5) or OASIS-374 (lane 6). Cells were treated or not with tunicamycin (Tm), thapsigargin (Tg; 1 μm for 4 h), dithiotreitol (DTT; 1 mm 30 min), or BFA (5 μm, 30 min) as indicated. Cell lysates (10 μg protein) were immunobloted for OASIS. F, INS-1 832/13 cells were treated or not with thapsigargin (Tg; 1 μm for 4 h) and lysates were immunobloted for OASIS. Con, Control.
To examine whether OASIS is ER stress responsive in β-cells, we transfected INS-1 832/13 cells with full-length OASIS and monitored sensitivity to ER stress-inducing compounds (Fig. 2E). The full-length protein was sensitive to thapsigargin and dithiothreitol, which resulted in reduced levels of the full-length protein and an increase in protein products at approximately 55 kDa (bottom arrows, Fig. 2E). Full-length OASIS was also sensitive to tunicamycin treatment, although clear cleavage products were not apparent in the experiment shown. The bands at approximately 55 kDa are likely to be cleavage products mediated by Golgi-localized proteases because treatment with brefeldin A, which redistributes Golgi proteins to the ER, caused the disappearance of full-length OASIS and the appearance of an intense band migrating at the same molecular weight as the transfected cytosolic domain of OASIS (Fig. 2E, compare lanes 5 and 6). Thus, OASIS appears to respond to ER stress by translocation from the ER to the Golgi followed by proteolysis as has been reported previously in astrocyte cells (7,16). Endogenous OASIS also appeared to be sensitive to thapsigargin in INS-1 832/13 cells, but no cleavage product was detected, likely due to the low endogenous levels of this protein (Fig. 2F).
OASIS induces genes involved in extracellular matrix formation but not classical ER stress response genes in pancreatic β-cells
To determine the complement of genes OASIS induces in pancreatic β-cells, we generated an inducible INS-1 stable clone expressing the nuclear-localized (active) form of OASIS. Doxycycline treatment induced the expression of myc-OASIS-374, which was localized to the nucleus (Fig. 3, A and B). Expression of myc-OASIS-374 did not appreciably affect the steady-state protein levels of GRP78 or PDI (Fig. 3B) or GRP78 mRNA (Fig. 3C) but caused a reduction in proinsulin levels (Fig. 3, B and D). GRP78 and PDI are classic target genes of the ER stress response and the observation that the levels were not changed was unexpected because OASIS has been shown to induce GRP78 in astrocytes (7,10).
We therefore used a luciferase reporter system to examine the transcriptional activity of OASIS on the human GRP78 promoter. Transfection of a version of human OASIS devoid of the transmembrane and luminal domains into INS-1 832/13 β-cells and C6 glioma cells resulted in the localization of myc-OASIS-374 primarily in the nucleus (Fig. 4A). The transcriptional activation efficiency of the OASIS construct was examined using a reporter plasmid containing the human GRP78 promoter region (−304 to +52) upstream of the luciferase gene (Fig. 4B). Human OASIS was able to induce transcriptional activation of the GRP78 promoter in C6 cells (Fig. 4C). This activity, although significant, was lower than active ATF6-p50, which induced a more robust response. However, no significant reporter activity was induced by OASIS in INS-1 832/13 cells, whereas ATF6-p50-induced promoter activity as expected (Fig. 4D). A similar result was observed in HeLa cells (not shown). These results indicate that the activity of OASIS on the GRP78 promoter is cell type specific.
To gain insight into the complement of genes affected by the expression of nuclear-localized OASIS, we performed microarray expression profiling experiments in the inducible β-cell line. Total RNA was extracted from the control uninduced cells and after doxycycline induction for 24 and 48 h. Global gene expression changes were compared using Affymetrix rat gene chip arrays. Expression of nuclear-localized OASIS for 24 h resulted in the induction of approximately 19 genes greater than 2-fold (Table 1), with an additional 16 genes induced after expression for 48 h (Supplemental Table 1). Although these genes are involved in a diverse range of functions (signaling, metabolism, transport, and ubiquitination), the most highly induced gene was Papss2 involved in sulfate conjugation. The most common were genes involved in protein transport (Kdelr3, COPζ2, and Sv2c), proline metabolism (P5cr1 and Oat), and extracellular matrix protein production (Papss2, Matn1, and Chst12). However, no classic UPR target genes were induced, which is consistent with the results shown in Figs. 3 and 4.
To verify some of the microarray results, we examined the expression of two genes (Papss2 and Matn1) by real-time PCR and Western blotting. As shown in Fig. 5, A and B, the mRNA and protein levels of Papss2 were significantly increased by OASIS-374 expression as were the mRNA levels of Matn1 (Fig. 5C). As predicted, given that OASIS induces some genes involved in extracellular matrix formation, induction of active OASIS-374 resulted in increased cellular chondroitin sulfate proteoglycan levels (Fig. 5D). Importantly, the expression levels of Papss2 and Matn1 were also induced by ER stress caused by tunicamycin in INS-1 832/13 cells (Fig. 5, E–G), suggesting that these genes may be up-regulated as part of the ER stress response in β-cells.
OASIS protein expression may be regulated at the translational level by micro-RNAs
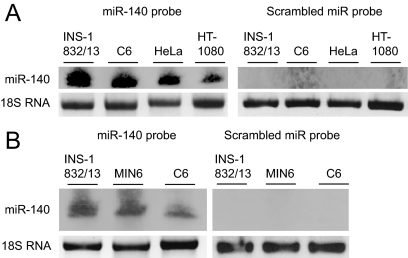
Paradoxically, although OASIS protein levels in adult islets and pancreatic β-cell lines appear to be low (Fig. 2), the mRNA levels appear to be reasonably abundant (Fig. 1). We hypothesized that OASIS protein expression may be regulated at the translational level, potentially by micro-RNAs. Recently micro-RNAs have been implicated in translational control by inhibiting translation of mRNAs (26). Interestingly, we discovered that the OASIS mRNA includes at least two regions with consensus sites for binding of the specific micro-RNA miR-140 in the 3′ untranslated region that is conserved between rat, mouse, and human (Supplemental Fig. 2). Indeed, we found abundant levels of miR-140 expressed in pancreatic β-cell lines and C6 astrocytes (Fig. 6). Whether this particular micro-RNA regulates OASIS translation in insulinoma cells and islets requires future study.
Figure 6.
miR-140 expression is detected in pancreatic β-cells. Total RNA was isolated from the indicated cell lines as described in Materials and Methods. The RNA was resolved on a 15% TBE-urea-acrylamide gel and analyzed using an miR-140-specific detection probe or a scrambled miR probe.
OASIS may play a role during pancreas development
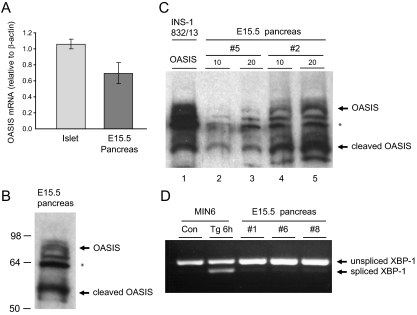
Although OASIS protein expression is detected in adult islets and pancreatic β-cell lines, the levels appear to be low. In addition, the fact that OASIS modulates the expression of genes encoding components of the extracellular matrix, the formation of which is important in development (27,28,29,30,31), led us to hypothesize that this ER stress sensor may play a prominent role in the developing pancreas as opposed to differentiated β-cells, in which it may have a lesser role. To examine whether OASIS is expressed during pancreas development, we obtained pancreas tissue from mouse embryos at E15.5. At this stage of pancreas development, endocrine cells that eventually form islets have begun to differentiate (29). The levels of OASIS mRNA were similar in E15.5 pancreas tissue compared with differentiated islets (Fig. 7A). However, whereas OASIS protein seemed to be expressed at low levels in islets (Fig. 2), it was readily detected in E15.5 pancreas tissue. Immunoblots from three independent E15.5 pancreas tissue samples are shown (Fig. 7, B and C). Although there was some variability in protein loading because the pancreas tissue was lysed directly in sample buffer due to limited material, OASIS protein was clearly detected in each sample. Even more striking was the fact that the levels of the cleaved (active) form of OASIS were even more prominent than the full-length (nonactive) form. This suggests that in the developing pancreas, there may be physiological ER stress, as has been detected in other developing tissues (32,33,34), and that OASIS is highly active during pancreas development. Indeed, low levels of spliced XBP-1 were detected in E15.5 pancreas tissue, indicative of some ER stress occurring at this stage of development (Fig. 7D).
Figure 7.
Full-length and cleaved (active) OASIS and spliced XBP-1 mRNA is detected in embryonic mouse pancreas tissue. A, Total RNA was isolated from adult mouse islets or pancreas tissue from E15.5 embryos. Analysis of OASIS mRNA levels was performed by real-time PCR and presented relative to β-actin levels. B, Pancreas tissue from E15.5 embryos was lysed directly in 40 μl 2× NuPAGE sample buffer, and 20 μl were immunoblotted for OASIS. The full-length and cleaved forms of OASIS are indicated. The asterisk marks the cross-reactive 64-kDa unknown protein detected with this antibody. C, Two additional pancreas preparations (no. 5 and no. 2) were prepared as in B, and 10 and 20 μl of samples were resolved. In lane 1, 10 μg of lysate from INS-1 832/13 cells transfected with full-length human OASIS was loaded. D, Total RNA was isolated from mouse MIN6 β-cells treated or not with thapsigargin (Tg; 1 μm, 6 h) or from E15.5 mouse pancreas tissue. Levels of unspliced and spliced XBP-1 were examined by RT-PCR.
Discussion
Although the basic ER stress response consisting of the prototypical ER stress sensors (PERK, IRE1α, and ATF6) is well described, more recently several other cell-specific ER stress sensors have been identified (6). In this study we were able to detect abundant OASIS mRNA in pancreatic β-cell lines and rodent islets by RT-PCR and real-time quantitative PCR (Fig. 1). Furthermore, at the mRNA level, ER stress-inducing compounds such as tunicamycin and thapsigargin caused an induction of OASIS mRNA levels in pancreatic β-cell lines and in the C6 glioma line. Thus, OASIS is ER stress inducible and this is cell type independent.
We were also able to detect protein expression in pancreatic β-cell lines and rodent islets, but the endogenous signal was weak, indicating that the protein may be expressed at low levels in differentiated β-cells (Fig. 2). It is possible that OASIS expression is stringently regulated at the translational level, potentially by micro-RNAs, which have been implicated in translational control (26). The OASIS mRNA includes regions with consensus sites for miR-140 binding (Supplemental Fig. 2) and this micro-RNA is detected in pancreatic β-cell lines (Fig. 6). A recent study has shown that miRNA-140 has the capability of inhibiting translation of certain proteins and is likely to have multiple targets (35). Future studies will explore whether miR-140 is responsible for regulating OASIS mRNA translation in pancreatic β-cells and other cell types.
In the pancreatic β-cell, OASIS is unlikely to contribute to inducing classic ER stress response genes based on two observations. First, the expression of an active version of OASIS failed to activate the GRP78 promoter in pancreatic β-cells, in contrast to C6 glioma cells (Fig. 4). Second, expression of an active nuclear-localized version of OASIS in an inducible pancreatic β-cell line failed to induce any classic ER stress response genes (Fig. 3 and Table 1, and Supplemental Table 1). Thus, although OASIS has the potential to induce ER stress response genes in astrocytes, it does not do so in the pancreatic β-cell lines. Cell-specific activation of the GRP78 promoter may reflect the fact that other endogenous cofactors may be required for promoter activity of certain target genes or the presence of inhibitors to OASIS activity in β-cells that are not present in astrocytes.
Interestingly, most of the genes induced by OASIS expression in INS-1 pancreatic β-cells have functions related to extracellular matrix formation and protein transport (Table 1 and Supplemental Table 1). The microarray data in the inducible pancreatic β-cell line is consistent with a recent study in OASIS knockout mice (18). A major phenotype of these mice is abnormal bone formation due to defects in osteoblast function, including reduced collagen production and secretion of bone matrix proteins. In this study the authors found no changes in ER stress response genes in osteoblasts but did identify that OASIS can activate the type 1 collagen gene Col1a1 (18). Interestingly, in the context of a pancreatic β-cell, OASIS expression induced Papss2, a gene encoding a 3′-phosphoadenosine 5′-phosphosulfate synthase required for normal bone, cartilage, and skeletal development (36). In addition, several genes involved in proline biosynthesis (P5cr1, Oat, P4ha1), protein transport (Kdelr3, COPζ2, Sv2c), and extracellular matrix protein production (Papss2, Matn1, Chst12) were up-regulated. Thus, our results are consistent with the knockout mouse phenotype and suggest that OASIS regulates genes broadly involved in extracellular matrix formation. In the pancreas it has been documented that genes involved in cell adhesion, cell migration, and extracellular matrix remodeling are important in controlling islet development (27,28,29,30,31). It remains to be determined whether pancreas and islet development is affected in OASIS knockout mice. Interpretation of a negative result, however, may be complicated by the fact that proteins homologous to OASIS exist, such as BBF2H7/CREB3L2 (8), which could possibly compensate for loss of OASIS function.
The high level of OASIS expression that we found in the developing pancreas suggests that it is likely playing a role in modulating extracellular matrix production during development. Indeed, even in mouse alveolar bone tissue, OASIS expression peaks at approximately E14.5–16.5 and decreases after E18.5 (13), suggesting that production of extracellular matrix during development is vital for normal differentiation. Physiological ER stress may be the trigger for OASIS activation in the developing pancreas. We observed evidence for ER stress in E15.5 pancreas tissue (Fig. 7D) and the PERK-eukaryotic initiation factor-2α ER stress pathway is essential for normal fetal/neonatal β-cell proliferation and differentiation, suggesting that there are physiological stimuli that induce the UPR in the developing pancreas (37,38).
Extracellular sulfated proteoglycans have been shown to regulate pancreatic endocrine differentiation during development by inhibiting endocrine cell (β and α cell) development (31). Thus, it is possible that OASIS is highly expressed in embryonic pancreas and plays a role during pancreas development by controlling extracellular matrix production, but it is repressed to reduce sulfated proteoglycan levels in fully differentiated β-cells. We hypothesize that this regulation may occur at the translational level, thus the rather low expression of OASIS protein in differentiated islets. The observation that the prolonged expression of active OASIS in a pancreatic β-cell line causes a reduction in proinsulin levels (Fig. 3) may indicate an alteration in the differentiated state of the cell, highlighting the need for stringently controlling OASIS expression. It is possible that in the differentiated β-cell, OASIS may become transiently induced when increased sulfated proteoglycan or other extracellular matrix proteins are required.
In summary, we show that endogenous OASIS protein expression is abundant in developing pancreas tissue but is expressed at low levels in differentiated islets or insulinoma cell lines. Expression of an active (nuclear localized) version of OASIS in immortalized β-cells results in the induction of various genes, some of which are involved in extracellular matrix formation, but not classic UPR target genes. OASIS-induced expression of such genes may be important in islet and β-cell development, and future studies will examine the role of OASIS in this process.
Supplementary Material
Acknowledgments
We thank Dr. Tianru Jin for comments on the manuscript and all the investigators who provided reagents and cell lines as mentioned in the Materials and Methods. We also thank Monika Sharma (University Health Network microarray facility) for help with the microarray analysis and Allison Naiquan Zhang for generating the GRP78 promoter luciferase construct.
Footnotes
This work was supported by Operating Grant MOP 86641 from the Canadian Institutes of Health Research (to A.V.); Grant NA6591 frome the Heart and Stroke Foundation of Ontario (to J.R.). M.G. is funded by Grant R01 DK071052 from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases and Grant 1-2007-548 from the Juvenile Diabetes Research Foundation International. A.V. is a recipient of a Tier II Canada Research Chair award. R.N.V. was funded by a postdoctoral fellowship from the Banting and Best Diabetes Centre (University of Toronto). M.A.G. was supported by the Vanderbilt Molecular Endocrinology Training Program 5-T32-DK07563.
Disclosure Summary: The authors have nothing to disclose.
First Published Online July 28, 2010
Abbreviations: ATF6, activating transcription factor 6; CREB, cAMP responsive element binding protein; CREB3L1, CREB 3-like 1; DIG, digoxigenin; E, embryonic day; ER, endoplasmic reticulum; GRP78, glucose-regulated protein 78; IRE1α, inositol-requiring enzyme-1α; OASIS, old astrocyte specifically induced substance; PDI, protein disulfide isomerase; PERK, RNA-dependent protein kinase-like ER kinase; TBE, Tris-borate EDTA; UPR, unfolded protein response; XBP, X-box binding protein.
References
- Lai E, Teodoro T, Volchuk A 2007 Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology 22:193–201 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P 2007 Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529 [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A 2006 Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7:880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuRose JB, Tam AB, Niwa M 2006 Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol Biol Cell 17:3095–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K 2007 How transmembrane proteins sense endoplasmic reticulum stress. Antioxid Redox Signal 9:2295–2303 [DOI] [PubMed] [Google Scholar]
- Bailey D, O'Hare P 2007 Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid Redox Signal 9:2305–2321 [DOI] [PubMed] [Google Scholar]
- Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K, Iseki K, Wanaka A, Imaizumi K 2005 OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat Cell Biol 7:186–194 [DOI] [PubMed] [Google Scholar]
- Kondo S, Saito A, Hino S, Murakami T, Ogata M, Kanemoto S, Nara S, Yamashita A, Yoshinaga K, Hara H, Imaizumi K 2007 BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol 27:1716–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ 2006 Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124:587–599 [DOI] [PubMed] [Google Scholar]
- Saito A, Hino S, Murakami T, Kondo S, Imaizumi K 2007 A novel ER stress transducer, OASIS, expressed in astrocytes. Antioxid Redox Signal 9:563–571 [DOI] [PubMed] [Google Scholar]
- Honma Y, Kanazawa K, Mori T, Tanno Y, Tojo M, Kiyosawa H, Takeda J, Nikaido T, Tsukamoto T, Yokoya S, Wanaka A 1999 Identification of a novel gene, OASIS, which encodes for a putative CREB/ATF family transcription factor in the long-term cultured astrocytes and gliotic tissue. Brain Res Mol Brain Res 69:93–103 [DOI] [PubMed] [Google Scholar]
- Hikake T, Mori T, Iseki K, Hagino S, Zhang Y, Takagi H, Yokoya S, Wanaka A 2003 Comparison of expression patterns between CREB family transcription factor OASIS and proteoglycan core protein genes during murine tooth development. Anat Embryol 206:373–380 [DOI] [PubMed] [Google Scholar]
- Nikaido T, Yokoya S, Mori T, Hagino S, Iseki K, Zhang Y, Takeuchi M, Takaki H, Takaki H, Kikuchi S, Wanaka A 2001 Expression of the novel transcription factor OASIS, which belongs to the CREB/ATF family, in mouse embryo with special reference to bone development. Histochem Cell Biol 116:141–148 [DOI] [PubMed] [Google Scholar]
- Schubert SW, Abendroth A, Kilian K, Vogler T, Mayr B, Knerr I, Hashemolhosseini S 2008 bZIP-type transcription factors CREB and OASIS bind and stimulate the promotor of the mammalian transcription factor GCMa/Gcm1 in trophoblast cells. Nucleic Acids Res 36:3834–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori Y, Imai J, Suzuki Y, Watanabe S, Tanigami A, Sugano S 2002 OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem Biophys Res Commun 293:470–477 [DOI] [PubMed] [Google Scholar]
- Murakami T, Kondo S, Ogata M, Kanemoto S, Saito A, Wanaka A, Imaizumi K 2006 Cleavage of the membrane-bound transcription factor OASIS in response to endoplasmic reticulum stress. J Neurochem 96:1090–1100 [DOI] [PubMed] [Google Scholar]
- Chihara K, Saito A, Murakami T, Hino S, Aoki Y, Sekiya H, Aikawa Y, Wanaka A, Imaizumi K 2009 Increased vulnerability of hippocampal pyramidal neurons to the toxicity of kainic acid in OASIS-deficient mice. J Neurochem 110:956–965 [DOI] [PubMed] [Google Scholar]
- Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, Saitoh M, Nishimura R, Yoneda T, Kou I, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Wanaka A, Imaizumi K 2009 Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol 11:1205–1211 [DOI] [PubMed] [Google Scholar]
- Laybutt DR, Preston AM, Akerfeldt MC, Kench JG, Busch AK, Biankin AV, Biden TJ 2007 Endoplasmic reticulum stress contributes to β cell apoptosis in type 2 diabetes. Diabetologia 50:752–763 [DOI] [PubMed] [Google Scholar]
- Marchetti P, Bugliani M, Lupi R, Marselli L, Masini M, Boggi U, Filipponi F, Weir GC, Eizirik DL, Cnop M 2007 The endoplasmic reticulum in pancreatic β cells of type 2 diabetes patients. Diabetologia 50:2486–2494 [DOI] [PubMed] [Google Scholar]
- Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB 1992 Establishment of 2-mercaptoehtnaol-dependent differentiated insulin-secreting cell lines. Endocrinology 130:167–178 [DOI] [PubMed] [Google Scholar]
- Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB 2000 Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and independent glucose-stimulated insulin secretion. Diabetes 49:424–430 [DOI] [PubMed] [Google Scholar]
- Lai E, Bikopoulos G, Wheeler MB, Rozakis-Adcock M, Volchuk A 2008 Differential activation of ER stress and apoptosis in response to chronically elevated free fatty acids in pancreatic β-cells. Am J Physiol Endocrinol Metab 294:E540–E550 [DOI] [PubMed] [Google Scholar]
- MacDonald PE, Ha XF, Wang J, Smukler SR, Sun AM, Gaisano HY, Salapatek AM, Backx PH, Wheeler MB 2001 Members of the Kv1 and Kv2 voltage-dependent K(+) channel families regulate insulin secretion. Mol Endocrinol 15:1423–1435 [DOI] [PubMed] [Google Scholar]
- Zhang L, Lai E, Teodoro T, Volchuk A 2009 GRP78, but not protein disulfide isomerase, partially reverses hyperglycemia-induced inhibition of insulin synthesis and secretion in pancreatic β-cells. J Biol Chem 284:5289–5298 [DOI] [PubMed] [Google Scholar]
- Meister G 2007 miRNAs get an early start on translational silencing. Cell 131:25–28 [DOI] [PubMed] [Google Scholar]
- Cirulli V, Beattie GM, Klier G, Ellisman M, Ricordi C, Quaranta V, Frasier F, Ishii JK, Hayek A, Salomon DR 2000 Expression and function of α(v)β(3) and α(v)β(5) integrins in the developing pancreas: roles in the adhesion and migration of putative endocrine progenitor cells. J Cell Biol 150:1445–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding Crawford L, Tweedie Ables E, Oh YA, Boone B, Levy S, Gannon M 2008 Gene expression profiling of a mouse model of pancreatic islet dysmorphogenesis. Plos ONE 3:e1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guney MA, Gannon M 2009 Pancreas cell fate. Birth Defects Res C Embryo Today 87:232–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Li J, Lyte K, Yashpal NK, Fellows F, Goodyer CG 2005 Role for β1 integrin and its associated α3, α5, and α6 subunits in development of the human fetal pancreas. Diabetes 54:2080–2089 [DOI] [PubMed] [Google Scholar]
- Zertal-Zidani S, Bounacer A, Scharfmann R 2007 Regulation of pancreatic endocrine cell differentiation by sulfated proteoglycans. Diabetologia 50:585–595 [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Pypaert M, Glimcher LH 2007 The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J Exp Med 204:2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH 2005 XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J 24:4368–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang KY, Chan D, Cheslett D, Chan WC, So CL, Melhado IG, Chan TW, Kwan KM, Hunziker EB, Yamada Y, Bateman JF, Cheung KM, Cheah KSE 2007 Surviving endoplasmic reticulum stress is coupled to altered chondrocyte differentiation and function. Plos Biol 5:e44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais H, Nicolas FE, Soond SM, Swingler TE, Clark IM, Chantry A, Moulton V, Dalmay T 2010 Analyzing mRNA expression identified Smad3 as a microRNA-140 target regulated only at protein level. RNA 16:489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam KV 2003 Human 3′-phosphoadenosine 5′-phosphosulfate (PAPS) synthase: biochemistry, molecular biology and genetic deficiency. IUBMB Life 55:1–11 [DOI] [PubMed] [Google Scholar]
- Zhang W, Feng D, Li Y, Iida K, McGrath B, Cavener DR 2006 PERK EIF2AK3 control of pancreatic β-cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab 4:491–497 [DOI] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ 2001 Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7:1165–1176 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.