Abstract
There is evidence for hypercortisolemia playing a role in the generation of psychiatric symptoms and for epigenetic variation within hypothalamic-pituitary-adrenal (HPA) axis genes mediating behavioral changes. We tested the hypothesis that expression changes would be induced in Fkbp5 and other HPA axis genes by chronic exposure to corticosterone and that these changes would occur through the epigenetic mechanism of loss or gain of DNA methylation (DNAm). We administered corticosterone (CORT) to C57BL/6J mice via their drinking water for 4 wk and tested for behavioral and physiological changes and changes in gene expression levels using RNA extracted from hippocampus, hypothalamus, and blood for the following HPA genes: Fkbp5, Nr3c1, Hsp90, Crh, and Crhr1. The CORT mice exhibited anxiety-like behavior in the elevated plus maze test. Chronic exposure to CORT also caused a significant decrease in the hippocampal and blood mRNA levels of Nr3c1 and a decrease in Hsp90 in blood and caused an increase in Fkbp5 for all tissues. Differences were seen in Fkbp5 methylation in hippocampus and hypothalamus. To isolate a single-cell type, we followed up with an HT-22 mouse hippocampal neuronal cell line exposed to CORT. After 7 d, we observed a 2.4-fold increase in Fkbp5 expression and a decrease in DNAm. In the CORT-treated mice, we also observed changes in blood DNAm in Fkbp5. Our results suggest DNAm plays a role in mediating effects of glucocorticoid exposure on Fkbp5 function, with potential consequences for behavior.
DNA methylation decreased, expression of the glucocorticoid signaling-related gene Fkbp5 increased, in response to corticosterone administration, suggesting epigenetic influences in the glucocorticoid–Fkbp5 interaction.
Strong evidence for a role for hypercortisolemia in the generation of psychiatric symptoms comes from observations of patients with Cushing syndrome, a condition characterized by hypersecretion of cortisol. The syndrome typically includes anxiety, depression, irritability, and insomnia (1). Psychiatric impairment in Cushing syndrome is directly correlated with cortisol concentrations (1,2) and symptoms improve in direct relationship to resolution of the hypercortisolemia (3).
The relationship between hypercortisolemia and psychiatric symptoms is mediated by the hypothalamic-pituitary-adrenal (HPA) axis, involving a number of critical proteins including the glucocorticoid receptor (GR), encoded by the gene Nr3c1, CRH, a neuropeptide regulating ACTH, and glucocorticoid secretion through activation of the CRH receptor-1. The GR protein interacts with a molecular complex that includes the chaperone heat shock protein 90 and the cochaperone FKBP5. Variations in the genes encoding these proteins may influence the response to glucocorticoids. Previous studies have found elevated levels of FKBP5 expression in response to glucocorticoid administration, and genetic variation in this gene has been associated with disorders characterized by abnormal glucocorticoid regulation, including posttraumatic stress disorder (4) and mood disorders (5,6,7).
Epigenetics plays a critical role in brain diseases. A dramatic example is Rett syndrome, in which loss of function of methyl-CpG-binding protein 2 (MeCP2), which recognizes DNA methylation (DNAm), leads to a failure to maintain gene silencing in the brain and a subsequent loss of developmental milestones (8). Whereas Rett syndrome results from a mutation, there is also ample evidence that the epigenetic machinery can be altered by the environment (9,10). For example, administration of the anticonvulsant and antibipolar disorder medication valproate can cause changes in DNAm (11). Epigenetics may be a mechanism through which GR signaling influences brain function and mood. Glucocorticoid hormones alter gene expression in brain (12,13), and these alterations may be related to epigenetic modifications. Several genes, including some central to the HPA axis, appear to be epigenetically modified by glucocorticoids. For example, in a paradigm in which a human lung cancer cell line was exposed to dexamethasone, FKBP5 showed elevated levels of histone H3 acetylation and histone H3-lysine 4 trimethylation, indicating chromatin activation (14).
Whereas expression and DNAm changes in the brain are more obviously relevant to glucocorticoid-induced changes in behavior, comparable changes in blood might provide a clinically valuable surrogate, given the easy access to this tissue in patients. Some earlier studies have shown partial correlation in gene expression between brain and blood (15,16). Furthermore, two papers have demonstrated that epigenetic marks in lymphocytes can distinguish patients from controls in two brain diseases, Rett syndrome (17) and Alzheimer’s disease (18). In the realm of HPA axis genes, previous work has shown that FKBP5 mRNA levels in lymphocytes have functional relevance because they have a positive correlation with plasma cortisol (5).
In this report we tested the hypothesis that differential exposure to high-dose corticosterone would cause behavioral abnormalities as well as expression changes in HPA axis genes mediated through DNAm changes in the promoter or other key regions. We used a paradigm in which corticosterone was added to the drinking water (100 μg/ml) and given to mice for 4 wk followed by a 4-wk recovery period in which corticosterone was not administered. We then tested the mice for behavioral and physiological changes as well as expression and methylation differences in mRNA and DNA extracted from hippocampus, hypothalamus, and blood for the following genes: Fkbp5, Nr3c1, Hsp90, Crh, and Crhr1. We followed up these results in two ways. First, we sought greater homogeneity within the brain to determine whether subtle differences in Fkbp5 methylation we observed in hypothalamus and hippocampus might be amplified by examining a narrower brain subregion and then a single neuronal cell line. Second, we sought to replicate the results we observed in blood of DNAm change in Fkbp5, using tissues derived from a second cohort.
Materials and Methods
Animals
Four-week-old male C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) were housed in conventional polycarbonate mouse cages in a temperature- and humidity-controlled room on a 12-h light, 12-h dark cycle with light onset at 0600 h. All animals received ad libitum access to water and standard laboratory chow (Harlan Teklad 2018, Indianapolis, IN) for 1 wk after arrival in the laboratory. At 5 wk of age, animals were given ad libitum access to solutions containing corticosterone [Sigma-Aldrich, St. Louis, MO; 100 μg/ml with 1% ethanol; corticosterone (CORT) group] or vehicle (1% ethanol; control group) in place of their normal drinking water, and this treatment continued for 4 wk (cohorts 1 and 2). The CORT and vehicle solutions were made fresh daily, and the animals were given ad libitum access to 0.9% saline solution to prevent symptoms of adrenal insufficiency. A second cohort was used in which the animals were treated with CORT for 4 wk followed by a recovery period during which CORT was not administered. All procedures were approved by the Institutional Animal Care and Use Committee at Johns Hopkins University School of Medicine and were performed in accordance with guidelines established in the National Research Council’s Guide for the Care and Use of Laboratory Animals.
Blood collection
Blood samples were collected weekly 3 h after onset of the light cycle (0900 h) to verify plasma corticosterone levels. Animals remained in a quiet room, and about 20 μl of whole blood was collected from each mouse into heparinized glass capillary tubes through a small nick at the tip of the tail. Blood glucose levels were determined simultaneously by a handheld glucometer (Freestyle, Alameda, CA). Blood samples were centrifuged at 4 C and plasma was collected and frozen at −80 C for later RIA. Total RNA was extracted from remaining blood cells at each time point. A second blood sample (∼70 μl whole blood) was collected immediately after the first for genomic DNA extraction.
Elevated plus maze (EPM)
Mice were tested for anxiety-like behavior in an elevated plus maze (arms: 6.0 × 29.5 cm; center: 6.0 × 6.0 cm; Harvard Apparatus, Holliston, MA) raised 55.0 cm above the ground. Two arms had 15.0 cm high opaque walls (closed), whereas the remaining two arms remained open (open). Animals were moved to a quiet room outside the testing room. Each animal was tested in the EPM for 5 min. The apparatus was thoroughly cleaned with 70% ethanol after testing of each animal. The test was video recorded and subsequently scored to measure time spent in the open and closed arms, using computerized behavioral scoring software Hindsight (Scott Weiss, UK), by observers blinded to the subject’s treatment group.
Tail suspension test
Animals were tested using the tail suspension test to assess depressive-like behavior. Mice were moved to a quiet testing room and tested in a separate behavioral room. Each mouse was suspended by its tail from an aluminum bar and secured with adhesive tape. Behavior was recorded for 6 min. Observers blinded to the subject’s treatment group scored video recordings for time spent struggling or immobile, using computerized behavioral scoring software (Hindsight; version 1.5).
Tissue acquisition
Animals were killed by decapitation. Brains were removed, frozen immediately on powdered dry ice, and stored at −80 C. Spleen, adrenal glands, and thymus were dissected and weighed. Whole carcasses were frozen for later body composition and fat distribution analysis.
Hippocampus, dentate gyrus, and hypothalamus dissection
Frozen mouse brains were sectioned using a cryostat, and 200-μm sections were mounted on plain glass slides. For the first cohort of mice, entire hippocampal and hypothalamic tissues were dissected. For the second cohort, a 19-gauge needle (inner diameter 0.686 mm × outer diameter 1.086 mm) was used to punch out the dentate gyrus from sections containing the hippocampus (bregma −1.22 through −2.30 mm). A similar approach was used to punch out the relevant regions from the hypothalamus (bregma −0.58 through −1.06 mm). Samples were stored at −80 C until processed for genomic DNA and mRNA.
Body composition by nuclear magnetic resonance (NMR)
In this established procedure (19), all of the skin and fat attached to the skin is gently removed from the carcass. The skin and attached sc fat (i.e. the pelt) are then analyzed for fat content separately from the rest of the body, which contains bone, muscle, organs, and visceral fat. Fat pads that are included in the pelt include all fat attached to the skin and outside the peritoneal cavity (i.e. including dorso-sc and inguinal fat pads). All fat within the rest of the body (i.e. inside the muscle layer of the body) is nonpelt, and it includes all of the visceral fat and intraorgan (e.g. liver, heart) fat. Because most of this is visceral fat, we use that term as a descriptor. The two samples were analyzed by NMR technology (EchoMRI, Waco, TX), which reports fat, lean, and water content of the animal. Samples (at room temperature) were placed into a plastic restrainer and inserted into the NMR machine. Use of the NMR method to determine body composition has a significant correlation (r = +0.98, P < 0.01) with adipose content determined by a chemical method (lyophilization and ether extraction) (19,20,21).
RIA
Plasma hormone levels were determined by commercially available RIA kits for corticosterone (MP Biomedicals, Costa Mesa, CA) according to the manufacturer’s instructions as previously described (22). All samples were run in duplicate and comparisons were made within assay with coefficient of variance of 3.9%.
Cell lines
HT-22 cells derived from the mouse hippocampus were cultured using DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) under standard conditions (5% CO2, 37 C). Cells were trypsinized and replated in six-well plates before treatment with corticosterone. Corticosterone (Sigma) was prepared by first making a stock solution at 50 mm concentration in ethyl alcohol (EtOH) and then diluting the stock a further 10-fold to make a 5-mm working solution. The working solution was directly added to DMEM to make 1 μm concentration (23). The control samples were treated with an equal volume of EtOH. Media were changed daily, and the collected media were used to verify levels of corticosterone by RIA. Cells were treated for 0 h, 6 h, 1 d, 3 d, 5 d, and 7 d. In addition, cells were cultured for an additional 7 d in the absence of corticosterone treatment. Cells were split every 2 d to maintain them in the log phase of growth. After each time point, cells were harvested for genomic DNA and mRNA.
Genes
Fkbp5 was chosen because of prior evidence for epigenetic regulation (14) as well as involvement in glucocorticoid-related psychiatric illness (4,5,6,7). Because of our prior interest in this gene, we chose regions with known glucocorticoid response elements (intron 5 region) (24,25) and/or methyl-CpG binding domains (intron 1 region) (26) to assay in addition to the promoter region. We also chose Nr3c1 because of its role in regulating glucocorticoid-negative feedback and because of prior evidence for DNAm variation (27), examining the sequence orthologous to that previously shown to be altered. We further studied Hsp90, Crh, and Crhr1 due to their association with the HPA axis and glucocorticoid signaling.
Expression
mRNA from blood, hippocampus, and HT-22 cell lines was obtained using the MasterPure RNA purification kit (Epicentre Biotechnologies, Madison, WI) or the RNeasy lipid tissue mini kit (QIAGEN, Valencia, CA). SuperScript III (Invitrogen) was used to generate cDNA for subsequent quantitative real-time PCR. Negative reverse transcriptase samples were used to ensure the absence of contaminating genomic DNA. All reactions were carried out in triplicate using 1× Taqman master mix (Applied Biosystems, Foster City, CA), 1× Taqman probes for each gene [Fkbp5, Nr3c1, Hsp90, Crh, Crhr1, and Actb (β-actin)], and 30 ng of cDNA template in a total volume of 20 μl. Real-time reactions were performed on an Applied Biosystems 7900HT fast real-time PCR system with standard PCR conditions (50 C for 2 min; 95 C for 10 min; and 60 C for 1 min for 40 cycles). Each set of triplicates was checked to ensure that the threshold cycle (Ct) values were all within 1 Ct of each other. To determine relative expression values, the −ΔΔCt method (Applied Biosystems) was used, where triplicate Ct values for each mouse sample were averaged and subtracted from those derived from the housekeeping gene Actb. The Ct difference for a calibrator sample was subtracted from those of the test samples, and the resulting −ΔΔCt values were raised to a power of 2 to determine normalized relative expression.
DNA extraction and bisulfite treatment
Blood and cell line genomic DNA was isolated with the MasterPure DNA purification kit (Epicentre Biotechnologies). Hippocampal genomic DNA was isolated by an overnight digestion in a sodium dodecyl sulfate lysis buffer (1% sodium dodecyl sulfate, 0.15M NaCl, and 0.5 mg/ml proteinase K in Tris/EDTA buffer) followed by two phenol extraction steps and a final phenol/chloroform/isoamyl alcohol extraction. DNA was precipitated with EtOH and sodium acetate buffer, and the subsequent pellet was resuspended in H2O. Bisulfite conversion of 500 ng of genomic DNA was achieved through use of the Epitect bisulfite kit (QIAGEN), according to the manufacturer’s instructions.
Bisulfite sequencing
Sequencing of bisulfite PCR products was used as a qualitative screen on many of the assays of interest. This was done using an ABI Prism 3100 genetic analyzer (Applied Biosystems), and data were analyzed using CodonCode Aligner (CodonCode Corporation, Dedham, MA). The chromatograms were analyzed to determine qualitative methylation differences and to ensure the absence of unconverted cytosines. PCR products were subsequently subjected to bisulfite pyrosequencing.
Bisulfite PCR and pyrosequencing
We measured DNAm by pyrosequencing of the PCR products, which measures methylation variation at greater than 90% precision (28). Primers were designed against the promoter and other regions of HPA axis genes. Regions covered were from 150 to 450 bp long. Thermocycling was carried out using Peltier Thermal Cycler 200 (MJ Research, Waltham, MA). Twenty-five nanograms of bisulfite-treated DNA were used for each PCR. An additional nested PCR was performed with 2 μl of the previous PCR and one biotinylated primer (other primer being unmodified). Amplification for both PCR steps consisted of 40 cycles (94 C for 1 min, 60 C for 30 sec, 72 C for 1 min). PCR products were confirmed on agarose gels. Pyro Gold reagents (QIAGEN) were used to prepare samples for pyrosequencing according to the manufacturer’s instructions. For each sample, biotinylated PCR product was mixed with streptavidin-coated Sepharose beads (GE Healthcare, Indianapolis, IN), binding buffer, and Milli-Q water and shaken at room temperature. The vacuum prep tool (QIAGEN) was used to isolate the Sepharose bead-bound single-stranded PCR products. The attached DNAs were released into a PSQ HS 96-plate containing sequencing primer in annealing buffer (QIAGEN). Pyrosequencing reactions were performed in a PyroMark MD system (QIAGEN). CpG methylation quantification was performed with Pyro Q-CpGt 1.0.9 software (QIAGEN). An internal quality-control step was used to disqualify any assays that contained unconverted DNA. Percentage of methylation at each CpG as determined by pyrosequencing was compared between DNA from CORT and control animals.
Analysis
Data were analyzed by t tests to determine the P values for each group of tissues examined (Simple Interactive Statistical Analysis statistics). P < 0.05 was considered statistically significant.
Results
For cohort 1 (n = 5 per treatment group), plasma corticosterone levels were 209 ± 40 ng/ml for corticosterone-treated and 83 ± 18 ng/ml for control mice at wk 4. For cohort 2 (n = 10 per treatment group), corticosterone levels were 290 ± 27 ng/ml for corticosterone-treated and 116 ± 23 ng/ml for control mice. Animals were killed after 4 wk of exposure, and blood and brain tissues were harvested for genomic DNA and total RNA. Only half of the animals in the second cohort were killed at wk 4, and the other half was allowed to recover in the absence of corticosterone. After 4 wk of recovery, plasma corticosterone levels were 75 ± 17 ng/ml for the CORT and 94 ± 21 ng/ml for the control group.
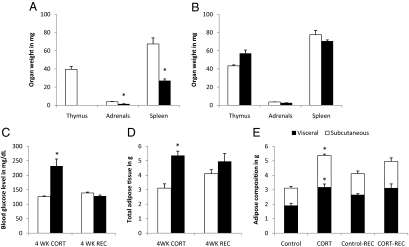
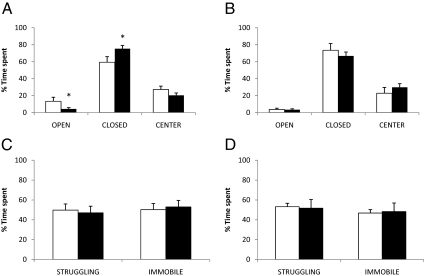
After 4 wk of treatment with corticosterone in the drinking water to mimic Cushing syndrome, we observed notable physiological changes in CORT mice, including significant reductions in the mass of the thymus, spleen, and adrenal glands (Fig. 1A) as well as increases in blood glucose levels and adipose tissue deposition (Fig. 1, C and D, left panels). The increase in adipose tissue mass reflected an increase in both visceral and sc fat depots (Fig. 1E, left panel). However, there were no significant differences in body weight between the two groups of animals (4 wk treatment: control 22.1 ± 0.4 g, CORT 22.2 ± 0.4 g; P = n.s.). All of these physiological parameters returned to levels comparable with control mice after 4 wk of recovery (Fig. 1, B–E, right panels). The CORT mice also exhibited anxiety-like behavior in the EPM test, spending more time in the closed arms (16% increase, P = 0.05) compared with control mice (Fig. 2A). This difference on the EPM test also diminished to near control levels once the CORT-treated mice were allowed 4 wk of recovery (Fig. 2B). On the other hand, no behavioral differences were observed between CORT and control mice in the tail suspension test after 4 wk of corticosterone treatment (Fig. 2C) nor at the conclusion of the 4-wk recovery period (Fig. 2D).
Figure 1.
Organ weights and physiological measurements. A, Thymus, spleen, and adrenal glands were weighed for corticosterone-treated (black bars) and vehicle-treated (white bars) mice. The absence of data for thymic mass of corticosterone-treated mice reflects a complete atrophy of the organ after 4 wk of treatment with corticosterone. B, Weight measurements of the same organs from corticosterone and vehicle-treated mice after the mice were allowed to recover for 4 wk in the absence of corticosterone in the drinking water. C and D, Effect of corticosterone (4 wk CORT) on the blood glucose level and total adipose tissue mass, respectively. The right panel of each graph shows corresponding measurements after the recovery period (4 wk REC). Adipose tissues were further analyzed by visceral (black bars) and sc (white bars) fat (E). Asterisks indicate differences that are statistically significant (P < 0.05) between the CORT and control groups. Organ weights were not normalized to body weights because mean body weight between the CORT-treated and the control groups were virtually identical.
Figure 2.
Mice were challenged with the elevated plus maze (EPM) and tail suspension test (TST) to test for anxiety-like and depressive-like symptoms, respectively. EPM: After 4 wk of treatment (A), corticosterone (black bars) and vehicle-treated (white bars) animals were each tested for a 10-min period, and the amount of time spent in each of the open arms (OPEN), closed arms (CLOSED), and the center (CENTER) is displayed as a percentage of the total time spent (10 min) on the apparatus. Same measurements were made at the conclusion of the 4-wk recovery period (B). TST: After 4 wk of treatment (C), corticosterone (black bars) and vehicle-treated (white bars) animals were tested for a 6-min period, and the amount of time spent struggling (STRUGGLING) or immobile (IMMOBILE) is displayed as a percentage of the total time spent (6 min) on the suspension apparatus. The same measurements were made at the conclusion of the 4-wk recovery period (D). Asterisks indicate differences that are statistically significant (P < 0.05).
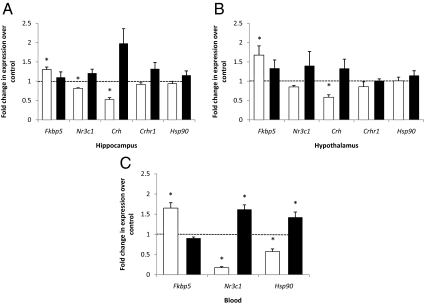
Relative hippocampal gene expression levels for cohort 1 in the CORT treatment phase and the recovery phase are shown in Fig. 3A. A significant increase in expression with treatment in CORT mice was observed for Fkbp5 (130%, P = 0.0063), whereas significant decreases were seen in Nr3c1 (18%, P = 0.006) and Crh (47%, P = 0.027). Relative expression levels for the hypothalamus are shown in Fig. 3B. Again, a significant increase in expression with CORT treatment was seen for Fkbp5 (168%, P = 0.043), whereas a significant decrease was noted in Crh (58%, P = 0.027). We also tested expression levels in blood to assess the feasibility of using blood mRNA as a surrogate for that in brain regions. We observed an increase in Fkbp5 with CORT treatment (165%, P = 0.007) and decreases in Nr3c1 (17%, P = 0.006) and Hsp90 (57%, P = 0.0003) in CORT mice (Fig. 3C). No significant expression differences were observed between CORT and control mice in the recovery phase for either brain region; differences were seen in blood only for Nr3c1 and Hsp90.
Figure 3.
Relative HPA axis gene expression levels in hippocampus (A), hypothalamus (B), and blood (C). The white bars show the gene expression levels of CORT treated mice compared with those of the vehicle solution-treated control mice at the conclusion of 4 wk of the treatment phase of the experiment (n = 5). The black bars show the gene expression levels of CORT-treated mice compared with those of the corresponding control mice at the conclusion of 4 wk of the recovery phase of the experiment, in the absence of corticosterone (n = 5). Expression levels were normalized to those of β-actin. The dotted line in each graph is used to indicate the normalized levels of gene expression for the control group. Asterisks represent statistically significant differences between the CORT group and the corresponding control group of the same time period. In blood, levels of Crh and Crhr1 were undetectable.
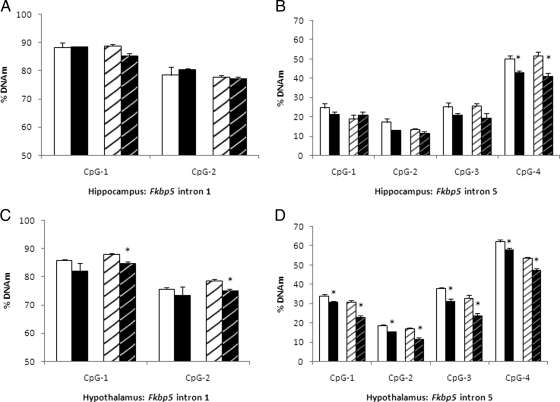
To better understand the mechanism behind CORT-induced changes in gene expression, we assayed for changes in DNAm in promoters (and additional regions for Fkbp5) in the same three tissues in which expression was measured. All sequences assayed in cohort 1 are depicted in Table 1. The results of our bisulfite sequencing for DNAm assessment in hippocampus, hypothalamus, and blood are seen in Tables 2–4. We detected notable DNAm changes in the functionally relevant intron 1 and 5 regions of Fkbp5. The intron 5 region, a distal enhancer containing two glucocorticoid response elements, showed modestly decreased DNAm in the hippocampus (CpG-4: CORT, 43% vs. control, 50%, P = 0.004), and the hypothalamus (greatest difference seen in CpG-3: CORT, 31% vs. control, 38%, P = 0.003). Interestingly, these differences persisted after 4 wk of recovery without CORT treatment (Fig. 4, B and D). There were no significant differences seen during treatment in these brain regions for the intron 1 region. By contrast, in blood, there were no significant differences in DNAm for the enhancer region in intron 5, but the region in intron 1 of Fkbp5 that binds a transcriptional repressor showed substantial differences (CpG-1: 61.0% CORT vs. 83.6% control, P = 0.0001 and CpG-2: 54.9% CORT vs. 76.8% control, P = 0.0001). Persistence of these DNAm differences was not seen at 4 wk of recovery.
Table 1.
CpGs assayed with bisulfite pyrosequencing
| Gene | Position | Genome Browser coordinates
|
Position relative to transcript
|
Size | Assaya | Sequencee | ||
|---|---|---|---|---|---|---|---|---|
| Start | End | Start | End | |||||
| Nr3c1 | Promoterb | 39649870 | 39650086 | −3201 | −2984 | 217 | Py1/Py2 | CCCGcCCCCCGcCCCAGCTCCGCGGCTTGGAACCGCTCGGGAGCCCGCCCCGTGGGTCTGCCGCCAGAGCCCCCGAGGGG |
| Crh | Promoter | 19595332 | 19595485 | −1010 | −856 | 154 | Py1/Py2 | CGACCCTCTTCAGAAAGCACGCAGCATTTGCCTAATAAGCTGACGCTCTCTTGACAGCTCGATTGCG |
| Crhr1 | Promoter | 103994167 | 103994612 | −665 | −219 | 446 | Py1/Py2 | CGGGCCGAACGAGCGCGGGACAGGCGTGGGCAGGGGTGCCGGAGGAGAGGATCGGGGAACCCGGCG |
| Hsp90 | Promoter | 111935054 | 111935176 | −449 | −571 | 122 | Py1 | CGCCTCCCGGACTGGGGTTCTGGTTGCG |
| Py2 | GGCTGACCCCGCTGGAGTGT | |||||||
| Py3 | CGGTGGGTCTTACAAATCCG | |||||||
| Fkbp5 | Promoterd | 28654990 | 28655200 | −32167 | −31957 | 210 | Py1 | CGCGGCGCTGCAAGACCG |
| Py2 | CCACATGGTCGGGTACG | |||||||
| Py3 | CGGAGTACCCTCCTCACTGTACCCACGCATGGGGAGAACCACGGAGATGCCG | |||||||
| Fkbp5 | Intron 1 | 28578186 | 28578235 | 44798 | 44847 | 49 | Py1 | TTCCCGTCTAGCCACGGTCCTAG |
| Fkbp5 | Intron 5 | 28557301 | 28557495 | 65732 | 65538 | 194 | Py1 | CAGGCGCTCACCGAGCTCA |
| Py2 | AATCGGAGAACAAATTGCTGTCAGCACATCGAGTT | |||||||
The assay column includes specific pyrosequencing primers used (e.g. Py1) to assess percent methylation.
This sequence is the promoter for transcript 7 (27).
These CpGs were implicated by Weaver et al. (27).
These CpGs are located in the promoter CpG island of the alternate exon.
CpG dinucleotides assayed by pyrosequencing are displayed in bold.
Table 2.
Bisulfite pyrosequencing results: percent DNAm in the treatment phase by individual CpG in CORT vs. control mice in the hippocampusa
| Gene | CpG position
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Nr3c1 | 3.7/5.4 | 2.5/3.4 | 1.5/1.4 | 4.6/1.5 | 1.5/1.4 | 2.4/3.1 | 2.4/2.4 | 2.4/2.4 | ||
| Crh | 10.0/10.0 | 30.5/29.4 | 37.0/36.6 | 16.7/16.2 | 19.4/18.4 | |||||
| Crhr1 | 1.4/1.2 | 3.1/3.3 | 2.3/1.7 | 2.8/3.8 | 3.0/3.1 | 3.6/2.9 | 5.3/5.4 | 4.0/4.3 | 4.3/4.2 | 0.7/1.8 |
| Hsp90 | 2.7/2.1 | 3.4/4.1 | 5.1/4.5 | 2.5/2.0 | 3.6/3.7 | 8.2/7.0 | ||||
| Fkbp5- promoter | 10.4/8.8 | 9.5/4.3 | 16.8/12.6 | 40.2/40.0 | 17.6/15.0 | 9.1/12.0 | 14.8/15.4 | 7.7/5.3 | 24.3/19.6 | 19.3/22.1 |
| Fkbp5-Int1 | 88.6/88.3 | 80.4/78.4 | ||||||||
| Fkbp5-Int5 | 21.4/24.8 | 12.9/17.2 | 20.7/25.2 | 43.1/50.2 | ||||||
Values that are significantly different between corticosterone-treated (first value) and control (second value) groups at a nominal P < 0.05 are in bold.
Table 3.
Bisulfite pyrosequencing results: percent DNAm in the treatment phase by individual CpG in CORT vs. control mice in the hypothalamusa
| Gene | CpG position
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Nr3c1 | 4.5/5.0 | 2.4/3.4 | 1.0/1.6 | 2.2/2.4 | 3.0/2.8 | 2.4/3.3 | 6.4/6.7 | 1.9/3.3 | ||
| Crh | 24.6/25.0 | 51.6/47.6 | 59.9/58.8 | 35.0/32.5 | 31.4/30.9 | |||||
| Crhr1 | 1.1/1.5 | 2.5/2.1 | 1.8/1.9 | 3.3/3.0 | 2.0/1.9 | 1.9/1.7 | 4.2/5.0 | 2.7/2.7 | 3.2/3.6 | 1.6/1.4 |
| Hsp90 | 2.1/2.2 | 3.7/3.4 | 5.4/5.4 | 1.9/2.0 | 3.8/3.9 | 9.1/9.3 | ||||
| Fkbp5- promoter | 5.0/5.1 | 5.5/4.8 | 11.2/10.9 | 46.2/48.6 | 18.1/17.0 | 6.3/6.0 | 12.7/16.5 | 7.5/7.8 | 17.3/20.3 | 22.7/19.4 |
| Fkbp5-Int1 | 82.0/85.9 | 73.4/75.6 | ||||||||
| Fkbp5-Int5 | 30.5/34.0 | 15.5/18.6 | 31.1/37.9 | 57.8/62.2 | ||||||
Values that are significantly different between corticosterone-treated (first value) and control (second value) groups at a nominal P < 0.05 are in bold.
Table 4.
Bisulfite pyrosequencing results: percent DNAm in the treatment phase by individual CpG in CORT vs. control mice in the blooda
| Gene | CpG position
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Nr3c1 | 3.0/3.4 | 2.4/2.4 | 1.3/1.5 | 1.2/1.7 | 1.2/1.6 | 1.8/2.6 | 1.8/2.4 | 1.5/2.3 | ||
| Crh | 18.2/18.7 | 49.3/50.0 | 54.0/59.7 | 30.0/33.3 | 21.1/24.0 | |||||
| Crhr1 | 6.7/8.0 | 10.2/11.5 | 5.1/6.0 | 10.8/11.7 | 7.5/7.6 | 8.6/8.4 | 17.1/22.0 | 6.4/7.7 | 6.8/6.3 | 3.4/4.2 |
| Hsp90 | 3.0/1.9 | 3.7/4.0 | 5.7/5.5 | 1.7/2.5 | 5.6/3.8 | 6.5/7.5 | ||||
| Fkbp5- promoter | 14.6/10.9 | 7.4/6.9 | 22.5/19.8 | 51.0/50.9 | 20.6/19.3 | 11.7/10.1 | 8.7/9.2 | 7.5/6.3 | 32.4/38.4 | 27.8/28.9 |
| Fkbp5-Int1 | 61.0/83.6 | 54.9/76.8 | ||||||||
| Fkbp5-Int5 | 11.4/12.6 | 15.5/20.7 | 11.7/10.5 | 24.0/26.9 | ||||||
Values that are significantly different between corticosterone-treated (first value) and control (second value) groups at a nominal P < 0.05 are in bold.
Figure 4.
Assessment of hippocampal and hypothalamic Fkbp5 methylation in the two intronic regions. Percent methylation at two CpG positions is shown for hippocampal Fkbp5 intron 1 region (A) and four CpG positions for the intron 5 region (B). Each intronic region was assayed for both CORT (black bars) and control (white bars) groups. Solid bars on the left indicate the treatment phase, whereas striped bars on the right indicate the recovery phase. The comparable data for the hypothalamus is shown in C and D. Asterisks represent statistically significant differences between the CORT and control groups.
We followed up these results in two ways. First, we sought greater homogeneity within the brain to determine whether we might find greater DNAm changes with such a strategy. Second, we sought to replicate the results we observed in blood.
Because of the functional diversity of subregions and cell types in the hippocampus, we reasoned that the small intron 5 region methylation differences we observed using this whole-brain region might be magnified if we focused on a narrower, more homogenous hippocampal unit. For this purpose, we examined the dentate gyrus, a hippocampal subregion. We observed lower DNAm in the CORT-treated group of a 2.7–10.2% magnitude across four CpGs, although these differences did not reach statistical significance (data not shown). We next studied a mouse hippocampal cell line, HT-22, which has previously been shown to express glucocorticoid receptors (23). Levels of corticosterone were measured at 259 ng/ml for corticosterone-treated media, a level quite comparable with that seen in the CORT mice, and 19 ng/ml for vehicle solution-treated media. We again observed increased expression levels of Fkbp5 in the corticosterone-treated samples. At 7 d, the expression difference was 2.4-fold (P = 0.036, Fig. 5A), and at the same time DNAm was as much as 21.3% (P = 0.014) lower in the treated group at the four CpG positions in the intron 5 region (Fig. 5B).
Figure 5.
Expression and methylation levels for Fkbp5 in the mouse hippocampal neuronal cell line HT-22. Cells were treated with 1 μm corticosterone for 7 d and cultured in the absence of the hormone for an additional 7 d. Media collected from corticosterone (black triangles with dashed lines) and vehicle-treated (white circles with solid lines) samples showed average hormone levels of 264 and 19.9 ng/ml, respectively. Cells were harvested for mRNA and genomic DNA to determine the expression levels of Fkbp5 (A) and methylation differences at intron 5, CpG position 3 (B) and intron 1, and CpG position 2 (C). Asterisks indicate differences that are statistically significant (P < 0.05).
In the recovery period, Fkbp5 expression returned to normal within a single day, whereas DNAm differences remained for another 7 d. We did not observe any significant changes in the intron 1 region of Fkbp5 in the cell line (Fig. 5C) throughout the treatment and recovery period.
To replicate our observation of expression and DNAm changes in Fkbp5 in blood, we examined a second cohort of mice. In our Fkbp5 intron 1 assay, we again saw a difference at wk 4 of CORT treatment in blood DNAm (CpG-1: 56.8% CORT vs. 80.2% control, P = 0.0001). After 4 wk of recovery, DNAm differences disappeared (CpG-1: 83.5% CORT vs. 85.2% control). Concomitantly, corticosterone-dependent changes in Fkbp5 expression returned to baseline (Fig. 6).
Figure 6.
Gene expression and DNAm for Fkbp5 in blood for cohort 2. A, A time-course change (weeks) in blood DNAm of Fkbp5 intron 1, CpG position 1 in the second cohort, in which significant methylation differences were observed between corticosterone-treated (black triangles with dashed lines) and vehicle-treated groups (white circles with solid lines) during the 4-wk treatment period and the subsequent 4-wk recovery period. B, Expression levels of Fkbp5 in blood during the 8-wk period that includes 4 wk of treatment with corticosterone and 4 wk of recovery. Asterisks indicate differences that are statistically significant (P < 0.05).
Discussion
This study was conducted to better understand the potential effects and mechanisms of chronic corticosterone action on expression of Fkbp5 and related stress genes and on DNAm in those genes as well as on mouse behavior. To this end, we performed real-time expression assays on Fkbp5 and four other genes. We observed significant differences in expression for three of the five genes studied. Assessment of promoter and intronic DNAm between corticosterone-treated and control groups showed several CpGs that underwent significant demethylation in blood, most strikingly within Fkbp5. In the hypothalamus and hippocampus, we observed modest levels of demethylation within Fkbp5, with this decrease being more pronounced in the HT-22 hippocampal neuronal cell line treated with corticosterone.
We had a prior interest in Fkbp5 because of work relating variation in the human ortholog to GR signaling and mood disorders. The FKBP5 protein regulates GR sensitivity as binding of the protein to the receptor complex reduces the latter’s affinity for cortisol and translocation to the nucleus. Genetic variation in FKBP5 is associated with enhanced expression after GR activation, leading to more GR resistance, diminished negative feedback, and a prolonged stress hormone activation after a stressor (29). The increase in Fkbp5 expression we observed in both brain regions and in blood is consistent with previous studies that have shown an increase in Fkbp5 in response to acute glucocorticoid administration in cell lines (30,31,32) and mouse brain (26). The expression changes, as well as the DNAm changes, we observed were associated with increased anxiety-like behavior in the CORT mice, although additional studies will be required to determine whether the relationship is causal.
Our report of DNAm changes in Fkbp5 is the first one of which we are aware. We saw decreased methylation in the intron 1 region of the gene in blood DNA and more subtle decreases in the intron 5 region of hypothalamic and hippocampal DNA. Functional differences exist within the hippocampus between subregions and between cell types. For example, laser-capture microdissection studies have shown expression differences between CA1, CA3, and dentate gyrus (33). Cell types within the hippocampus similarly show differences in gene expression patterns (34). We hypothesized that DNAm changes might be cell type specific, and thus, we took an approach that would allow us to examine neurons in particular, examining a HT-22 mouse hippocampal neuronal cell line.
The potential correlation we observed between DNAm in brain and blood for Fkbp5 could have implications for clinical applications of epigenetics research. However, our findings also suggest that distinct regions of the gene may be affected by DNAm changes in these differing tissues, given that the intron 1 region showed DNAm change in blood, whereas the intron 5 region showed DNAm change in hypothalamus, hippocampus, and the hippocampal neuronal cell line. Interestingly, the intron 5 region is similar to the intron 1 region, which has been shown to bind MeCP2, because both regions contain hormone response elements (25), and all of the four CpG dinucleotides of the intron 5 region bioinformatically meet criteria for predicted MeCP2 binding (35). It is plausible that NR3C1 binding to hormone/glucocorticoid response elements and subsequent DNA demethylation may initially decrease MeCP2 occupancy, cause euchromatinization of the intronic region, and allow for the binding of sequence and tissue-specific transcription factors that together with the activated NR3C1 coordinate the transactivation of the Fkbp5 gene. However, it is unclear at this time how the activated NR3C1 complexes are targeted to the specific intronic regions in the first place and whether DNA demethylation is a prerequisite, rather than the result, of NR3C1 occupancy. We now have an excellent cell line model system in which to answer such questions.
Decreased Fkbp5 DNAm in response to corticosterone persisted for 1–4 wk after the cessation of hormone application. By contrast, elevated Fkbp5 expression returned to baseline more quickly. Despite a significant reduction in methylation of nearby CpGs, it is unlikely that the enhancer or glucocorticoid response element would function in the absence of an activated NR3C1 and hence the observed rapid reduction in Fkbp5 expression after corticosterone cessation. However, in the period of persistent demethylation, the hormone response elements could still be occupied by transcription factors such as forkhead box A1 and CCAAT/enhancer binding protein that are essential cofactors for NR3C1-mediated transactivation (36,37). This raises the possibility of a biological consequence of these demethylated regions. If the region has become euchromatinized, as suggested by diminished binding to MeCP2, and if it were still occupied by many of the essential cofactors, then any additional NR3C1 binding might produce a faster and more robust signal than it would otherwise.
In contrast to increased expression of Fkbp5 after corticosterone treatment, we observed decreased expression of Nr3c1 and Crh in the hippocampus and decreased Crh expression in the hypothalamus. Prior studies of chronic stress in animals have yielded varying assessments of glucocorticoid receptor expression changes in these and other brain regions with some reporting down-regulation (38,39), others showing no significant change (38,39), and one showing an increase (40). Fewer studies have examined Crh and Crhr1 expression changes in the brain in a chronic glucocorticoid exposure paradigm. One study showed no change in Crh or Crhr1 (41), whereas a second observed a nonsignificant increase in Crhr1 (39) and a third reported a significant decrease in Crhr1 (42) after chronic treatment with corticosterone. Factors that might explain these variations include experimental differences in species, hormone exposure/stress paradigms, and regions of the brain assayed. Of course, corticosterone exposure can yield quite different results from a stress paradigm. For example, the former decreases CRH in hypothalamus, whereas the latter generally increases it.
For Nr3c1, we focused our attention mainly on the two CpG dinucleotides implicated in binding the transcription factor nerve growth factor-inducible protein A (27). Contrary to our expectation, we did not find a difference in methylation levels between CORT-treated and control mice. One explanation for this might be that we treated adolescents, whereas the study by Weaver et al. (27) reported on effects of maternal behavior on offspring during the early neonatal period. Furthermore, methylation patterns might differ in the mice we used compared with the rats used by Weaver et al. We note that one study found no DNAm at this site in rat pups subjected to maternal separation (43). It is also possible that the effect on methylation found by Weaver et al. is mediated by factors other than varying corticosterone levels between differentially reared groups of pups.
Limitations include the following. First, the EPM test was done 3 days before the animals were killed. The anxiety on EPM could have caused changes in gene expression rather than being the result of it, given that CORT mice became anxious and control mice did not. Second, we have assayed only small representative regions of a few of the potential genes and other genomic regions in which DNAm may potentially be influenced by corticosterone. Given the fact that many of the glucocorticoid response elements are located at nonpromoter regions, use of a genome-wide assay such as the Comprehensive High-throughput Arrays for Relative Methylation (CHARM) (44) would greatly expand the opportunity for discovery. The difficulty in pinpointing DNAm differences that can account for the expression changes observed in most of the chosen candidate HPA axis genes further highlights the need for such a comprehensive genome-wide approach. Third, changes accompanying chronic corticosterone exposure mimic those in patients suffering from endogenous or iatrogenic Cushing syndrome, in which anxiety is frequently present; however, our paradigm may not reflect physiological changes due to stress in the animal. A model such as a social defeat stress or chronic variable stress procedure (45,46) in mice might allow for comparable DNAm experiments with a closer tie to human stress-related illness. Fourth, a causal relationship between decreased DNAm and increased expression of Fkbp5 is only suggested, but not proven, by our data. Future studies will further explore causality through the use of DNA methylation inhibitors and reporter constructs. Finally, it is plausible that other epigenetic mechanisms such as chromatin modifications might account for the expression differences. Lending support to this, a recent paper shows a dexamethasone-induced increase in expression of human FKBP5 resulting from chromatin structure changes in the distal intronic enhancer homologous to the mouse region assayed in our study (14). Additional studies should explore the potential interaction of changes in methylated/acetylated lysine residues of histone proteins with the DNAm changes we observed.
In summary, we tested the hypothesis that expression changes would be induced in HPA axis genes by differential, chronic exposure to corticosterone and that these changes would occur through loss or gain of DNAm. Although we saw significant decreases in the mRNA levels of Nr3c1 and Crh and an increase in Fkbp5, only in the latter gene did we detect substantial changes in DNAm. A decrease in methylation was observed in blood DNA, and to a lesser extent, in hippocampal DNA. The same pattern of increase in expression and decrease in methylation of Fkbp5 was more pronounced in a mouse hippocampal neuronal cell line exposed to corticosterone. Our results suggest the need for additional study of the mechanisms underlying the impact of corticosterone on Fkbp5 methylation and expression. They further suggest that additional study of epigenetic influences on Fkbp5 expression in other excess glucocorticoid paradigms, including those induced by stress, is warranted.
Acknowledgments
We thank Akira Sawa for generously providing HT-22 cell lines.
Footnotes
This work was supported by National Institutes of Health Grants AA10158 (to G.S.W.), HD055030 (to K.L.K.T.), and T32MH015330 (to R.S.L.); the Kenneth Lattman Foundation (to G.S.W.); and the Margaret Ann Price Investigatorship (to J.B.P. and R.S.L.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online July 28, 2010
Abbreviations: Ct, Threshold cycle; DNAm, DNA methylation; EPM, elevated plus maze; EtOH, ethyl alcohol; GR, glucocorticoid receptor; HPA, hypothalamic-pituitary-adrenal; MeCP2, methyl-CpG-binding protein 2; NMR, nuclear magnetic resonance.
References
- Starkman MN, Schteingart DE, Schork MA 1981 Depressed mood and other psychiatric manifestations of Cushing’s syndrome: relationship to hormone levels. Psychosom Med 43:3–18 [DOI] [PubMed] [Google Scholar]
- Cohen SI 1980 Cushing’s syndrome: a psychiatric study of 29 patients. Br J Psychiatry 136:120–124 [DOI] [PubMed] [Google Scholar]
- Starkman MN, Schteingart DE, Schork MA 1986 Cushing’s syndrome after treatment: changes in cortisol and ACTH levels, and amelioration of the depressive syndrome. Psychiatry Res 19:177–188 [DOI] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, Schwartz AC, Cubells JF, Ressler KJ 2008 Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA 299:1291–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Pütz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Künzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, Zill P, Bondy B, Rupprecht R, Messer T, Köhnlein O, Dabitz H, Brückl T, Müller N, Pfister H, Lieb R, Mueller JC, Lõhmussaar E, Strom TM, Bettecken T, Meitinger T, Uhr M, Rein T, Holsboer F, Muller-Myhsok B 2004 Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet 36:1319–1325 [DOI] [PubMed] [Google Scholar]
- Lekman M, Laje G, Charney D, Rush AJ, Wilson AF, Sorant AJ, Lipsky R, Wisniewski SR, Manji H, McMahon FJ, Paddock S 2008 The FKBP5-gene in depression and treatment response—an association study in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Cohort. Biol Psychiatry 63:1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willour VL, Chen H, Toolan J, Belmonte P, Cutler DJ, Goes FS, Zandi PP, Lee RS, MacKinnon DF, Mondimore FM, Schweizer B, DePaulo Jr JR, Gershon ES, McMahon FJ, Potash JB 2009 Family-based association of FKBP5 in bipolar disorder. Mol Psychiatry 14:261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY 1999 Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188 [DOI] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA 1998 Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 12:949–957 [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M 2005 Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102:10604–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovic S, D'Alessio AC, Detich N, Szyf M 2007 Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis 28:560–571 [DOI] [PubMed] [Google Scholar]
- Datson NA, van der Perk J, de Kloet ER, Vreugdenhil E 2001 Identification of corticosteroid-responsive genes in rat hippocampus using serial analysis of gene expression. Eur J Neurosci 14:675–689 [DOI] [PubMed] [Google Scholar]
- Sato H, Horikawa Y, Iizuka K, Sakurai N, Tanaka T, Shihara N, Oshima A, Takeda J, Mikuni M 2008 Large-scale analysis of glucocorticoid target genes in rat hypothalamus. J Neurochem 106:805–814 [DOI] [PubMed] [Google Scholar]
- Paakinaho V, Makkonen H, Jääskeläinen T, Palvimo JJ 2010 Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol Endocrinol 24:511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST 2001 Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107:477–487 [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM 2006 Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet 141B:261–268 [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Jarrar MH, Wang JS, Lee YJ, Reddy S, Bibat G, Naidu S 2005 Histone modifications in Rett syndrome lymphocytes: a preliminary evaluation. Brain Dev 27:331–339 [DOI] [PubMed] [Google Scholar]
- Wang SC, Oelze B, Schumacher A 2008 Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS ONE 3:e2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg DJ, Brown LM, Woods SC, Benoit SC 2006 Gonadal hormones determine sensitivity to central leptin and insulin. Diabetes 55:978–987 [DOI] [PubMed] [Google Scholar]
- Taicher GZ, Tinsley FC, Reiderman A, Heiman ML 2003 Quantitative magnetic resonance (QMR) method for bone and whole-body-composition analysis. Anal Bioanal Chem 377:990–1002 [DOI] [PubMed] [Google Scholar]
- Künnecke B, Verry P, Bénardeau A, von Kienlin M 2004 Quantitative body composition analysis in awake mice and rats by magnetic resonance relaxometry. Obes Res 12:1604–1615 [DOI] [PubMed] [Google Scholar]
- Yang X, Wang S, Rice KC, Munro CA, Wand GS 2008 Restraint stress and ethanol consumption in two mouse strains. Alcohol Clin Exp Res 32:840–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behl C, Skutella T, Lezoualc'h F, Post A, Widmann M, Newton CJ, Holsboer F 1997 Neuroprotection against oxidative stress by estrogens: structure-activity relationship. Mol Pharmacol 51:535–541 [PubMed] [Google Scholar]
- U M, Shen L, Oshida T, Miyauchi J, Yamada M, Miyashita T 2004 Identification of novel direct transcriptional targets of glucocorticoid receptor. Leukemia 18:1850–1856 [DOI] [PubMed] [Google Scholar]
- Magee JA, Chang LW, Stormo GD, Milbrandt J 2006 Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology 147:590–598 [DOI] [PubMed] [Google Scholar]
- Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, Bird A 2005 Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet 14:2247–2256 [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ 2004 Epigenetic programming by maternal behavior. Nat Neurosci 7:847–854 [DOI] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R 2003 Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques 35:146–150 [DOI] [PubMed] [Google Scholar]
- Binder EB 2009 The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology 34(Suppl 1):S186–S95 [DOI] [PubMed] [Google Scholar]
- Rees-Unwin KS, Craven RA, Davenport E, Hanrahan S, Totty NF, Dring AM, Banks RE, J Morgan G, Davies FE 2007 Proteomic evaluation of pathways associated with dexamethasone-mediated apoptosis and resistance in multiple myeloma. Br J Haematol 139:559–567 [DOI] [PubMed] [Google Scholar]
- Billing AM, Fack F, Renaut J, Olinger CM, Schote AB, Turner JD, Muller CP 2007 Proteomic analysis of the cortisol-mediated stress response in THP-1 monocytes using DIGE technology. J Mass Spectrom 42:1433–1444 [DOI] [PubMed] [Google Scholar]
- Hubler TR, Scammell JG 2004 Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones 9:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JG, Borges K, Dingledine R 2009 Quantitative transcriptional neuroanatomy of the rat hippocampus: evidence for wide-ranging, pathway-specific heterogeneity among three principal cell layers. Hippocampus 19:253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamme F, Salunga R, Yu J, Tran DT, Zhu J, Luo L, Bittner A, Guo HQ, Miller N, Wan J, Erlander M 2003 Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J Neurosci 23:3607–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP 2005 DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell 19:667–678 [DOI] [PubMed] [Google Scholar]
- Schoneveld OJ, Gaemers IC, Das AT, Hoogenkamp M, Renes J, Ruijter JM, Lamers WH 2004 Structural requirements of the glucocorticoid-response unit of the carbamoyl-phosphate synthase gene. Biochem J 382:463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008 FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132:958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez F, Lahmame A, de Kloet ER, Armario A 1996 Hypothalamic-pituitary-adrenal response to chronic stress in five inbred rat strains: differential responses are mainly located at the adrenocortical level. Neuroendocrinology 63:327–337 [DOI] [PubMed] [Google Scholar]
- Raone A, Cassanelli A, Scheggi S, Rauggi R, Danielli B, De Montis MG 2007 Hypothalamus-pituitary-adrenal modifications consequent to chronic stress exposure in an experimental model of depression in rats. Neuroscience 146:1734–1742 [DOI] [PubMed] [Google Scholar]
- Mizoguchi K, Ishige A, Aburada M, Tabira T 2003 Chronic stress attenuates glucocorticoid negative feedback: involvement of the prefrontal cortex and hippocampus. Neuroscience 119:887–897 [DOI] [PubMed] [Google Scholar]
- Pournajafi-Nazarloo H, Partoo L, Sanzenbacher L, Paredes J, Hashimoto K, Azizi F, Sue Carter C 2009 Stress differentially modulates mRNA expression for corticotrophin-releasing hormone receptors in hypothalamus, hippocampus and pituitary of prairie voles. Neuropeptides 43:113–123 [DOI] [PubMed] [Google Scholar]
- Urigüen L, Arteta D, Díez-Alarcia R, Ferrer-Alcón M, Diaz A, Pazos A, Meana JJ 2008 Gene expression patterns in brain cortex of three different animal models of depression. Genes Brain Behav 7:649–658 [DOI] [PubMed] [Google Scholar]
- Daniels WM, Fairbairn LR, van Tilburg G, McEvoy CR, Zigmond MJ, Russell VA, Stein DJ 2009 Maternal separation alters nerve growth factor and corticosterone levels but not the DNA methylation status of the exon 1(7) glucocorticoid receptor promoter region. Metab Brain Dis 24:615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP 2008 Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 18:780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams N, Blizard DA 1986 Effects of social defeat on acute cardiovascular response in salt-sensitive and salt-resistant rats. Behav Neural Biol 46:325–336 [DOI] [PubMed] [Google Scholar]
- Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, Graham D, Tsankova NM, Bolanos CA, Rios M, Monteggia LM, Self DW, Nestler EJ 2006 Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science 311:864–868 [DOI] [PubMed] [Google Scholar]