Abstract
Covalent adduction of a nitrosyl group to cysteines [S-nitrosylation (S-NO)] is emerging as a key route for nitric oxide (NO) to directly modulate protein functions. Here, we studied the effects of estrogens on endothelial protein S-NO and analyzed the nitrosyl-proteomes by biotin/CyDye switch technique combined with two-dimensional fluorescence difference gel electrophoresis and identified nitrosoproteins by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Estradiol-17β (E2) rapidly stimulated protein S-NO in human umbilical vein endothelial cells, maximizing within 10- to 30-min post-E2 (10 nm) exposure. E2-BSA also rapidly stimulated protein S-NO. Both E2 and E2-BSA-induced protein S-NO was blocked by ICI 182,780 and N-nitro-l-arginine-methylester. Human umbilical vein endothelial cells expressed estrogen receptor (ER)α and ERβ; both seemed to be required for E2 stimulation of protein S-NO because: 1) neither ERα or ERβ agonist alone, but their combination, stimulated protein S-NO; and 2) either ERα or ERβ antagonist blocked E2-induced protein S-NO. Numerous nitrosoproteins (spots) were observed on two-dimensional fluorescence difference gel. One hundred spots of interest were picked up; 58 were identified and, of which 15 were novel nitrosoproteins, 28 were up-regulated, 11 were decreased, and the rest were unchanged by E2. Pathway analysis suggested that nitrosoproteins are involved in regulating various endothelial functions, including apoptosis, cell structure and metabolism, redox homeostasis, etc. Thus, estrogens stimulate dynamic endothelial protein S-NO via mechanisms linked to specific ERs possibly on the plasma membrane and endogenous NO. These findings signify a critical next step for the understanding of the biological targets of enhanced NO production by estrogens.
Estradiol-17β stimulation of specific receptor and endogenous nitric oxide dependent dynamic S-nitrosylation of an array of proteins in human umbilical vein endothelial cells implicates a critical pathway for estrogens to elicit their cardiovascular effects.
Endothelial cells, lining the luminal surface of all blood vessels throughout the body, play a key role in vascular health largely by synthesizing nitric oxide (NO) via endothelial NO synthase (eNOS), because the liable gas possesses potent antiinflammatory, antiapoptotic, antithrombotic, and antioxidant effects (1,2,3). When administrated in vivo, estradiol-17β (E2) potently dilates blood vessels within 5–30 min (4). Intensive investigations during the last decade have shown that estrogen-induced vasodilatation is endothelium and NO dependent (5,6). E2 rapidly stimulates endothelial NO production in association with increased eNOS activity but not eNOS expression in vitro (7,8). Rapid eNOS activation by estrogens involve complex protein kinase(s)-mediated phosphorylation and dynamic interactions with cofactors and other regulatory proteins, including heat shock protein 90 and caveolin-1 (9,10,11). These mechanisms have greatly advanced the concept of rapid “nongenomic” estrogen action via specific receptors localized on the plasma membrane. In addition, endothelial cells are direct targets for long-term estrogen actions, including transcription of genes, such as eNOS, through binding to classic estrogen receptors (ERs) (i.e. ERα and ERβ) in the nucleus (12).
Formation of cyclic GMP (cGMP) is the classical route for many NO biological functions, including vascular remodeling, vessel relaxation, platelet aggregation, etc. (13). However, many NO bioactivities are cGMP independent (14). NO is a short-lived gaseous molecule (15). Once formed, it can rapidly converted into other reactive nitrogen species, such as nitrogen trioxide, peroxinitrite, and nitrosoglutathione (GSNO) (16). These reactive nitrogen species can directly donate a nitrosyl (-NO) group to cysteines in peptides or proteins thereby forming nitrosothiols. This reaction is referred as to S-nitrosylation (S-NO), which has emerged as a crucial cGMP-independent NO signaling pathway (17). This rapid, reversible, and redox-sensitive posttranslational modification regulates the functions of a plethora of proteins in nearly all major biological pathways; thus, its biological importance has been compared with O-phosphorylation (18). However, only until the biotin switch technique (BST) was invented in 2001 (19) can the fragile S-NO bond be accurately measured. In this method, the SNO group is selectively reduced by ascorbate and then labeled with biotin, thus allowing nitrosoproteins to be readily displayed, affinity purified, and identified.
Estrogens stimulation of endothelial NO production has been well established (7,8,9,20,21). Enhanced endothelial NO production conveys significantly to the vascular effects of estrogens. However, little is known regarding the direct endothelial protein targets of NO on estrogens stimulation. S-NO represents a major route that NO directly modulates protein functions (18). In this study, experiments were designed to specifically test the hypothesis that E2 stimulates specific ER and endogenous NO-dependent dynamic protein S-NO in endothelial cells. We also developed a BST based novel proteomic approach and analyzed basal and estrogen responsive endothelial nitrosyl-proteomes by using primary human umbilical vein endothelial cells (HUVEC) as the model.
Materials and Methods
Chemicals and antibodies
E2, β-estradiol 6-(O-carboxymethyl)oxime-BSA (E2-BSA), sodium ascorbate, HEPES, fatty acid-free BSA, N-nitro-l-arginine-methylester (l-NAME), N-nitro-d-arginine-methylester (d-NAME), methyl methanethiosulfonate (MMTS), sodium dodecyl sulfate (SDS), and all other chemicals unless specified, were from Sigma (St. Louis, MO). N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide were from Thermo Scientific (Rockford, IL). ICI 182,780, 4,4′,4′-(4-propyl-[1H]-pyrazole-1,3,5-triyl)trisphenol (PPT), diarylpropionitrile (DPN), 1,3-bis(4-hydroxyphenyl)-4-methyl-5,[4-(2 piperidinylethoxy)phenol]-1H-pyrazole dihydrochloride dihydro-chloride (MPP), 4-[2-Phenyl-5, 7-bis(trifluoromethyl)pyrazolo[1,5-a]pyri midin-3-yl]phenol (PHTPP) were from Tocris (Ellisville, MO). GSNO was from Cayman (Ann Arbor, MI). Antibiotin antibody was from Cell Signaling (Beverly, MA). The 2-(6-biotinoyl-amino-hexanoyl amino) ethylmethanethiosulfonate (MTSEA)-Texas Red was from Toronto Research Chemicals (Downsview, Ontario, Canada). CyDye difference gel electrophoresis (DIGE) fluors were from GE Healthcare (Buckinghamshire, UK). Anti-β-actin antibody was from Ambion (Austin, TX). Anti-ERα antibody was from Neomarkers (Fremont, CA). Anti-ERβ antibody was from Affinity Bioreagents (Rockford, IL). Prolong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) and M-199 medium was from Invitrogen (Carlsbad, CA).
Cell culture
HUVEC was isolated from healthy term placentas by collagenase digestion as described (20). Cord collection was from University of California San Diego Medical Center Hospital (San Diego, CA) and approved by the Institutional Review Boards. Cells were cultured on gelatin-coated dishes in endothelial cell medium (ScienCell, Carlsbad, CA) containing 5% fetal bovine serum (ScienCell, Carlsbad, CA) and supplemented with 1% antibiotics and 1% endothelial cell growth supplement and used within five passages. When grown to approximately 70% confluence, the medium was replaced with phenol red-free M199 containing 0.1% BSA, 25 mm HEPES for 6 h. After 1-h equilibration with fresh M199-0.1% BSA-25 mm HEPES, the cells were treated with E2 for various times with or without pharmacological inhibitors. Ethanol (<0.01%) was used to dissolve E2 and other drugs, which did not alter cellular responses. GSNO was used as a NO donor for a positive control. Primary uterine artery endothelial cells (UAEC) were isolated by collagenase digestion from late pregnant (120–130 d of gestation, term ≈145 d) ewes as described previously (22). The animal use protocol was approved by the University of California San Diego Animal Subjects Committee, and we followed the National Research Council’s Guide for the Care and Use of Laboratory Animals.
BST method
BST was performed as previously described (19). Briefly, HUVEC or UAEC (∼4 × 106 cells) was lysed in HEN buffer [25 mm HEPES (pH 7.7), 1 mm EDTA, and 0.1 mm neocuproine] containing 0.4% 3-(3-cholamidopropyl) dimethylammonio-1-propanesulfonate (CHAPS), 20 mm MMTS, and 2.5% SDS. Protein content of all samples was adjusted to 0.6 mg/ml using HEN buffer. The samples (50 μg/group) were transferred to amber Eppendorf tubes and incubated at 50 C in dark for 30 min. After centrifugation (12,000 × g, 10 min), the supernatants were transferred into 2-ml tubes and mixed with cooled (−20 C) acetone (1:4, vol/vol) and incubated at −20 C for 2 h. Acetone precipitated proteins were resuspended in HEN buffer containing 1 mm sodium ascorbate, 0.1 μm CuCl2, 1 mm N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide, and 1% SDS. The samples (0.6 mg/ml) were incubated at 37 C in dark for 2 h with occasional agitation, and finally mixed with SDS sample buffer for SDS-PAGE.
SDS-PAGE and immunoblotting
Proteins were separated on 8–12% SDS-PAGE and transferred onto polyvinylidene fluoride membranes as described previously (23). Total nitrosoproteins were detected by immunoblotting with antibiotin antibody. The level of all nitrosoproteins for each lane was summed and normalized as a ratio to β-actin.
Detection of nitrosoproteins in intact cells by fluorescence microscopy
Nitrosoproteins were detected in intact cells by a modified BST protocol as previously described (24) and modified as below. HUVEC grown on gelatin-coated glass coverslips were serum starved and treated with E2 or GSNO, washed with cold PBS, and then fixed in methanol at −20 C for 30 min. Free thiols were blocked with 20 mm MMTS in HEN buffer containing 80% methanol at 50 C in dark for 30 min. After three washings with HEN buffer, the cells were incubated with 0.2 m ascorbate and 0.2 mm MTSEA-Texas Red in HEN/methanol in dark at room temperature for 1 h. After extensive washing with HEN buffer, the samples were mounted with Prolong Gold antifade reagent with DAPI and examined under a Leica fluorescence microscopy for image acquisition by a Hamamatsu high-resolution charge-coupled device camera (Leica, Wetzlar, Germany) using the SimplePCI image analysis software.
CyDye switch and two-dimensional fluorescence difference gel electrophoresis (2D-DIGE)
CyDye switch was performed as described below, which was recently published by others (25). Briefly, after blocking free thiols in cell lysates (100 μg protein/sample) in HEN buffer containing 20 mm of MMTS, acetone precipitated proteins were resuspended in 35 μl of reducing buffer [30 mm Tris-HCl (pH 8.0), 7 m urea, 2 m thiourea, and 4% CHAPS] containing 1 mm sodium ascorbate and 0.1 μm copper choloride and incubated in dark at 37 C for 1 h. CyDye DIGE Fluor Cy3 (or Cy5) saturation dye (4 μl, 2 mm) were added into control or E2-treated samples, respectively. The samples were mixed and incubated in dark at 37 C for 30 min. The samples were then mixed with 2D sample buffer (7 m urea, 2 m thiourea, 4% CHAPS, 2% pharmalytes 3–10, and 130 mm dithiothreitol) and stored at −80 C for 2D-DIGE within 2 wk.
2D-DIGE was performed by Applied Biomics, Inc. (Hayward, CA). Just before 2D-DIGE, equal amounts of Cy3- and Cy5-labeled samples (50 μg each) were mixed with rehydration buffer. After adding destreak solution (GE Healthcare) and 1% pH 3–10 pharmalyte (GE Healthcare), the samples were loaded onto an isoelectric focusing (IEF) strip (pH 3–10 linear range; GE Healthcare). IEF was done for a total of 25,000 V/h with standard conditions using Ettan IPGPhore II. After the IEF, electrophoresis was performed at 16 C on 10% SDS-PAGE. The resulting 2D gel was scanned using a Typhoon Trio scanner (GE Healthcare) with excitation and emission wavelengths for Cy3 (548/560 nm)- and Cy5 (641/660 nm)-labeled proteins with settings that the same samples labeled with Cy3 or Cy5 resulted in similar relative red or green fluorescence intensities. Image analysis for intensity measurement of spots of interest was performed using the ImageQuant and DeCyder softwares (GE Healthcare).
These BST-based methods are summarized diagrammatically in Supplemental Fig. 1 published on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org.
Protein identification by matrix-assisted laser desorption/ionization (MALDI)-time of flight (TOF) and tandem MS (mass spectrometry) (MS/MS)
Protein identification was performed by Applied Biomics, Inc. After analyses of the 2D-DIGE image, selected spots of interest (based on intensity and visibility) were picked up. Each sample was washed twice with 25 mm ammonium bicarbonate and 50% acetonitrile to remove staining dye, once with water and once with 100% acetonitrile. The samples were dried, rehydrated in digestion buffer [25 mm ammonium bicarbonate, 2% acetonitrile, and 0.5% Promega sequencing grade trypsin (Promega, Madison, WI)]. Proteins were digested in-gel at 37 C overnight. Digested peptides were extracted and desalted using C-18 Zip-tips (Millipore, Billerica, MA). The samples were mixed with α-cyano-4-hydroxycinnamic acid matrix and spotted into a MALDI plate. Mass spectra of each sample were obtained by using an Applied Biosystems 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA). Ten to 20 of the most abundant peptides in each sample were further subjected to fragmentation and MS/MS analysis. Identification of each sample (spot) was searched based on peptide fingerprinting MS and fragmentation MS/MS spectra. The spectra were submitted for database search using GPS Explorer software (Applied Biosystems, Foster City, CA) equipped with the MASCOT search engine to identify proteins from National Center for Biotechnology Information nonredundant Homo sapiens amino acid sequence database with oxidation and carbamidomethy and phosphorylation as variable modifications. The highest scoring hit with a protein score confidence interval more than 95% for each spot was accepted as positive identification.
Pathway analysis and statistics
Ingenuity pathway analysis (Ingenuity Systems, Redwood City, CA) tool was used to perform pathway analysis of the identified endothelial S-nitrosoproteins. Positive result was defined as -log (P value) more than 1.30 (P < 0.05). Statistics were performed using SigmaStat version 3.5 (Systat Software, Inc., San Jose, CA). For comparison of data between estrogen treatment and control, we used Student’s t test. Significance was defined as P < 0.05.
Results
E2 stimulates protein S-NO in endothelial cells
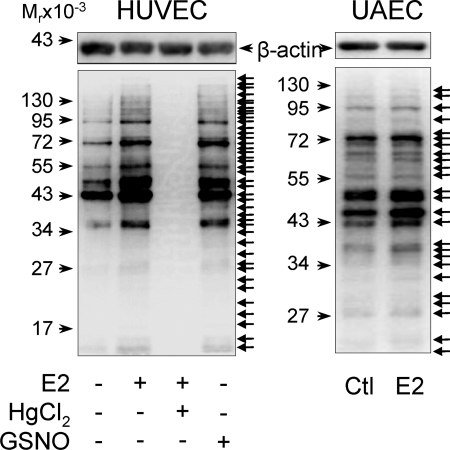
To determine the effects of E2 on protein S-NO in endothelial cells, endothelial cells were pre-starved in phenol red-free medium without serum; thus the cells were presumptively deprived of estrogens before E2 treatment. We first measured total levels of nitrosoproteins in HUVEC treated with or without E2 or a NO donor GSNO. S-NO of various proteins was readily detectable in untreated control HUVEC. Treatment with 10 nm E2 or 1 mg/ml GSNO for 30 min stimulated S-NO of various proteins in HUVEC. All bands were lost in the presence of HgCl2, a reagent selectively displaces NO from S-NO bonds (26), implicating specificity of the assay. Similarly, E2 increased the levels of nitroso-proteins in UAEC (Fig. 1).
Figure 1.
Total nitrosoprotein profiles in E2 and GSNO-treated HUVEC and ovine UAEC on SDS-PAGE. HUVEC and UAEC were treated with E2 (10 nm), an NO donor (GSNO, 1 mg/ml), or vehicle control (Ctl) for 30 min. Whole-cell lysates were prepared and subjected to biotin switch reaction with or without 0.2% HgCl2. The biotin-labeled nitrosoproteins were analyzed by 10% SDS-PAGE and detected by Western blot analysis with an antibiotin antibody. β-Actin was measured for monitoring protein loading. Images shown depict a typical experiment of similar results of three independent experiments using HUVEC from different placentas and an experiment of UAEC from three different pregnant ewes. Arrows on right point to visible bands representing S-nitrosylated proteins.
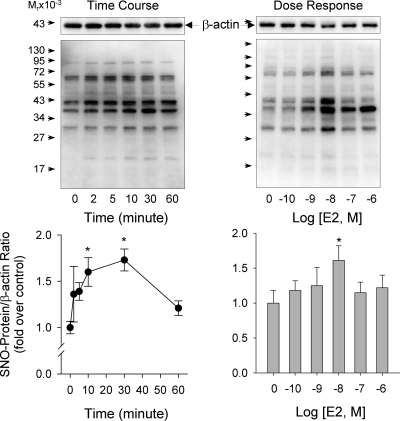
In HUVEC, total levels of nitrosoproteins began to increase by 10 nm E2 at 2 min, maximized around 10–30 min, and returned to baseline at 60 min (Fig. 2A). When treated with 0.1 nm to 1 μm E2 for 30 min, only 10 nm E2 significantly stimulated total levels of nitrosoproteins in HUVEC. However, several individual proteins were clearly S-nitrosylated by 1 nm to 1 μm E2 (Fig. 2B). These data suggest that E2 stimulates protein S-NO in a time and concentration-dependent manner.
Figure 2.
Time courses and dose responses of the effects of E2 treatment on protein S-NO in HUVEC. Subconfluent HUVEC were treated with or without 10 nm E2 for the indicated times (up to 1 h) or with increasing concentration of E2 (0.1 nm to 1 μm) for 30 min. Total protein extracts were harvested for determining the total nitrosoproteins. Representative blots of nitrosoproteins and β-actin of one typical experiment are shown. Lower graphs summarize data (mean ± sem, n = 3) from three independent experiments using HUVEC from different placentas. *, P < 0.05 vs. control.
E2 stimulation of protein S-NO is mediated by specific ERs
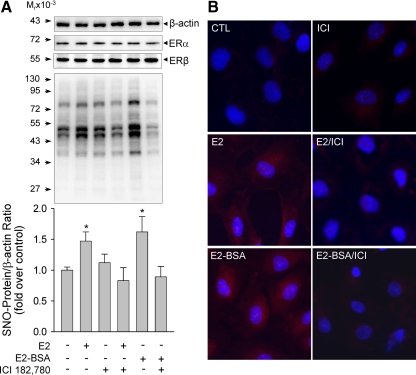
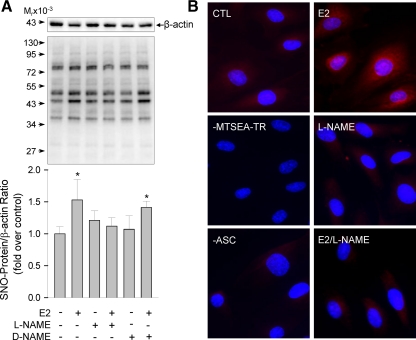
The biological function of estrogens in endothelial cells is mediated by specific ERs, including ERα and ERβ (27). We detected immunoreactive ERα and ERβ proteins HUVEC, confirming a previous report (28) and showing that HUVEC are direct estrogen target cells. To test whether estrogens stimulation of protein S-NO in HUVEC is mediated by its specific receptors, we pretreated HUVEC with or without a pure ER antagonist ICI 182,780 (1 μm) for 1 h and then determined the effects of E2 and E2-BSA treatments on protein S-NO in HUVEC. As shown in Fig. 3, in comparison with control cells, ICI 182,780 alone did not alter total nitrosoprotein levels. In the presence of ICI 182,780, the levels of total nitrosoproteins in E2 (10 nm, 20 min)-treated cells were significantly decreased. Estrogen-induced protein S-NO seemed to be atypical rapid nongenomic action presumptively via receptors on the plasma membrane as this occurred within minutes (Fig. 2A). We then tested the effects of E2-BSA in the presence or absence of ICI 182,780 on protein S-NO. Treatment with 10 nm E2-BSA for 20 min significantly stimulated total levels of S-NO; similar to E2, the E2-BSA-induced S-NO was blocked by ICI 182,780 (Fig. 3A). E2-BSA is an impermeable estrogen compound that has been widely used as a membrane ER agonist (29). Thus, these findings suggest that E2 stimulation of endothelial protein S-NO is possibly mediated by ER localized on the plasma membrane. We confirmed the findings with a modified BST method by using a fluorescent tag to replace the biotin tag for visualizing nitrosoproteins in intact cells. As shown in Fig. 3B, baseline Texas Red-labeled nitrosoproteins were detected in methanol-fixed HUVEC. Treatment with 10 nm E2 or E2-BSA for 20 min dramatically increased the fluorescence labeling intensities of both cytosol and nuclear nitrosoproteins. Pretreatment with ICI 182,780 drastically decreased the fluorescently labeled nitrosoproteins by E2 or E2-BSA stimulation.
Figure 3.
Effects of ICI 182,780 on E2-induced S-NO in HUVEC. HUVEC were pretreated with or without ICI 182,780 (1 μm) for 1 h, then treated with 10 nm of E2 or E2-BSA for 20 min. A, Nitrosoproteins (SNO-protein) were detected by the BST method. Representative blots of nitrosoproteins and β-actin of one typical experiment are shown. Lower graphs summarize data (mean ± sem, n = 3) from three independent experiments using HUVEC from different placentas. *, P < 0.05 vs. control (Ctl). B, Nitrosoproteins were detected in intact cells by a modified BST using MTSEA-Texas Red as the labeling reagent. Nuclei were labeled by DAPI (blue). Fluorescence micrographs shown depict one of four separate experiments using cells from different placentas.
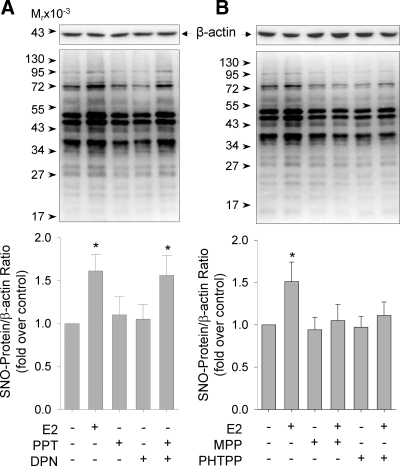
Specific agonists and antagonists of ERα and ERβ were then used to distinguish the specific role of subtype ERs in E2 stimulation of HUVEC protein S-NO. Neither the ERα agonist PPT nor the ERβ agonist DPN alone stimulated endothelial protein S-NO. However, their combination stimulated protein S-NO comparable with E2 (Fig. 4A). Further, treatment with either one of the specific antagonists of ERα and ERβ, MPP and PHTPP, respectively, effectively attenuated E2-induced protein S-NO (Fig. 4B). These data suggest that E2 stimulation of endothelial protein S-NO requires both receptors.
Figure 4.
Specific roles of ERα and ERβ in E2-induced S-NO in HUVEC. HUVEC were (A) treated by E2 (10 nM) or the specific agonists of ERα (PPT, 10 nm) or ERβ (DPN, 10 nm) or their combination for 20 min, or (B) pretreated without or with the specific antagonists of ERα (MPP, 1 μm) or ERβ (PHTPP, 1 μm) for 60 min, followed by E2 (10 nm) for 20 min. Nitrosoproteins were detected by the BST method. Representative blots of nitrosoproteins and β-actin of one typical experiment are shown. Bar graphs summarize data (mean ± sem, n = 3) from three independent experiments using HUVEC from different placentas. *, P < 0.05 vs. control.
E2 stimulation of protein S-NO is mediated by endogenous NO
Estrogens stimulation of endothelial cell NO production is well established (7,8,9,20,21). This easily raise a key question whether E2-induced endothelial protein S-NO is mediated by endogenous NO. To address this, we tested the effects of E2 on protein S-NO in HUVEC pretreated with or without a specific NOS inhibitor l-NAME or its inactive form d-NAME. In the presence of 1 mm l-NAME, but not d-NAME, E2 stimulation of S-NO was abolished (Fig. 5A). We confirmed these findings with fluorescence labeling of nitrosoproteins in intact cells. Baseline Texas Red-labeled nitrosoproteins were detected in the cytosol and nucleus in methanol-fixed HUVEC. Treatment with E2 (10 nm, 20 min) dramatically increased the fluorescence intensities of both cytosol and nuclear nitrosoproteins. Pretreatment with l-NAME drastically decreased the nitrosoprotein fluorescence labeling by E2 stimulation. As assay controls, cells displayed basal fluorescence labeling when ascobate was omitted in the BST and cells incubated without MTSEA-Texas Red showed no labeling (Fig. 5B).
Figure 5.
Role of endogenous NO in E2-induced S-NO in HUVEC. HUVEC were pretreated with or without l-NAME or d-NAME (1 mm) for 1 h, then treated with or without E2 (10 nm) for 20 min. A, Nitrosoproteins were detected by the BST method. Representative blots of nitrosoproteins and β-actin of one typical experiment are shown. Lower graphs summarize data (mean ± sem, n = 3) from three independent experiments using HUVEC from different placentas. *, P < 0.05 vs. control (CTL). B, Nitrosoproteins were detected in intact cells by a modified BST using MTSEA-Texas Red as the labeling reagent. Nuclei were labeled by DAPI (blue). Fluorescence micrographs shown depict one of four separate experiments using cells from different placentas.
CyDye switch, 2D-DIGE analysis of estrogen-regulated endothelial nitrosyl-proteome
Because only a limited number (∼20) of nitrosoproteins can be visualized by BST and immunoblotting with antibiotin antibody, to fully analyze estrogen-regulated nitrosyl-proteome, we developed a more powerful proteomic approach 2D-DIGE to separate the nitrosoproteins. First, in the BST procedure, fluorescent CyDye flours were used to replace the biotin tag for labeling S-nitrosoproteins in control (labeled with green Cy3 fluor)- and E2 (labeled with red Cy5 fluor)-treated HUVEC, respectively. After this modified BST with CyDye (termed as CyDye switch), protein concentrations in the samples were redetermined. Equal amounts of total proteins (50 μg/sample) from control and E2-treated cells were mixed and then separated on a 2D analytic gel. After 2D electrophoresis, the resulting gel was scanned in both Cy3 and Cy5 channels, and relative fluorescence intensities for the spots of interest, based on intensity/visibility and estrogen responsiveness, were calculated as a ratio between control and E2-treated HUVEC (Table 1).
Table 1.
E2-regulated nitrosoproteins in HUVEC identified by CyDye switch and 2D-DIGE and MALDI-TOF and MS/MS
| Spot no., protein name | Swiss-protein ID | MW (Da)/PI | No. peptides matched | Protein score | Ratio (E2/Ctl) (mean ± sem) | P valuea |
|---|---|---|---|---|---|---|
| 1) Ubiquitin C (UBC)b | P62988 | 25,742/6.86 | 5 | 100 | 2.98 ± 0.23 | <0.001 |
| 2) Peptidylprolyl isomerase A (PPIA) | P62937 | 18,000/7.68 | 9 | 248 | 2.11 ± 0.11 | <0.001 |
| 3) Enolase 1 (ENO1) | P06733 | 47,139/7.01 | 13 | 162 | 2.06 ± 0.15 | <0.001 |
| 4) Proteasome (prosome, macropain) subunit, β type, 3 (PSMB3) | P49720 | 22,933/6.14 | 7 | 114 | 2.06 ± 0.11 | <0.001 |
| 5) Adenine phosphoribosyl transferase (APRT)b | P07741 | 19,595/5.78 | 6 | 102 | 1.98 ± 0.05 | <0.001 |
| 6) Peptidylprolyl isomerase A (PPIA) | P62937 | 18,000/7.68 | 9 | 164 | 1.94 ± 0.17 | <0.001 |
| 7) Triosephosphate isomerase 1 (TPI1) | P60174 | 26,625/6.45 | 14 | 398 | 1.93 ± 0.11 | <0.001 |
| 8) Peroxiredoxin 6 (PRDX6) | P30041 | 24,904/6.00 | 13 | 243 | 1.93 ± 0.02 | <0.001 |
| 9) Transketolase (TKT)b | P29401 | 67,751/7.90 | 18 | 214 | 1.88 ± 0.23 | <0.001 |
| 10) Peroxiredoxin 2 (PRDX2) | P32119 | 21,843/6.84 | 7 | 142 | 1.85 ± 0.27 | <0.001 |
| 11) Eukaryotic translation elongation factor 1 γ (EEF1G) | P26641 | 49,814/6.27 | 16 | 244 | 1.83 ± 0.41 | <0.001 |
| 12) Eukaryotic translation elongation factor (EEF2) | P13639 | 95,277/6.41 | 21 | 205 | 1.77 ± 0.30 | <0.001 |
| 13) Cofilin 1 (CFL1) | P23528 | 18,490/8.22 | 7 | 159 | 1.75 ± 0.17 | <0.001 |
| 14) Annexin A2 (ANXA2) | P07355 | 38,551.8/7.57 | 17 | 261 | 1.75 ± 0.11 | <0.001 |
| 15) Parkinson disease (autosomal recessive, early onset) 7 (PARK7) | Q99497 | 19,834/6.33 | 7 | 125 | 1.68 ± 0.01 | <0.001 |
| 16) Glutathione S-transferase omega 1 (GSTO1) | P78417 | 27,548/6.23 | 11 | 130 | 1.61 ± 0.14 | <0.001 |
| 17) Lectin, galactoside-binding, soluble, 3 (LGALS3)b | P17931 | 31,774/4.84 | 10 | 121 | 1.58 ± 0.07 | <0.001 |
| 18) Transgelin 2 (TAGLN2)b | P37802 | 24,438/8.41 | 12 | 148 | 1.56 ± 0.17 | 0.001 |
| 19) RNA binding motif protein 8A (RBM8A) | Q9Y5S9 | 19,889/5.50 | 5 | 218 | 1.55 ± 0.04 | 0.002 |
| 20) Glutathione S-transferase pi 1 (GSTP1) | P09211 | 23,327/5.43 | 6 | 87 | 1.54 ± 0.11 | 0.002 |
| 21) Eukaryotic translation initiation factor 5A (EIF5A) | P63241 | 20,157/6.50 | 7 | 186 | 1.51 ± 0.07 | 0.004 |
| 22) Nucleoside phosphorylase (NP)b | P00491 | 32,097/6.4 5 | 15 | 271 | 1.47 ± 0.17 | 0.007 |
| 23) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | P04406 | 36,031/8.26 | 9 | 95 | 1.46 ± 0.01 | 0.008 |
| 24) Proteasome (prosome, macropain) subunit, β type, 6 (PSMB6) | P28072 | 25,341/4.80 | 5 | 62 | 1.45 ± 0.14 | 0.010 |
| 25) Annexin A1 (ANXA1) | P04083 | 22,741/5.39 | 10 | 199 | 1.43 ± 0.01 | 0.014 |
| 26) Chloride intracellular channel 1 (CLIC1) | O00299 | 26167/4.95 | 8 | 106 | 1.42 ± 0.12 | 0.016 |
| 27) ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d (ATP5H) | O75947 | 18,480/5.21 | 11 | 427 | 1.40 ± 0.12 | 0.022 |
| 28) Phosphofructokinase, platelet (PFKP)b | Q01813 | 85,542/7.50 | 18 | 125 | 1.35 ± 0.09 | 0.044 |
| 29) Eukaryotic translation initiation factor 6 (EIF6) | P56537 | 26,332/4.56 | 5 | 71 | 1.32 ± 0.11 | 0.065 |
| (Continued) | ||||||
Table 1A.
Continued
| Spot no., protein name | Swiss-protein ID | MW (Da)/PI | No. peptides matched | Protein score | Ratio (E2/Ctl) (mean ± sem) | P valuea |
|---|---|---|---|---|---|---|
| 30) Staphylococcal nuclease and tudor domain containing 1 (SND1)b | Q7KZF4 | 99,628/6.62 | 25 | 199 | 1.32 ± 0.11 | 0.065 |
| 31) DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (DDX1)b | Q92499 | 77,812/8.27 | 11 | 64 | 1.30 ± 0.07 | 0.083 |
| 32) Heat shock 27-kDa protein 1 (HSPB1) | P04792 | 22,313/7.83 | 11 | 185 | 1.24 ± 0.12 | 0.165 |
| 33) Transketolase (TKT)b | P29401 | 67,751/7.90 | 11 | 120 | 1.19 ± 0.05 | 0.271 |
| 34) Proliferating cell nuclear antigen (PCNA)b | P12004 | 28,730/4.53 | 7 | 130 | 1.13 ± 0.12 | 0.451 |
| 35) Far upstream element (FUSE) binding protein 1 (FUBP1)b | Q96AE4 | 67,519/7.18 | 22 | 205 | 1.00 ± 0.03 | 1.000 |
| 36) Proteasome (prosome, macropain) subunit, α type, 2 (PSMA2) | P25787 | 25,882/6.92 | 11 | 120 | 0.98 ± 0.02 | 0.907 |
| 37) Pyruvate kinase (PKM2) | P14618 | 57,877/7.95 | 18 | 192 | 0.93 ± 0.04 | 0.684 |
| 38) Annexin A5 (ANXA5) | P08758 | 35,783/4.94 | 15 | 322 | 0.89 ± 0.06 | 0.523 |
| 39) Cofilin 1 (CFL1) | P23528 | 18,490/8.22 | 4 | 87 | 0.85 ± 0.06 | 0.417 |
| 40) Heat shock protein 90-kDa α (cytosolic), class A member 1 (HSP90AA1) | P07900 | 84,607/4.94 | 19 | 185 | 0.86 ± 0.11 | 0.384 |
| 41) Thrombospondin 1 (THBS1)b | P07996 | 129,300/4.71 | 20 | 216 | 0.83 ± 0.05 | 0.324 |
| 42) Heat shock 70-kDa protein 5 (glucose-regulated protein, 78 kDa) (HSPA5) | P11021 | 72,288/5.07 | 26 | 607 | 0.82 ± 0.05 | 0.297 |
| 43) Prolyl 4-hydroxylase, β polypeptide (P4HB)b | P07237 | 57,069/4.82 | 17 | 256 | 0.78 ± 0.04 | 0.203 |
| 44) Heat shock protein 90-kDa β (Grp94), member 1 (HSP90B1) | P14625 | 92,411/4.76 | 22 | 273 | 0.74 ± 0.04 | 0.165 |
| 45) Heat shock 27-kDa protein 1 (HSPB1) | P04792 | 22,313/7.83 | 11 | 156 | 0.76 ± 0.09 | 0.133 |
| 46) Heterogeneous nuclear ribonucleoprotein C (C1/C2) (HNRNPC) | P07910 | 32,318/4.94 | 7 | 147 | 0.71 ± 0.02 | 0.094 |
| 47) Tropomyosin 4 (TPM4) | P67936 | 28,505/4.67 | 14 | 578 | 0.68 ± 0.06 | 0.065 |
| 48) Actin, β (ACTB) | P60709 | 40,194/5.55 | 12 | 128 | 0.63 ± 0.04 | 0.033 |
| 49) Keratin 7 (KRT7) | P08729 | 51,333/5.41 | 25 | 671 | 0.59 ± 0.01 | 0.019 |
| 50) SEC13 homolog (S. cerevisiae) (SEC13)b | P55735 | 35,518/5.22 | 4 | 81 | 0.57 ± 0.02 | 0.014 |
| 51) Annexin A1 (ANXA1) | P04083 | 38,690/6.57 | 15 | 381 | 0.57 ± 0.03 | 0.014 |
| 52) Stathmin 1/oncoprotein 18 (STMN1) | P16949 | 17,326/5.76 | 10 | 412 | 0.56 ± 0.09 | 0.012 |
| 53) Lectin, galactoside-binding, soluble, 3 (LGALS3)b | P17931 | 31,774/4.84 | 9 | 304 | 0.54 ± 0.02 | 0.008 |
| 54) Non-metastatic cells 1, protein (NM23A) expressed in (NME1)b | P15531 | 19,641/5.42 | 9 | 281 | 0.58 ± 0.15 | 0.016 |
| 55) Eukaryotic translation initiation factor 5 (EIF5A) | P63241 | 20,157/6.52 | 8 | 180 | 0.53 ± 0.04 | 0.007 |
| 56) Chloride intracellular channel 1 (CLIC1) | O00299 | 26,167/4.95 | 8 | 106 | 0.52 ± 0.01 | 0.006 |
| 57) Vimentin (VIM) | P08670 | 20,024/4.84 | 7 | 65 | 0.52 ± 0.07 | 0.006 |
| 58) Tumor protein, translationally controlled 1 (TPT1)b | P13693 | 21,512/5.34 | 6 | 227 | 0.42 ± 0.02 | <0.001 |
Statistical analysis was performed using one-way ANOVA (Fisher LSD test), and significance was defined as P < 0.05.
Proteins which were first time identified as nitrosoproteins.
MW, Molecular weight; Da, dalton; pl, isoelectric point; Ctl, control.
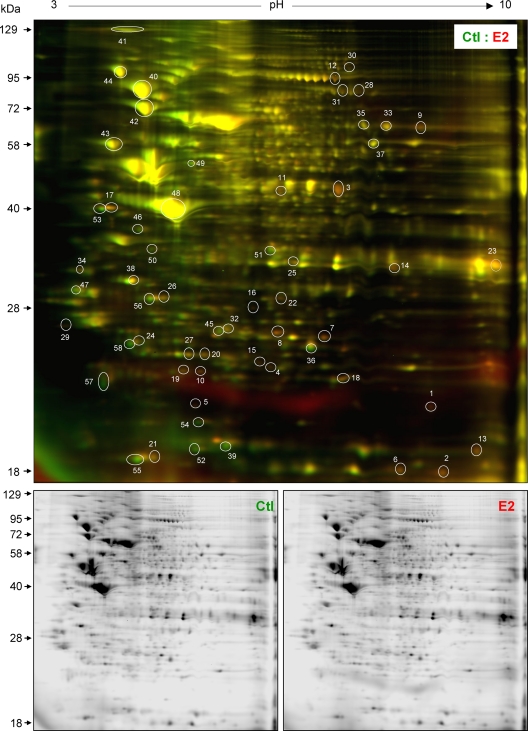
We obtained very similar results of the nitrosyl-proteomes in control and E2-treated HUVEC isolated from three different placentas (Supplemental Fig. 2). A representative merged 2D-DIGE image of control and E2-treated cells is shown in Fig. 6. There are approximately 1000 fluorescent spots that should be all nitrosoproteins. Many proteins, as expected, were readily nitrosylated under resting conditions. Treatment with E2 (10 nm, 20 min) significantly altered the nitrosyl-proteome in HUVEC. Treatment with E2 increased the levels of many nitrosoproteins (red spots) but also unexpectedly decreased some others (green spots) that were not detectable on SDS-PAGE. The rest (yellow spots) were not altered by E2 treatment.
Figure 6.
CyDye switch, 2D-DIGE analysis of nitrosyl-proteomes in control (Ctl) and estrogen-treated HUVEC. HUVEC were treated with or without E2 (10 nm) for 20 min. Total cell extracts (50 μg/treatment) were incubated with ascorbate. Control samples were labeled with Cy3 (green), and E2-treated samples were labeled with Cy5 (red). The samples were then mixed (total 100 μg) and then separated on analytical 2D-DIGE. The gel was scanned with a fluorescence scanner in green (Cy3, 548/560 nm) and red (Cy5, 641/660 nm) channels. A merged fluorescence image shown represents one of three separate experiments using cells from different placentas. Red, green, and yellow spots represent nitrosoproteins that were increased, decreased or unchanged by E2, respectively. The spots circled and numbered represents 58 nitrosoproteins as listed in Table 1, which were identified by MALDI-TOF and MS/MS. The lower black and white images shown represent fluorescent signals obtained from the red and green channels of one of three experiments.
Identification of nitrosoproteins in HUVEC by MALDI-TOF MS/MS
According to the intensity and visibility of the spots on the analytic 2D-DIGE image, as well as estrogen responsiveness of the nitrosoproteins, we focused on 100 spots of interest. The ratios in the levels of these nitrosoproteins between E2-treated and control cells were determined offline using the ImageQuant and DeCyder softwares and then calculated. Ratios greater than 1 and with P < 0.05 represented spots up-regulated by E2, whereas ratios ranging from 0 to 1 and with P < 0.05 represented spots decreased by E2 treatment. Based on these data, 200 μg total proteins of both control and E2-treated samples were prepared by CyDye switch and run on a new preparative 2D-DIGE. After image acquisition, spots are matched with the analytic gel. The same 100 spots of interest on the new preparative gel were picked up. Each of the 100 spots picked up was digested with trypsin and then subjected to MALDI-TOF and MS/MS. Spot identification was performed based on the peptide fingerprinting MS/MS spectra with the MASCOT algorithm. With confidence interval greater than 95%, we identified 58 of the 100 spots subjected to MS/MS analysis. All peptides matched for each of these spots were listed in Supplemental Table 1. The identifications of these nitrosoproteins were summarized in Table 1, in which ratios between the relative levels of all 58 identified nitrosoproteins in control and E2 treated cells were listed.
Pathway analysis of nitrosoproteins in HUVEC
Ingenuity pathway analysis was used to explore the potential biological functions of the identified nitrosoproteins in control and E2-treated HUVEC. Collectively, among the 58 nitrosoproteins identified, levels of 28 targets were significantly increased, 11 targets were decreased, and the rest were unchanged by E2 treatment. Of note, 15 of the nitrosoproteins identified are novel targets that have never been reported to date. Pathway analysis suggested that the nitrosoproteins identified are linked to various basic cellular physiologies, including viability, metabolism, cell cycle, signaling, gene expression, DNA replication/recombination/repair, energy production, molecular transport, migration/morphology, redox balance, apoptosis, etc. (see Table 2). Some housekeeping proteins, such as β-actin, Hsp90, cofilin-1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), vimentin, and annexin, are identified as S-nitrosoproteins, implicating that S-NO is involved in the maintenance of basic cell function. Many nitrosoproteins are enzymes critical for protein synthesis, folding, posttranslational modification, and degradation. Of note, these targets identified here are only a portion (less than ∼1/10) of the endothelial cellular nitrosyl-proteome visible on 2D gel. According to the many nitrosoproteins identified in the resting and E2-treated HUVEC, S-NO apparently plays a critical role in the maintenance of normal cell physiology and is also important for cells to respond to extracellular stimuli.
Table 2.
Pathway analysis of nitrosoproteins in human umbilical vein endothelial cells: potential functionality
| Biological functions | -log (P value)a | S-NO-proteins identified |
|---|---|---|
| Cell viability/apoptosis | 7.854–5.246 | ACTBb, APRTb, ANXA1b, ANXA5, ATP5Hb, EEF2b, EIF5Ab, ENO1b, FUBP1, GAPDHb, GSTP1b, HNRNPC, HSP90AA1, HSPA5, HSPB1, LGALS3b, NME1b, NPb, P4HB, PARK7b, PCNA, PPIAb, PRDX2b, PRDX6b, SND1, STMN1b, THBS1, TPT1b |
| Cellular assembly and organization | 4.216–1.325 | ACTBb, ANXA1b, ANXA2b, ANXA5, CFL1b, GAPDHb, LGALS3b, VIMb, SEC13b, NME1b, STMN1b, PRDX6b, HSP90B1, NPb, PRDX2b, HSP90AA1, HSPB1, THBS1 |
| Cellular function and maintenance | 4.152–1.457 | NME1b, THBS1, ANXA1b, VIMb, HSP90AA1, HSP90B1, HSPA5, HSPB1 |
| Molecular transport | 3.921–1.304 | ANXA1b, HSP90AA1, PRDX6b, STMN1b, THBS1, PARK7b, APRTb, LGALS3b, TPT1b, NME1b, NPb, GSTO1b, GAPDHb, ANXA5, PRDX2b |
| Protein synthesis | 3.347–1.544 | HSP90B1, HSPA5, PARK7b, PRDX6b, HSP90AA1, P4HB, ANXA2b, PARK7b, PPIAb, NME1b, THBS1, EIF5Ab, HSPB1, EEF2b, KRT7b, PSMB3b, UBCb |
| Small molecule biochemistry | 3.347–1.325 | ANXA1b, TKTb, PRDX6b, ANXA2b, EIF6, THBS1, STMN1b, GSTP1b, APRTb, PCNA, P4HB, NME1b, PARK7b, PPIAb, PRDX2b, ANAX5, NPb, TPT1b, GSTO1b, GAPDHb |
| Cell cycle | 2.492–1.386 | THBS1, VIMb, FUBP1, PCNA, STMN1b, PPIAb, NME1b, GSTP1b, HSPB1, PRDX6b, KRT7b |
| Metabolism | 2.492–1.325 | TKTb, PRDX6b, THBS1, GSTO1b, ANXA5, ANXA2b, ENO1b, PKM2, GAPDHb, ANXA1b, PRDX6b, GSTP1b, THBS1, PRDX2b, STMN1b, NME1b, HSPA5, GSTO1b, P4HB, APRTb, PCNA, NME1b, NPb, TPT1b |
| Cardiovascular system development and function | 2.294–1.420 | THBS1, LGALS3b, PRDX6b, TPT1b, TKTb, VIMb, ANXA1b, ANXA5 |
| Gene expression | 2.191–1.355 | SND1, PCNA, FUBP1, ENO1b, LGALS3b |
| Cell signaling | 2.191–1.325 | HSPA5, STMN1b, NPb, PRDXb, THBS1, NME1b, HSPB1, PPIAb |
| Antigen presentation | 2.156–1.420 | LGALS3b, ANXA1b, THBS1, ANXA2b, HSP90B1 |
The significance value associated with Functional Analysis for a dataset is a measure of the likelihood that the association between a set of molecules and a given process or pathway is due to random chance. The smaller the P value the less likely that the association is random and the more significant the association is. In general, P values less than 0.05 [-log (P) > 1.3] indicates statistically significant, nonrandom association. The P value is calculated using the right-tailed Fisher Exact test.
Proteins that nitrosylation was significantly altered by E2.
Discussion
Estrogen protection against cardiovascular diseases (CVDs) has been deduced for decades from a lower risk in CVDs in premenopausal women than age-matched men (30). Historically, this important phenomenon has led to hormone (estrogen/progesterone) replacement therapy for improving peri- and postmenopausal women’s health. This clinical practice has dramatically changed after several recent large clinical trials showed that hormone (estrogen/progesterone) replacement therapy increased cardiovascular complications and other side effects (31,32,33). However, post hoc analyses of the enrolled subjects in these trials suggest that estrogens may prevent progression but not regression of vascular lesions (34). This idea is supported by a primate atherosclerosis study, in which a 70% reduction in lesion formation was observed when estrogen replacement was initiated at the time of ovariectomy but with no lesion reduction if estrogens were given 2 yr after ovariectomy (35). Regardless, current knowledge favors a notion that estrogens are important for the maintenance of vascular health but unlikely provides a cue of established CVDs.
The mechanisms by which estrogens protect against CVD and many other diseases are complex, and the details remain largely unknown. Estrogens have the ability to improve serum lipid profiles (36). However, enhanced endothelial NO production seems to be a major mechanism underlying the protective effect of estrogens in the cardiovascular system. It is clear that estrogens stimulate endothelial NO production via acute nongenomic action to increase eNOS activity and/or by long-term nuclear actions to up-regulate eNOS expression (27,37). However, very little is known regarding the direct targets of enhanced NO production in endothelial cells. S-NO represents an emerging mechanism that NO and its derivatives directly denotes a -NO group to cysteines, thereby providing a novel route by which NO affects target proteins directly. Although S-NO has increasingly recognized as an important posttranslational modification of proteins on cysteines, its detection has been troublesome because of its low level and stability. Antibodies directed against the S-NO moiety have suffered from lack of specificity and loss of sensitivity during immunodetections, because the S-NO bond is redox sensitive and cleaves even during in vitro assay (38,39). Because there is no appropriate radiolabel of the S-NO group or reliable antibody against specific nitrosothiols to date, detection of specific nitrosoproteins has been heavily relying on indirect methodology. Jaffrey and Snyder (19) developed the three-step BST method to selectively convert the nitrosylated cysteines into stable biotinylated ones, which can then be detected immunologically by antibiotin antibody or affinity purified. With this method, over 100 nitrosoproteins have been identified (40). More recently, several BST variants have been developed by replacing the biotin tag with fluorescent tags (25,41,42). When combined with 2D-DIGE, this offers a more powerful proteomic approach for analyzing and identifying nitrosoproteins.
In this study, we have first used the BST and shown that E2 and E2-BSA regulates dynamic protein S-NO that is linked to specific receptors and endogenous NO-mediated mechanisms in HUVEC. The findings signify a novel pathway for estrogen signaling. We have consolidated these observations by a BST variant, in which MTSEA-Texas Red replaced the biotin tag (24) for localizing protein S-NO in intact cells. We have further analyzed the cellular nitrosyl-proteomes in resting and E2-stimulated HUVEC by another BST variant CyDye switch combined with the powerful 2D-DIGE method. Interestingly, E2 rapidly stimulates S-NO of many proteins and also decreased S-NO of some others. The inhibitory effects of E2 on protein S-NO are intriguing and very much unexpected, because at the time point (20–30 min) tested, E2 should have theoretically increased S-NO due to the stimulatory effects of estrogens on NO production (7,8,9,20,21). However, similar findings have recently reported in cardiomyocytes; E2 and a selective ERβ agonist DPN increase the levels of many nitrosoproteins, whereas they decrease some other proteins (43). Clearly, these findings have raised a critical question as to why endothelial S-NO is decreased by E2 stimulation. Although completely elusive at the moment, trans-nitrosylation may be one likely possibility as the observed “de-nitrosylation” of some proteins may be due to donation of their -NO moiety for nitrosylating the others (44). Another possibility is that estrogens may activate one or more of the enzymes important for trans-nitrosylation or de-nitrosylation, such as GSNO reductase (45,46) and thioredoxin reductase (47), which in turn regulate dynamic protein S-NO. However, these ideas need to be further investigated.
Endothelial action of estrogens has been well linked to specific ERα and ERβ, which constitute for a major portion of the protective effects of estrogens in the cardiovascular system (6). The ER antagonist ICI 182,780 inhibits E2 and E2-BSA stimulation of total levels of endothelial nitrosoproteins, clearly demonstrating ER-dependent mechanisms in this process. More recently, Lin et al. (43) have shown that chronic treatment with E2 or DPN stimulates S-NO of many proteins involved in cardioprotection against ischemia/reperfusion injury in mouse heart, implicating a critical role of ERβ activation-mediated protein S-NO in cardiomyocytes. More recently, Chakrabarti et al. (48) reported the effects of E2 and selective ERα and ERβ agonists (PPT and DPN) on HUVEC protein S-NO, which was determined extensively by immunofluorescence labeling with an antinitrosylcysteine antibody. In that study, they showed that E2 stimulated endothelial protein S-NO via ERα. They also found enhanced in vivo endothelial S-NO in aorta endothelium in ovareictomized rats chronically treated with E2 (48). Our data suggest that E2 stimulation of endothelial protein S-NO requires both ERα and ERβ as specific agonist of neither one, but their combination was able to do so, and specific antagonist of either one blocked E2-induced response. The reason for this discrepancy is unknown but possibly due to the methods used for detection of protein S-NO in these two studies. We also have shown that the membrane impermeable estrogen ligand E2-BSA also rapidly stimulates S-NO in HUVEC, which was restrained by ICI 182,780. These findings suggest that E2 stimulation of endothelial protein S-NO is mediated by ER possibly localized on the plasma membrane. Moreover, inhibition of NO production by l-NAME attenuated E2 stimulation of protein S-NO, suggesting a critical role of NOS-derived endogenous NO, possibly via activation of eNOS in endothelial cells as suggested recently (48).
Numerous nitrosoproteins (spots) are visible on 2D-DIGE in comparison with approximately 20 bands on SDS-PAGE. Among the 58 nitrosoproteins indentified, 39 proteins are E2 responsive and heterogeneous in biological functions. Among these, peroxiredoxins are a ubiquitous family of antioxidant enzymes that also control intracellular peroxide levels and thereby participating signal transduction (49). S-NO of peroxiredoxin 2 inhibited its enzymatic activity and protects it from oxidative stress (50). Annexins belong to a multigene family of calcium-dependent phospholipid-binding proteins that regulate membrane-associated events, including exocytosis, endocytosis, ion transport, etc. Annexin A1 is related to multiple cellular processes, including cell cycle, proliferation, antiapoptosis, cell motion, surface receptor-linked signal transduction, etc. Incubation of purified annexin A2 tetramer with GSNO led to the inhibition of annexin A2 tetramer-mediated liposome aggregation (51). Cofilin-1 (15–20 kDa) belongs to a family of small actin binding proteins that regulates actin dynamics by depolymerizing actin filaments at their pointed ends or by creating new filament barbed ends for filamentous actin assembly through their severing activity (52). Although the functional consequence of coffin-1 S-NO was unclear, NO causes translocation of cofilin-1 (53). GAPDH, enolase1, and phosphofructokinase are metabolic enzymes required for glycolysis. Their functions can be modulated by NO. S-NO of GAPDH has been linked to NO-induced apoptotic death (54). Loss of neuronal enolase activity in the presence of N-methyl-d-aspartate has been found to be restored by a competitive substrate inhibitor of NOS activity (55). NO suppresses phosphofructokinase activity both in vitro and in vivo (56,57). Glutathione transferases are a superfamily of enzymes involved in the detoxication of cells against toxic and carcinogenic compounds. Rapid and stable S-NO of glutathione transferase P1-1 has been reported and regarded as a possible NO carrier protein (58). Ubiquitin C and proteasome are key components in protein degradation system. Peptidylprolyl isomerase A, also named as cyclophilin A, is a catalyze for the interconversion of cis- and trans-peptide bonds and is important for protein folding. Adenine phosphoribosyltransferase is a salvage enzyme of purine and pyridine nucleotides and thus is involved in purine metabolism. Triosephosphate isomerase catalyzes the isomerization of glyceraldehydes 3-phosphate and dihydroxy-acetone phosphate in glycolysis and gluconeogenesis. Transketolase is an enzyme of both the pentose phosphate pathway in animals and the Calvin cycle of photosynthesis. Nucleoside phosphorylase is one enzyme of the nucleotide salvage pathways, which allow the cell to produce nucleotide monophosphates when the de novo synthesis pathway has been interrupted or is nonexistent. Eukaryotic translation elongation factor 1γ is a subunit of the elongation factor-1 complex responsible for the enzymatic delivery of aminoacyl tRNAs to the ribosome. Eukaryotic translation elongation factor 2 is a member of the GTP-binding translation elongation factor family. RNA binding motif protein 8A is preferentially associated with mRNAs produced by splicing, including both nuclear mRNAs and newly exported cytoplasmic mRNAs. Eukaryotic translation initiation factor 5A regulates gene expression. These four proteins are essential for protein synthesis. Chloride intracellular channel 1 is a nuclear protein that regulates fundamental cellular processes, including stabilization of cell membrane potential, trans-epithelial transport, and maintenance of intracellular pH. Galectin-3 (also known as lectin, galactoside-binding, soluble, 3) regulates cell differentiation. Parkinson disease 7, also known as DJ-1, protects against neuronal damage from oxidative stress. These proteins all play important roles in cellular functions. However, alterations of their functions by S-NO have yet to be determined.
Among the 11 proteins that are significantly de-nitrosylated by E2, vimentin, β-actin, keratin 7, stathmin1, and tumor protein (translationally controlled 1) are cytoskeleton proteins; nonmetastatic cells 1 protein regulates cell differentiation. However, the functional consequences of S-NO of these proteins await further investigation. Also of note is that the levels of the majority of endothelial nitrosoproteins visible on 2D-DIGE are unchanged by E2, implicating that these proteins are constitutively nitrosylated in HUVEC. The levels of 19 nitrosoproteins identified in present study did not change upon estrogen treatment. Among these proteins, some (far upstream element binding protein 1, pyruvate kinase, and heterogeneous nuclear ribonucleoprotein C) have been reported as stable S-nitrosoprtein (59). Their S-nitrosylated residues are embedded inside the proteins or protein complexes, thereby rendering their S-NO status barely changeable by estrogen. Moreover, to the best of our knowledge, 15 out of the 58 identified proteins are novel nitrosoproteins (see Table 1).
Of note, different spots on 2D-DIGE may be the same protein. For example, spots 17 and 53 have been identified as Galectin-3. However, each responded differently in S-NO to E2 stimulation. S-NO of spot 17 is significantly increased by 1.58-fold, whereas spot 53 was significantly decreased to ratio of 0.54 by estrogen. Similar scenario also applies to annexin A1 (spots 25 and 51), cofilin-1 (spots 13 and 39), Chloride intracellular channel 1 (spots 26 and 56), Eukaryotic translation initiation factor 5A (spots 21 and 55), and transketolase (spots 9 and 33). The position shift of proteins on 2D-DIGE is usually due to a change in isoelectric point as a result of various posttranslation modifications. Thus, our data show that S-NO of the same protein with different isoelectric point responds to E2 stimulation differently, demonstrating that protein posttranslational modifications may lead to different responses in S-NO to stimulation by estrogens.
In sum, estrogens stimulate S-NO of an array of endothelial proteins via mechanisms linked to specific ERα/β and endogenous eNOS-derived NO. Identification and analysis of the cellular nitrosyl-proteomes in resting and E2-treated HUVEC have revealed that estrogens not only increase S-NO of many proteins but also, surprisingly, inhibit S-NO of some other proteins. Pathway analysis of the endothelial cellular nitrosyl-proteomes implicates that S-NO regulates many critical biological pathways in endothelial cells. Although the functional sequelae of the identified nitrosoproteins especially these novel ones awaits for further investigation, dynamic protein S-NO offers a novel avenue for estrogens to elicit their cardiovascular effects.
Supplementary Material
Footnotes
This work was supported in part by National Institutes of Health R21 Grant HL98746 and RO1 Grants HL70562 and HL74947 (to D.-b.C).
Results from this work were presented in part as Abstract 191 at the 55th Annual Scientific Meeting of the Society for Gynecologic Investigation, San Diego, California, March 26–29, 2008.
Disclosure Summary: The authors have nothing to disclose.
First Published Online June 2, 2010
Abbreviations: BST, Biotin switch technique; CHAPS, 3-(3-cholamidopropyl) dimethylammonio-1-propanesulfonate; cGMP, cyclic GMP; CVD, cardiovascular disease; DAPI,4′,6-diamidino-2-phenylindole; 2D-DIGE, two-dimensional fluorescence DIGE; DIGE, difference gel electrophoresis; d-NAME, N-nitro-d-arginine-methylester; DPN, diarylpropionitrile; E2, estradiol-17β; E2-BSA, β-estradiol 6-(O-carboxymethyl)oxime-BSA; eNOS, endothelial NO synthase; ER, estrogen receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GSNO, nitrosoglutathione; HEN buffer, 25 mm HEPES (pH 7.7), 1 mm EDTA, and 0.1 mm neocuproine; HUVEC, human umbilical vein endothelial cells; IEF, isoelectric focusing; l-NAME, N-nitro-l-arginine-methylester; MALDI, matrix-assisted laser desorption ionization; MMTS, methyl methanethiosulfonate; MPP, 1,3-bis(4-hydroxyphenyl)-4-methyl-5,[4-(2 piperidinylethoxy)phenol]-1H-pyrazole dihydrochloride dihydro-chloride; MS, mass spectrometry; MS/MS, tandem MS; MTSEA, 2[hyhen](6-biotinoyl-amino-hexanoyl amino) ethylmethanethiosulfonate; NO, nitric oxide; PHTPP, 4-[2-phenyl-5,7-bis(trifluoromethyl)pyrazolo[1,5-a]pyri midin-3-yl]phenol; PPT, 4,4′,4′-(4-Propyl-[1H]-pyrazole-1,3,5-triyl)trisphenol; SDS, sodium dodecyl sulfate; S-NO, S-nitrosylation; TOF, time of flight; UAEC, uterine artery endothelial cells.
References
- Praticò D 2005 Antioxidants and endothelium protection. Atherosclerosis 181:215–224 [DOI] [PubMed] [Google Scholar]
- Napoli C, de Nigris F, Williams-Ignarro S, Pignalosa O, Sica V, Ignarro LJ 2006 Nitric oxide and atherosclerosis: an update. Nitric Oxide 15:265–279 [DOI] [PubMed] [Google Scholar]
- Taylor EL, Megson IL, Haslett C, Rossi AG 2003 Nitric oxide: a key regulator of myeloid inflammatory cell apoptosis. Cell Death Differ 10:418–430 [DOI] [PubMed] [Google Scholar]
- Gilligan DM, Badar DM, Panza JA, Quyyumi AA, Cannon 3rd RO 1994 Acute vascular effects of estrogen in postmenopausal women. Circulation 90:786–791 [DOI] [PubMed] [Google Scholar]
- Rosenfeld CR, Cox BE, Roy T, Magness RR 1996 Nitric oxide contributes to estrogen-induced vasodilation of the ovine uterine circulation. J Clin Invest 98:2158–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn ME, Karas RH 2005 Molecular and cellular basis of cardiovascular gender differences. Science 308:1583–1587 [DOI] [PubMed] [Google Scholar]
- Chen Z, Yuhanna IS, Galcheva-Gargova Z, Karas RH, Mendelsohn ME, Shaul PW 1999 Estrogen receptor α mediates the nongenomic activation of endothelial nitric oxide synthase by estrogen. J Clin Invest 103:401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DB, Bird IM, Zheng J, Magness RR 2004 Membrane estrogen receptor-dependent extracellular signal-regulated kinase pathway mediates acute activation of endothelial nitric oxide synthase by estrogen in uterine artery endothelial cells. Endocrinology 145:113–125 [DOI] [PubMed] [Google Scholar]
- Russell KS, Haynes MP, Caulin-Glaser T, Rosneck J, Sessa WC, Bender JR 2000 Estrogen stimulates heat shock protein 90 binding to endothelial nitric oxide synthase in human vascular endothelial cells. Effects on calcium sensitivity and NO release. J Biol Chem 275:5026–5030 [DOI] [PubMed] [Google Scholar]
- Gratton JP, Fontana J, O'Connor DS, Garcia-Cardena G, McCabe TJ, Sessa WC 2000 Reconstitution of an endothelial nitric-oxide synthase (eNOS), hsp90, and caveolin-1 complex in vitro. Evidence that hsp90 facilitates calmodulin stimulated displacement of eNOS from caveolin-1. J Biol Chem 275:22268–22272 [DOI] [PubMed] [Google Scholar]
- Chambliss KL, Yuhanna IS, Mineo C, Liu P, German Z, Sherman TS, Mendelsohn ME, Anderson RG, Shaul PW 2000 Estrogen receptor α and endothelial nitric oxide synthase are organized into a functional signaling module in caveolae. Circ Res 87:E44–E52 [DOI] [PubMed] [Google Scholar]
- MacRitchie AN, Jun SS, Chen Z, German Z, Yuhanna IS, Sherman TS, Shaul PW 1997 Estrogen upregulates endothelial nitric oxide synthase gene expression in fetal pulmonary artery endothelium. Circ Res 81:355–362 [DOI] [PubMed] [Google Scholar]
- Murad F 1986 Cyclic guanosine monophosphate as a mediator of vasodilation. J Clin Invest 78:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanstall JC, Homer KL, Doggrell SA 2005 Evidence for, and importance of, cGMP-independent mechanisms with NO and NO donors on blood vessels and platelets. Curr Vasc Pharmacol 3:41–53 [DOI] [PubMed] [Google Scholar]
- Smith KM, Moore LC, Layton HE 2003 Advective transport of nitric oxide in a mathematical model of the afferent arteriole. Am J Physiol Renal Physiol 284:F1080–F1096 [DOI] [PubMed] [Google Scholar]
- Radi R, Peluffo G, Alvarez MN, Naviliat M, Cayota A 2001 Unraveling peroxynitrite formation in biological systems. Free Radic Biol Med 30:463–488 [DOI] [PubMed] [Google Scholar]
- Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J 1992 S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci USA 89:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane P, Hao G, Gross SS 2001 S-nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation. Sci STKE 2001:RE1 [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH 2001 The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001:PL1 [DOI] [PubMed] [Google Scholar]
- Caulin-Glaser T, García-Cardeña G, Sarrel P, Sessa WC, Bender JR 1997 17β-Estradiol regulation of human endothelial cell basal nitric oxide release, independent of cytosolic Ca2+ mobilization. Circ Res 81:885–892 [DOI] [PubMed] [Google Scholar]
- Goetz RM, Thatte HS, Prabhakar P, Cho MR, Michel T, Golan DE 1999 Estradiol induces the calcium-dependent translocation of endothelial nitric oxide synthase. Proc Natl Acad Sci USA 96:2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian XX, Mata-Greenwood E, Liao WX, Zhang H, Zheng J, Chen DB 2007 Transcriptional regulation of endothelial nitric oxide synthase expression in uterine artery endothelial cells by c-Jun/AP-1. Mol Cell Endocrinol 279:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WX, Feng L, Zhang H, Zheng J, Moore TR, Chen DB 2009 Compartmentalizing VEGF-induced ERK2/1 signaling in placental artery endothelial cell caveolae: a paradoxical role of caveolin-1 in placental angiogenesis in vitro. Mol Endocrinol 23:1428–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Loscalzo J 2005 S-nitrosoprotein formation and localization in endothelial cells. Proc Natl Acad Sci USA 102:117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Chen SC, Wang DL 2009 Shear flow increases S-nitrosylation of proteins in endothelial cells. Cardiovasc Res 83:536–546 [DOI] [PubMed] [Google Scholar]
- Cook JA, Kim SY, Teague D, Krishna MC, Pacelli R, Mitchell JB, Vodovotz Y, Nims RW, Christodoulou D, Miles AM, Grisham MB, Wink DA 1996 Convenient colorimetric and fluorometric assays for S-nitrosothiols. Anal Biochem 238:150–158 [DOI] [PubMed] [Google Scholar]
- Chambliss KL, Shaul PW 2002 Estrogen modulation of endothelial nitric oxide synthase. Endocr Rev 23:665–686 [DOI] [PubMed] [Google Scholar]
- Wagner AH, Schroeter MR, Hecker M 2001 17β-Estradiol inhibition of NADPH oxidase expression in human endothelial cells. FASEB J 15:2121–2130 [DOI] [PubMed] [Google Scholar]
- Taguchi Y, Koslowski M, Bodenner DL 2004 Binding of estrogen receptor with estrogen conjugated to bovine serum albumin (BSA). Nucl Recept 2:5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett-Connor E 1997 Sex differences in coronary heart disease. Why are women so superior? The 1995 Ancel Keys Lecture. Circulation 95:252–264 [DOI] [PubMed] [Google Scholar]
- Grady D, Herrington D, Bittner V, Blumenthal R, Davidson M, Hlatky M, Hsia J, Hulley S, Herd A, Khan S, Newby LK, Waters D, Vittinghoff E, Wenger N 2002 Cardiovascular disease outcomes during 6.8 years of hormone therapy: heart and estrogen/progestin replacement study follow-up (HERS II). JAMA 288:49–57 [DOI] [PubMed] [Google Scholar]
- Hulley S, Grady D, Bush T, Furberg C, Herrington D, Riggs B, Vittinghoff E 1998 Randomized trial of estrogen plus progestin for secondary prevention of coronary heart disease in postmenopausal women. Heart and estrogen/progestin replacement study (HERS) research group. JAMA 280:605–613 [DOI] [PubMed] [Google Scholar]
- Naftolin F, Taylor HS, Karas R, Brinton E, Newman I, Clarkson TB, Mendelsohn M, Lobo RA, Judelson DR, Nachtigall LE, Heward CB, Hecht H, Jaff MR, Harman SM 2004 The Women’s Health Initiative could not have detected cardioprotective effects of starting hormone therapy during the menopausal transition. Fertil Steril 81:1498–1501 [DOI] [PubMed] [Google Scholar]
- Brinton EA, Hodis HN, Merriam GR, Harman SM, Naftolin F 2008 Can menopausal hormone therapy prevent coronary heart disease? Trends Endocrinol Metab 19:206–212 [DOI] [PubMed] [Google Scholar]
- Mikkola TS, Clarkson TB 2002 Estrogen replacement therapy, atherosclerosis, and vascular function. Cardiovasc Res 53:605–619 [DOI] [PubMed] [Google Scholar]
- Bush TL, Barrett-Connor E, Cowan LD, Criqui MH, Wallace RB, Suchindran CM, Tyroler HA, Rifkind BM 1987 Cardiovascular mortality and noncontraceptive use of estrogen in women: results from the lipid research clinics program follow-up study. Circulation 75:1102–1109 [DOI] [PubMed] [Google Scholar]
- Mendelsohn ME 2000 Nongenomic, ER-mediated activation of endothelial nitric oxide synthase: how does it work? What does it mean? Circ Res 87:956–960 [DOI] [PubMed] [Google Scholar]
- Smith JN, Dasgupta TP 2000 Kinetics and mechanism of the decomposition of S-nitrosoglutathione by l-ascorbic acid and copper ions in aqueous solution to produce nitric oxide. Nitric Oxide 4:57–66 [DOI] [PubMed] [Google Scholar]
- Dicks AP, Williams DL 1996 Generation of nitric oxide from S-nitrosothiols using protein-bound Cu2+ sources. Chem Biol 3:655–659 [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS 2005 Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6:150–166 [DOI] [PubMed] [Google Scholar]
- Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E 2007 Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res 101:1155–1163 [DOI] [PubMed] [Google Scholar]
- Torta F, Usuelli V, Malgaroli A, Bachi A 2008 Proteomic analysis of protein S-nitrosylation. Proteomics 8:4484–4494 [DOI] [PubMed] [Google Scholar]
- Lin J, Steenbergen C, Murphy E, Sun J 2009 Estrogen receptor-β activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation 120:245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Nudelman R, Stamler JS 2001 S-nitrosylation: spectrum and specificity. Nat Cell Biol 3:E46–E49 [DOI] [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS 2001 A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410:490–494 [DOI] [PubMed] [Google Scholar]
- Lima B, Lam GK, Xie L, Diesen DL, Villamizar N, Nienaber J, Messina E, Bowles D, Kontos CD, Hare JM, Stamler JS, Rockman HA 2009 Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci USA 106:6297–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Hess DT, Stamler JS 2008 Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320:1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Lekontseva O, Peters A, Davidge ST 2009 17β-Estradiol induces protein S-nitrosylation in the endothelium. Cardiovasc Res 2010 85:796–805 [DOI] [PubMed] [Google Scholar]
- Rhee SG, Chae HZ, Kim K 2005 Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med 38:1543–1552 [DOI] [PubMed] [Google Scholar]
- Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA 2007 S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci USA 104:18742–18747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Enright E, Sun P, Tsai SY, Mehta P, Beckman DL, Terrian DM 2002 Inactivation of annexin II tetramer by S-nitrosoglutathione. Eur J Biochem 269:4277–4286 [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P 2005 Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell 16:649–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi R, Matsui S, Kinoshita M, Nagaishi K, Sasaki H, Kasahara T, Suzuki K 2000 Nitric oxide induces chemotaxis of neutrophil-like HL-60 cells and translocation of cofilin to plasma membranes. Int J Immunopharmacol 22:855–864 [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A 2005 S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7:665–674 [DOI] [PubMed] [Google Scholar]
- Kollegger H, McBean GJ, Tipton KF 1993 Reduction of striatal N-methyl-D-aspartate toxicity by inhibition of nitric oxide synthase. Biochem Pharmacol 45:260–264 [DOI] [PubMed] [Google Scholar]
- Tsuura Y, Ishida H, Hayashi S, Sakamoto K, Horie M, Seino Y 1994 Nitric oxide opens ATP-sensitive K+ channels through suppression of phosphofructokinase activity and inhibits glucose-induced insulin release in pancreatic β cells. J Gen Physiol 104:1079–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuura Y, Ishida H, Shinomura T, Nishimura M, Seino Y 1998 Endogenous nitric oxide inhibits glucose-induced insulin secretion by suppression of phosphofructokinase activity in pancreatic islets. Biochem Biophys Res Commun 252:34–38 [DOI] [PubMed] [Google Scholar]
- Lo Bello M, Nuccetelli M, Caccuri AM, Stella L, Parker MW, Rossjohn J, McKinstry WJ, Mozzi AF, Federici G, Polizio F, Pedersen JZ, Ricci G 2001 Human glutathione transferase P1-1 and nitric oxide carriers; a new role for an old enzyme. J Biol Chem 276:42138–42145 [DOI] [PubMed] [Google Scholar]
- Paige JS, Xu G, Stancevic B, Jaffrey SR 2008 Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol 15:1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.