Abstract
The loss of chromosomes 1p–19q is the only prognostic molecular alteration identified in low-grade gliomas (LGGs) to date. Search for loss of heterozygosity (LOH) on chromosomes 1p, 9p, 10q, and 19q was performed in a series of 231 LGGs. Loss of chromosomes 1p–19q was strongly correlated with prolonged progression-free survival (PFS) and overall survival (OS) in univariate and multivariate analyses. LOH on 9p and 10q were associated with shortened PFS (P = .01 and .03, respectively) on univariate analysis. On multivariate analysis, LOH on 9p remained significant for PFS (P = .05), whereas LOH on 10q had a significant effect on OS (P = .02). Search for LOH 9p and 10q appears to be a useful complement to analysis of chromosomes 1p–19q in LGGs.
Keywords: chromosomes 9p and 10q, low-grade gliomas, prognosis
The prognosis of low-grade gliomas (LGGs) varies widely, with overall survival (OS) ranging from a few years to decades. Defining reliable prognostic factors in LGGs has been difficult, but the importance of clinical factors, including gender, age, Karnofsky performance status (KPS), symptoms at diagnosis, tumor size, and extent of tumor resection, has been recently recognized.1–4 To date, the only identified molecular alteration of prognostic importance in LGGs is the 1p–19q co-deletion.5,6 Loss of heterozygosity (LOH) on chromosomes 9p and 10q is rare in LGGs. These alterations are classically associated with high-grade tumors (anaplastic gliomas and glioblastomas)7 and have been found to be of prognostic significance in anaplastic gliomas8 but not in glioblastomas.9 In this study, we analyzed the prognostic impact of LOH on 9p and 10q in a series of LGGs.
Patients and Methods
Selection of Patients
This work is based on the analysis of a database created in January 1997 that collects clinical information on patients treated in the department of Neurologie 2-Mazarin, Groupe Hospitalier Pitié-Salpêtrière, Paris for primary brain tumor.6 The following inclusion criteria were selected: (1) age of 18 years or more at the time of surgery; (2) histological diagnosis of a cerebral LGG according to World Health Organization (WHO) classification (WHO grade II, 2007), including astrocytomas, oligodendrogliomas, and mixed oligoastrocytomas; (3) detailed clinical information at diagnosis and during follow-up; (4) follow-up of >24 months; and (5) availability of paired blood and tumor samples, obtained after informed consent, for molecular analysis.
Patients with anaplastic pathology features such as dysembryoplastic neuroepithelial tumors, pilocytic astrocytomas, gangliogliomas, or gliomatosis, according to the last WHO classification, were excluded.
Molecular Analysis
DNA from both blood and tumor tissue was extracted using a commercial kit (Qiagen, QIAmp DNA mini Kit) according to the manufacturer's instructions. LOH on chromosomes 1p, 19q, 9p, and 10q was detected by microsatellite analysis of blood and tumor DNA, as previously reported.10
Statistical Methods
Frequency distribution and summary statistics were calculated for all clinical, histological, and molecular variables. The χ2 test was used to test the association between molecular alterations. Progression-free survival (PFS) and OS were both used to study the prognostic impact of the analyzed variables. PFS was defined as the time from the date of surgery until the first unequivocal clinical or radiological sign of progressive disease, independent of treatment received. Probability estimates for PFS and OS were calculated using the Kaplan–Meier method. The log-rank test was used to test for equality of the PFS and OS distributions. Factors that were significant in univariate analysis were entered as candidate variables in the multivariate Cox proportional hazard regression model analysis: gender, KPS (>80 versus ≤80), type of surgery (biopsy and partial resection versus total), histological subtype, and 1p, 19q, 9p, and 10q status as well as age (>40 versus <40 years). Two-sided P-values <.05 were considered significant.
Results
Clinical Data
Two hundred thirty-one patients from the database fulfilled the inclusion criteria. The median age at surgery was 39 years (range, 18–78), the sex ratio (men/women) was 1.5, and the median preoperative KPS was 90 (range, 70–100). One hundred thirty-four patients (59%) underwent biopsy or partial resection, whereas 94 patients (41%) had gross total resection. In 202 of 231 patients (87%), tumor samples were reviewed by consensus between two neuropathologists expert in primary brain tumors (K.M. and A.R.), whereas 29 samples (13%) were reviewed by only one of them. Overall, 19% (43 of 231) of the patients had an astrocytoma, 25% (58 of 231) had an oligoastrocytoma, and 56% (130 of 231) had an oligodendroglioma. Adjuvant treatment consisted of radiotherapy (postoperative radiotherapy in 45 patients and delayed radiotherapy at tumor progression in 74 patients) and chemotherapy (postoperative chemotherapy in 47 patients and delayed chemotherapy at tumor progression in 115 patients).
The median follow-up time was 95.1 months (95% confidence interval [CI] 82.6–107.3). At the time of analysis, 70.1% (162 of 231) of patients had at least one relapse, and the median PFS was 39.6 months (95% CI, 35.8–44.5). In addition, 29% (67 of 231) of patients died, and the median OS was 175.8 months (95% CI, 150.1–261).
Molecular Analysis
LOH on chromosome arms 1p, 19q, 9p, and 10q was found in 37% (82 of 221), 48% (105 of 221), 25% (55 of 217), and 14% (30 of 217) of patients, respectively.
LOH on 1p and 19q was strongly associated (P = 8 × 10−20). Otherwise, there was no significant association between the molecular parameters.
Correlations between Clinical and Molecular Data
There was a tight correlation between an oligodendroglioma phenotype and LOH on chromosome arms 1p and 19q (P = 1.2 × 10−11), but there was no association between the histopathological subtype and the LOH on chromosome arm 9p or 10q.
There was no significant correlation between loss on 9p or 10q and age >40 years, Ki-67 expression >5%, or contrast enhancement in magnetic resonance images at the time of diagnosis. The time between first symptoms and surgery was not increased in patients with LOH on 9p or 10q when compared with patients without these alterations (134 days with LOH on 9p versus 188 days and 92 days with LOH on 10q versus 196 days).
In the univariate analysis (Table 1), several clinical and pathological factors were associated with increased PFS and OS: female gender, KPS >80, absence of neurologic deficit at diagnosis, gross total resection of the tumor, and histopathological diagnosis of oligodendroglioma.
Table 1.
Correlations between clinical or molecular factors and PFS or OS: univariate analysis. (*: p < 0.05)
| Number of Observations | Median PFS (months) | P-value | Median OS (months) | P-value | |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 139 | 41.4 | 0.59 | 150.1 | 0.04* |
| Female | 92 | 38.9 | 192.6 | ||
| Age at diagnosis | |||||
| <40 years | 122 | 42.6 | 0.79 | Not reached | 0.75 |
| >40 years | 109 | 36.8 | 163.3 | ||
| Preoperative KPS | |||||
| ≤80 | 28 | 25.7 | 0.03* | 98.1 | 0.001* |
| >80 | 184 | 41.4 | 242.6 | ||
| No neurological deficit | 129 | 41.3 | 0.01* | 163.3 | 0.04* |
| Neurological deficit | 16 | 19.5 | 98.1 | ||
| No contrast enhancement | 179 | 39.9 | 0.24 | 175.8 | 0.54 |
| Contrast enhancement | 36 | 25.2 | Not reached | ||
| Biopsy/partial resection | 134 | 35.9 | 0.02* | 163.3 | 0.07 |
| Gross total resection | 94 | 47.3 | Not reached | ||
| Astrocytoma | 43 | 29.3 | 0.03* | 113.2 | 0.15 |
| Oligodendroglioma | 130 | 41.4 | 175.8 | ||
| Ki-67 | |||||
| ≤5% | 46 | 44.5 | 0.33 | Not reached | 0.71 |
| >5% | 39 | 35.8 | |||
| Postoperative radiotherapy | 45 | 56.8 | <0.0001* | 163.3 | 0.7 |
| Delayed radiotherapy | 74 | 23.1 | 152.4 | ||
| Postoperative chemotherapy | 47 | 35.9 | 0.52 | Not reached | 0.46 |
| Delayed chemotherapy | 115 | 35.6 | 154.7 | ||
| No LOH 1p–19q | 149 | 35.8 | 0.002* | 138.5 | <0.0001* |
| LOH 1p–19q | 71 | 49.5 | 242.6 | ||
| No LOH 9p | 162 | 41.4 | 0.01* | 196.2 | 0.49 |
| LOH 9p | 55 | 31.5 | 150.1 | ||
| No LOH 10q | 187 | 42.6 | 0.03* | 192.6 | 0.1 |
| LOH 10q | 30 | 25.7 | 106.4 | ||
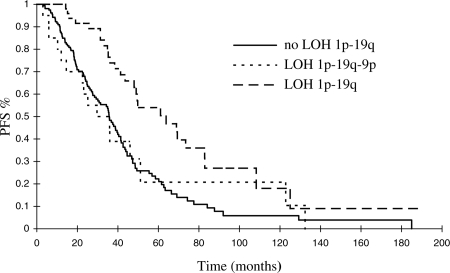
Among molecular markers (Table 1), LOH on 1p–19q was strongly associated with increased PFS and OS (P = .002 and <.0001, respectively). LOH on both 9p and 10q was correlated with shorter PFS (P = .01 and .03, respectively). In the subgroup of patients with LOH on 1p–19q, LOH on 9p was also associated with shorter PFS (P = .03). Moreover, the PFS curve of patients with LOH on 1p–19q–9p was almost superimposed onto that of the patients without LOH on 1p–19q (Fig. 1).
Fig. 1.
The prognostic advantage conferred by deletion of chromosomes 1p–19q on PFS is lost in patients with combined chromosome 9p loss.
In a multivariate analysis, including the main clinical and molecular parameters (Table 2), LOH on 1p–19q remained the main factor predicting a good prognosis. Gender, KPS, and the extent of tumor resection were still associated with survival. LOH on 9p was significantly associated with shorter PFS but not with shorter OS. LOH on 10q was associated with shorter OS.
Table 2.
Correlations between clinical or molecular factors and PFS or OS: multivariate analysis. (*: p < 0.05)
| Covariate | PFS |
OS |
||
|---|---|---|---|---|
| P-value | Hazard Ratio | P-value | Hazard Ratio | |
| Age >40 | 0.35 | 1.19 | 0.91 | 1.04 |
| Female gender | 0.97 | 0.99 | 0.01* | 0.45 |
| KPS >80 | 0.22 | 0.73 | 0.009* | 0.40 |
| Biopsy, partial resection (versus total resection) | 0.01* | 0.64 | 0.03* | 0.51 |
| Histology | 0.39 | 0.89 | 0.83 | 0.96 |
| LOH 1p + 19q | 0.0006* | 0.50 | 0.0001* | 0.16 |
| LOH 9p | 0.05* | 1.46 | 0.95 | 0.98 |
| LOH 10q | 0.11 | 1.49 | 0.02* | 2.53 |
Discussion
This study of a large series of LGGs confirms the favorable prognostic role of 1p–19q codeletion and shows that losses of chromosomes 9p and 10q are negative prognostic factors.
LGGs are known to progress to high-grade gliomas within several years,11 a process associated with the accumulation of molecular alterations, including the loss of chromosomes 9p or 10q.12–15 In the literature, the reported frequency of 9p and 10q losses in LGGs ranges from 0% to 25%,16–18 but these data were obtained in small series that rarely exceeded 10–20 patients. Since histopathologic diagnosis and grading of gliomas are notoriously difficult,19 one might hypothesize that tumors presenting with LOH on 9p or 10q were high-grade lesions misclassified as LGGs. However, several features lead us to consider that this hypothesis is very unlikely: first, almost 90% of our tumor samples (202 of 231) were reviewed by consensus between two neuropathologists expert in primary brain tumors. Second, LGG patients with losses of 9p or 10q did not exhibit the classic variables associated with a higher-grade tumor, such as older age, increased Ki-67 immunolabeling index, contrast enhancement, or any other clinical, radiological, or histological findings suggestive of anaplastic behavior. Finally, the OS of 106.4 and 150.1 months observed in our LGG patients whose tumors contained LOH on 9p and LOH on 10q is within the usual range for LGGs and is not compatible with the expected median survival of misdiagnosed anaplastic gliomas.8 One could also argue that LOH on 9p and 10q is only present in long-standing tumors. However, the time elapsed between the first symptoms and the surgical resection was not longer for tumors with 9p and/or 10q LOH than for tumors without these genetic alterations. These data suggest that loss of chromosome arm 9p or 10q may occur early in the course of the disease and confer a more aggressive biological phenotype at a time when clinical and histological features of anaplasia are absent.
In this series, LOH on 9p and 10q was associated with shorter PFS on univariate analysis. In multivariate analysis, LOH on 9p was an independent predictor of shorter PFS, whereas LOH on 10q was a predictor of shorter OS. These differences might be explained by an insufficient follow-up in this population with a very long OS time (median survival, 175 months).
A considerable amount of time and many patients are indeed needed to identify prognostic factors in LGGs. This is illustrated by our finding on the prognostic importance of the 1p–19q codeletion. In a previous study on a smaller cohort (131 versus 231 patients) with a shorter follow-up (63.3 versus 95.1 months), only LOH on 1p was a significant favorable prognostic predictor. LOH on 9p and 10q was not identified as a potential prognostic factor at this stage.6 A larger series with longer follow-up time strongly strengthens the data, confirming the major favorable impact of 1p–19q LOH not only on PFS but also on OS,20–22 in addition to detecting the role of 9p and 10q deletions. Analyzing these alterations together seems useful. Indeed, as illustrated in Fig. 1, the benefit of the 1p–19q deletion on PFS was lost when deletion of chromosome 9p was also present.
This study confirms the important prognostic role of several clinical factors, including preoperative KPS, neurological deficit at time of diagnosis, extent of tumor resection, and histological subtype.1–4 We did not confirm the prognostic value of age, possibly because of the low number of older patients in our series.
In summary, this study indicates that the search for LOH on 9p and 10q is a useful complement to clinical evaluation and analysis of 1p–19q status in the prognostic work-up of LGGs.
Conflict of interest statement. None declared.
Funding
This study was supported by the Ligue Nationale Contre le Cancer and the Délégation à la Recherche Clinique (AP-HP; grant MUL-03012). The authors gratefully thank Anne-Marie Lekieffre, Marie-Aline Renard, and Muriel Brandel from the ARTC for their assistance.
References
- 1.Karim AB, Maat B, Hatlevoll R, et al. A randomized trial on dose–response in radiation therapy of low-grade cerebral glioma. European Organization for Research and Treatment of Cancer (EORTC) Study 22844. Int J Radiat Oncol Biol Phys. 1996;36:549–556. doi: 10.1016/s0360-3016(96)00352-5. [DOI] [PubMed] [Google Scholar]
- 2.Pignatti F, van den Bent M, Curran D, et al. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. 2002;20:2076–2084. doi: 10.1200/JCO.2002.08.121. [DOI] [PubMed] [Google Scholar]
- 3.Shaw E, Arusell R, Scheuthauer B, et al. Prospective randomized trial of low- versus high-dose radiation therapy in adults with supratentorial low-grade glioma: initial report of a North Central Cancer Treatment Group/Radiation Therapy Oncology Group/Eastern Cooperative Oncology Group study. J Clin Oncol. 2002;20:2267–2276. doi: 10.1200/JCO.2002.09.126. [DOI] [PubMed] [Google Scholar]
- 4.Claus EB, Black PM. Survival rates and patterns of care for patients diagnosed with supratentorial low-grade gliomas: data from the SEER program, 1973–2001. Cancer. 2006;106:1358–1363. doi: 10.1002/cncr.21733. [DOI] [PubMed] [Google Scholar]
- 5.Kaloshi G, Benouaich-Amiel A, Diakite F, et al. Temozolomide for low-grade gliomas: predictive impact of 1p/19q loss on response and outcome. Neurology. 2007;68:1831–1836. doi: 10.1212/01.wnl.0000262034.26310.a2. [DOI] [PubMed] [Google Scholar]
- 6.Kujas M, Lejeune J, Benouaich-Amiel A, et al. Chromosome 1p loss: a favorable prognostic factor in low-grade gliomas. Ann Neurol. 2005;58:322–326. doi: 10.1002/ana.20543. [DOI] [PubMed] [Google Scholar]
- 7.von Deimling A, Louis DN, Wiestler OD. Molecular pathways in the formation of gliomas. Glia. 1995;15:328–338. doi: 10.1002/glia.440150312. [DOI] [PubMed] [Google Scholar]
- 8.Dehais C, Laigle-Donadey F, Marie Y, et al. Prognostic stratification of patients with anaplastic gliomas according to genetic profile. Cancer. 2006;107:1891–1897. doi: 10.1002/cncr.22211. [DOI] [PubMed] [Google Scholar]
- 9.Houillier C, Lejeune J, Benouaich-Amiel A, et al. Prognostic impact of molecular markers in a series of 220 primary glioblastomas. Cancer. 2006;106:2218–2223. doi: 10.1002/cncr.21819. [DOI] [PubMed] [Google Scholar]
- 10.He J, Mokhtari K, Sanson M, et al. Glioblastomas with an oligodendroglial component: a pathological and molecular study. J Neuropathol Exp Neurol. 2001;60:863–871. doi: 10.1093/jnen/60.9.863. [DOI] [PubMed] [Google Scholar]
- 11.McCormack BM, Miller DC, Budzilovich GN, et al. Treatment and survival of low-grade astrocytoma in adults—1977–1988. Neurosurgery. 1992;31:636–642. doi: 10.1227/00006123-199210000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Hulsebos TJ, Oskam NT, Troost D, et al. Dynamics of genetic alterations associated with glioma recurrence. Genes Chromosomes Cancer. 1998;23:153–158. [PubMed] [Google Scholar]
- 13.Idbaih A, Carvalho Silva R, Crinière E, et al. Genomic changes in progression of low-grade gliomas. J Neurooncol. 2008;90:133–140. doi: 10.1007/s11060-008-9644-z. [DOI] [PubMed] [Google Scholar]
- 14.Weber RG, Sabel M, Reifenberger J, et al. Characterization of genomic alterations associated with glioma progression by comparative genomic hybridization. Oncogene. 1996;13:983–994. [PubMed] [Google Scholar]
- 15.Hulsebos TJ, Troost D, Leenstra S. Molecular-genetic characterisation of gliomas that recur as same grade or higher grade tumours. J Neurol Neurosurg Psychiatry. 2004;75:723–726. doi: 10.1136/jnnp.2003.025031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitange G, Misra A, Law M, et al. Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer. 2005;42:68–77. doi: 10.1002/gcc.20108. [DOI] [PubMed] [Google Scholar]
- 17.Trost D, Ehrler M, Fimmers R, et al. Identification of genomic aberrations associated with shorter overall survival in patients with oligodendroglial tumors. Int J Cancer. 2007;120:2368–2376. doi: 10.1002/ijc.22574. [DOI] [PubMed] [Google Scholar]
- 18.Thiessen B, Maguire JA, McNeil K, et al. Loss of heterozygosity for loci on chromosome arms 1p and 10q in oligodendroglial tumors: relationship to outcome and chemosensitivity. J Neurooncol. 2003;64:271–278. doi: 10.1023/a:1025689004046. [DOI] [PubMed] [Google Scholar]
- 19.Coons SW, Johnson PC, Scheithauer BW, et al. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79:1381–1393. doi: 10.1002/(sici)1097-0142(19970401)79:7<1381::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 20.Mariani L, Deiana G, Vassella E, et al. Loss of heterozygosity 1p36 and 19q13 is a prognostic factor for overall survival in patients with diffuse WHO grade 2 gliomas treated without chemotherapy. J Clin Oncol. 2006;24:4758–4763. doi: 10.1200/JCO.2006.05.9238. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 22.Iwamoto FM, Nicolardi L, Demopoulos A, et al. Clinical relevance of 1p and 19q deletion for patients with WHO grade 2 and 3 gliomas. J Neurooncol. 2008;88:293–298. doi: 10.1007/s11060-008-9563-z. [DOI] [PubMed] [Google Scholar]