Abstract
O6-Methylguanine DNA methyltransferase (MGMT) is implicated as a major predictive factor for treatment response to alkylating agents including temozolomide (TMZ) of glioblastoma multiforme (GBM) patients. However, whether the MGMT status in GBM patients should be detected at the level of promoter methylation or protein expression is still a matter of debate. Here, we compared promoter methylation (by methylation-specific polymerase chain reaction) and protein expression (by Western blot) in tumor cell explants with respect to prediction of TMZ response and survival of GBM patients (n = 71). Methylated MGMT gene promoter sequences were detected in 47 of 71 (66%) cases, whereas 37 of 71 (52%) samples were scored positive for MGMT protein expression. Although overall promoter methylation correlated significantly with protein expression (χ2 test, P < .001), a small subgroup of samples did not follow this association. In the multivariate Cox regression model, a significant interaction between MGMT protein expression, but not promoter methylation, and TMZ therapy was observed (test for interaction, P = .015). In patients treated with TMZ (n = 42), MGMT protein expression predicted a significantly shorter overall survival (OS; hazard ratio [HR] for death 5.53, 95% confidence interval [CI] 1.76–17.37; P = .003), whereas in patients without TMZ therapy (n = 29), no differences in OS were observed (HR for death 1.00, 95% CI 0.45–2.20; P = .99). These data suggest that lack of MGMT protein expression is superior to promoter methylation as a predictive marker for TMZ response in GBM patients.
Keywords: O6-Methylguanine DNA methyltransferase, glioblastoma multiforme, protein expression, temozolomide
Glioblastoma multiforme (GBM) represents the most frequent form of primary brain tumor and is characterized by poor prognosis. For many years, the standard care for high-grade glioma patients was resection followed by radiotherapy (RT). An EORTC-NCIC multi-center trial published in 20051 revealed a statistically significant survival benefit for GBM patients treated with RT plus temozolomide (TMZ). Consequently, concomitant TMZ therapy/RT currently represents the standard treatment for newly diagnosed GBM patients.2
O6-Methylguanine DNA methyltransferase (MGMT) is a key enzyme in the DNA repair network that removes mutagenic, cytotoxic adducts from O6-guanine in DNA, the preferred point of attack of alkylating agents.3–5 This transfer irreversibly inactivates MGMT.6 Accordingly, MGMT knockout mice are hypersensitive against alkylating drugs,7 including TMZ,8 and depletion of the enzyme by the substrate analog O6-benzylguanine increased the sensitivity of, for example, glioma cells against those drugs.9–12 Thus, MGMT is believed to function as a major resistance factor against alkylating agent–based chemotherapy.4,13,14
The proportion of GBM tissues lacking MGMT expression is relatively high,15,16 suggesting that a subgroup of GBM patients should benefit from the treatment with TMZ. Indeed, several reports have indicated a significant correlation between MGMT status and response to alkylating agents, including TMZ.15,17–20 Consequently, intense efforts have been made to define an ideal MGMT-related marker to predict benefit from TMZ treatment in GBM patients.2,21,22 As silencing of the MGMT gene is one of the most common epigenetic changes in gliomas,23 detection of MGMT promoter methylation has, beside immunostaining, been in the focus of interest. However, conflicting data have been reported and thus the choice of the ideal MGMT-related marker and the appropriate detection method is still a matter of debate.17,22,24–28
In this study, we used, for the first time, primary tumor cell explants derived from GBM surgical specimens instead of tissue samples to determine tumor cell–specific protein expression of MGMT and methylation of the respective gene promoter. Using this approach, we demonstrate that MGMT protein expression has a strong predictive value with regard to TMZ response and survival in GBM patients.
Patients and Methods
Patient Characteristics and Tissue Samples
A total of 71 patients who underwent surgery between 2003 and 2007 at the Department of Neurosurgery, Wagner Jauregg Hospital of Linz, were included. The patient group was composed of 39 men and 32 women with a median age of 64.5 years (range 13–81). Patients had primary glioblastoma (n = 69) and gliosarcoma (n = 2) according to WHO criteria. Written informed consent was obtained from all patients for molecular analyses. Subsequent to surgery, 42 patients received combined RT/chemotherapy (60 Gy and daily TMZ at 75 mg/m2; 7 days per week over a 40-day period). Twenty-nine patients did not receive chemotherapy, out of which 15 had only RT and 14 did not have further therapy because of low performance status (5 of 14) or rapid worsening (9 of 14). Because of tumor progression, RT had to be ceased ahead of time in one patient. Patient characteristics in relation to MGMT promoter methylation and MGMT protein expression are outlined in Table 1.
Table 1.
Characteristics of the patients according to MGMT promoter methylation and protein expression status
| Characteristic | No. of Patients (%) n = 71 | MGMT Promoter Methylation Status, No. of Patients (%) |
P-valuea | MGMT Protein Expression, No. of Patients (%) |
P-valuea | ||
|---|---|---|---|---|---|---|---|
| Unmethylated, n = 24 (34) | Methylated, n = 47 (66) | Negative, n = 34 (48) | Positive, n = 37 (52) | ||||
| Age (years) | |||||||
| <45 | 9 (13) | 4 (17) | 5 (11) | .47 | 4 (12) | 5 (14) | .83 |
| ≥45 | 62 (87) | 20 (83) | 42 (89) | 30 (88) | 32 (87) | ||
| Gender | |||||||
| Female | 32 (45) | 13 (54) | 19 (40) | .27 | 19 (56) | 13 (35) | .08 |
| Male | 39 (55) | 11 (46) | 28 (60) | 15 (44) | 24 (65) | ||
| Performance status | |||||||
| <90 | 33 (47) | 13 (54) | 20 (43) | .35 | 16 (47) | 17 (46) | .93 |
| ≥90 | 38 (53) | 11 (46) | 27 (57) | 18 (53) | 20 (54) | ||
| TMZ therapy | |||||||
| Yes | 42 (59) | 12(50) | 30 (64) | .26 | 20 (59) | 22 (60) | .96 |
| No | 29 (41) | 12 (50) | 17(36) | 14 (41) | 15 (41) | ||
Percentages may not total 100 because of rounding.
aTwo-sided χ2 test.
Primary Tumor Cell Cultures
All of the analyzed cell cultures were established from surgery specimens, as published previously,29 and analyzed between passages 2 and 6. The presence of predominantly malignant cells in primary cell cultures was proved by comparative genomic hybridization and/or fluorescent in situ hybridization analyses.29 All cell cultures were periodically checked for Mycoplasma contamination (Mycoplasma Stain Kit, Sigma, St. Louis, MO).
DNA Modification and Methylation-Specific Polymerase Chain Reaction
DNA was extracted from tumor cell cultures or frozen surgery tissues with QIAmp DNA Blood Mini Kit (QIAGEN) and DNA bisulfite conversion performed with Epitect Bisulfite Kit (QIAGEN) according to the manufacturer's instructions. Methylation-specific PCR (MSP) was performed with primers specific for either methylated or modified unmethylated DNA16 as described previously.29 CpGenome Universal methylated and Universal unmethylated DNA (Chemicon International, Temecula, CA, USA) were used as controls. PCR products were separated by 6% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), stained with ethidium bromide, and visualized under UV illumination (ChemiDoc, BioRad, Hercules, CA, USA). Expression levels were quantified by Quantity One Quantitation software (BioRad) and calculated relative to the indicated controls. All results were confirmed in at least two independent experiments.
Immunoblot Analysis
Protein extracts were prepared, separated by SDS–PAGE (12%), and blotted onto nitrocellulose membranes (Hybond ECL, Amersham, Aylesbury, UK) as described previously.30 Blots were probed with the monoclonal mouse antibody anti-human MGMT (DakoCytomation, Carpinteria, CA, USA). Visualization and quantification were performed using the ChemiDoc System (BioRad). Data obtained by the densitometric evaluation of immunoblots for MGMT were expressed relative (arbitrary units) to the MGMT-overexpressing glioblastoma cell line GL80 that is included as a positive control in each blot and set arbitrarily as 1. All experiments were performed in triplicate.
Statistical Analyses
Baseline characteristics of patients were compared to MGMT promoter methylation or MGMT protein expression using χ2 tests. The correlation between MGMT promoter methylation and protein expression was assessed either by χ2 test or by Mann–Whitney U test. Overall survival (OS) was defined as the period between the time of surgery and death. Survival times of patients still alive were censored with the date of the last follow-up. Survival probabilities were estimated by means of the Kaplan–Meier method, and the survival rates were compared using the log-rank test. To describe the unadjusted effects of covariates on OS, univariate Cox proportional hazards regression models were used. The independent prognostic and predictive values of MGMT promoter methylation as well as protein expression were studied using Cox models that were adjusted for age (as a continuous variable), gender (female or male), performance status (as a continuous variable), and TMZ therapy (yes or no). These models were also applied to assess interactions between treatment and other covariates. All reported P values are 2-sided. All analyses were performed with the use of SPSS software, version 15.0 (SPSS).
Results
MGMT Promoter Methylation, Gene Expression, and Patient Characteristics
Primary in vitro tumor cell culture was successful in all (n = 71) cases, allowing DNA and protein extraction at sufficient amounts and subsequent MGMT promoter and protein expression analyses, respectively. Methylated MGMT gene promoter sequences were detected in 47 of 71 (66%) GBM samples. Out of the 47 cases with methylated sequences, 14 (30%) lacked any unmethylated DNA, whereas the remaining 33 samples contained a mixed profile with varying proportions of methylated and unmethylated sequences (Fig. 1A). In 24 of 71 (34%) GBM patients, the MGMT promoter was completely unmethylated. To investigate the potential impact of in vitro cell culture, MGMT promoter methylation was analyzed in both snap-frozen tumor tissues and the respective cell cultures in 17 patients. In all cases, the methylation status agreed well (Table 2), and semi-quantification of MSP products for methylated DNA sequences revealed a highly significant correlation (linear regression analysis, P < .0001). MGMT protein levels were successfully determined by Western blot in all tumor cell cultures (Fig. 1B). Thirty-seven of the 71 tumors (52%) were scored positive for MGMT protein expression based on a clearly visible band at the respective molecular size (25 kDa) and corresponding to a relative expression level >0.1. Neither MGMT promoter methylation nor MGMT protein expression was significantly associated with any of the clinical variables listed in Table 1.
Fig. 1.
Detection of the MGMT status in glioblastoma-derived cell cultures. Representative examples are shown. (A) MGMT promoter methylation was detected by MSP. Amplification products with specific primers for methylated (m, 81 bp) and unmethylated (u, 93 bp) DNA sequences are shown, and scoring as methylated (m), unmethylated (u), and mixed (u/m) is indicated. Positive controls for methylated (pcm) and unmethylated (pcu) sequences were included. M, size marker. (B) MGMT protein expression was determined by Western blot (band at 25 kDa), and relative expression levels were obtained by the densitometric evaluation of immunoblots compared with the MGMT-overexpressing glioblastoma cell line GL80 included as positive control (pc) and set arbitrarily as 1. Probing with β-actin served as loading control. (C) Scatter gram analysis of MGMT protein expression in subgroups with methylated (m) and unmethylated (u) MGMT promoter.
Table 2.
MGMT promoter methylation of DNA extracted from cell cultures and tumor tissues
| Patient | Methylated MGMT Promoter Sequencesa |
|
|---|---|---|
| Primary Cell Culture | Tumor Tissue | |
| 1 | 0.7 | 0.2 |
| 2 | 0.8 | 0.3 |
| 3 | 0 | 0 |
| 4 | 0 | 0 |
| 5 | 0.5 | 0.2 |
| 6 | 1.2 | 0.7 |
| 7 | 0 | 0 |
| 8 | 0.4 | 0.2 |
| 9 | 0.2 | 0.4 |
| 10 | 0.7 | 0.9 |
| 11 | 0.9 | 0.2 |
| 12 | 1.8 | 0.8 |
| 13 | 0 | 0 |
| 14 | 1.6 | 1.0 |
| 15 | 0.3 | 0.2 |
| 16 | 0 | 0 |
| 17 | 0.8 | 0.2 |
aMGMT promoter methylation status was analyzed by MSP as described in Patients and Methods. MSP amplification products were evaluated relative to the respective positive controls set as 1.
Relation between MGMT Promoter Methylation and Protein Expression
For all patients, both MGMT promoter methylation and MGMT protein expression data were available. The median MGMT expression level was significantly higher in cell cultures harboring only unmethylated MGMT promoter sequences when compared with the methylated subgroup (p < .0001) (Fig. 1C). Accordingly, the MGMT promoter methylation status was significantly associated with protein expression when analyzed by 2-sided χ2 test (p = .0004). Despite this clear-cut association, protein expression levels in the subgroups, according to promoter methylation, were strongly overlapping (Fig. 1C). Data for 4 representative examples not following the correlation between promoter methylation status and protein expression are shown in Fig. 2. These observations included samples with completely methylated promoter but high protein expression as well as lack of MGMT expression, despite exclusively unmethylated promoter sequences.
Fig. 2.
Discrepancy between MGMT promoter methylation status and protein expression determined by methylation specific PCR (MSP) and Western blot (WB). Representative examples for promoter methylation and protein expression (BTL183, BTL35) as well as lack of promoter methylation and protein expression (BTL298, BTL365) are given. (A) Polyacrylamide gel showing amplification products of methylated (m, 81 bp) and unmethylated (u, 93 bp) DNA sequences. Positive controls for methylated (pcm) and unmethylated (pcu) sequences were included. (B) Western blot analysis for the corresponding samples. Data obtained by MSP and WB were expressed as relative (expression) levels as described in Patients and Methods.
Relation between MGMT Status and OS of GBM Patients with/without TMZ Treatment
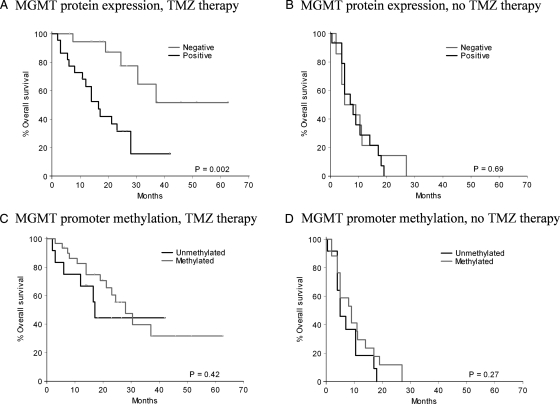
At a median follow-up time of 26.9 months (95% CI 24.7–29.1), 48 (68%) of 71 patients had died (20 of 42 patients who received TMZ therapy and 28 of 29 patients without TMZ therapy). The OS of GBM patients was compared to MGMT promoter methylation status or protein expression by univariate analyses (Table 3). Besides the well-known prognostic factors such as age, performance status, and TMZ therapy, MGMT protein expression (hazard ratio [HR] of death 2.02, 95% CI 1.12–3.66; P = .02), but not lack of MGMT promoter methylation (HR of death 0.61, 95% CI 0.33–1.11; P = .1), was significantly associated with shorter OS in the univariate analyses. In multivariate Cox regression analysis (Table 4), only MGMT protein expression, but not promoter methylation, remained as an independent prognostic factor (HR of death 1.99, 95% CI 1.07–3.70; P = .03) in addition to age, performance status, and TMZ therapy. To determine a possible interaction between the MGMT parameters and TMZ therapy, an interaction term, the product of MGMT parameter and TMZ therapy, was incorporated into the Cox models. These analyses revealed a strong interaction between MGMT protein expression and TMZ therapy (p = .005), whereas this association was not significant in the case of promoter methylation. Because the interaction term of MGMT protein expression and TMZ treatment was statistically significant, we determined the association of MGMT expression with OS in TMZ-treated and untreated subgroups (Table 5, Fig. 3A and B). In the cohort of TMZ-treated patients, lack of MGMT protein expression was strongly associated with longer OS (HR of death 5.53, 95% CI 1.76–17.37; P = .003). In contrast, in patients who received no TMZ therapy, no differences in OS according to MGMT protein expression (HR of death 1.0, 95% CI 0.45–2.2; P = .99) were observed. A similar analysis using patient subgroups according to the MGMT promoter methylation status revealed no significance with OS in the TMZ-treated subgroup (Fig. 3C and D).
Table 3.
Univariate survival analyses
| Variable | HRa | 95% CI | P-value |
|---|---|---|---|
| Age | 1.07 | 1.03–1.10 | <.001 |
| Gender | 0.72 | 0.40–1.30 | .28 |
| Performance status | 0.96 | 0.94–0.98 | <.001 |
| TMZ therapy | 0.2 | 0.11–0.38 | <.001 |
| MGMT promoter methylation | 0.61 | 0.33–1.11 | .11 |
| MGMT protein expression | 2.02 | 1.22–3.66 | .02a |
Variables were coded as described in Patients and Methods.
aHazard ratio for death.
Table 4.
Multivariate survival analysis
| Promoter Methylation |
Protein Expression |
|||||
|---|---|---|---|---|---|---|
| HRa | 95% CI | P-value | HRa | 95% CI | P-value | |
| Variable without interaction term | ||||||
| Age | 1.05 | 1.02–1.08 | .003 | 1.05 | 1.02–1.08 | .003 |
| Gender | 0.79 | 0.42–1.49 | .47 | 0.92 | 0.49–1.74 | .81 |
| Performance status | 0.96 | 0.94–0.98 | .001 | 0.96 | 0.94–0.98 | .001 |
| TMZ therapy | 0.25 | 0.13–0.49 | <.001 | 0.25 | 0.13–0.48 | <.001 |
| MGMT promoter methylation | 0.6 | 0.32–1.14 | .12 | |||
| MGMT protein expression | 1.99 | 1.07–3.70 | .03 | |||
| Variable with interaction terms | ||||||
| Age | 1.05 | 1.02–1.08 | .003 | 1.05 | 1.02–1.09 | .001 |
| Gender | 0.8 | 0.42–1.53 | .5 | 0.99 | 0.52–1.85 | .99 |
| Performance status | 0.96 | 0.94–0.99 | .002 | 0.96 | 0.94–0.99 | .001 |
| TMZ therapy | 0.28 | 0.10–0.80 | .02 | 0.07 | 0.02–0.22 | <.001 |
| MGMT promoter methylation | 0.64 | 0.60–2.86 | .3 | |||
| MGMT protein expression | 0.94 | 0.43-2.04 | .88 | |||
| MGMT promoter methylation × TMZ therapy | .81 | |||||
| MGMT protein expression × TMZ therapy | .005 | |||||
Variables were coded as described in Patients and Methods.
aAdjusted hazard ratio for death.
Table 5.
Multivariate survival analysis in subgroups of patients regarding TMZ therapy
| TMZ Therapy |
No TMZ Therapy |
|||||
|---|---|---|---|---|---|---|
| Variable | HRa | 95% CI | P-value | HR | 95% CI | P-value |
| Age | 1.07 | 1.02–1.12 | .006 | 1.05 | 1.00–1.09 | .04 |
| Gender | 0.41 | 0.14–1.23 | .11 | 1.78 | 0.79–4.03 | .17 |
| Performance status | 0.95 | 0.92–0.98 | .01 | 0.97 | 0.94–1.00 | .02 |
| MGMT protein expression | 5.53 | 1.76–17.37 | .003 | 1.00 | 0.45–2.20 | .99 |
Variables were coded as described in Patients and Methods.
aAdjusted hazard ratio for death.
Fig. 3.
MGMT promoter methylation and protein expression when compared with overall survival (OS) of patients. Results of Kaplan–Meier analyses of OS according to MGMT protein expression (A and B) or MGMT promoter methylation (C and D) of GBM patients treated (A and C) and untreated (B and D) with TMZ are shown.
Discussion
In this study, we have investigated the relation between MGMT promoter methylation status and protein expression and the association of these parameters with TMZ therapy as well as OS of glioma patients. To focus exclusively on the malignant cell compartment, we analyzed MGMT expression in primary tumor cells explanted from the surgical specimens instead of tissue extracts or tumor sections. Using this new approach, we prove that lack of MGMT protein expression is a strong predictive indicator for TMZ treatment response and the related survival benefit. Despite a significant correlation between the two MGMT parameters, protein expression was of superior predictive value in our patient cohort. This indicates that reliable MGMT protein detection methods have to be developed to identify GBM patients likely to benefit from TMZ therapy.
The optimal method for MGMT status determination and its predictive quality on TMZ response in GBM patients is a matter of debate.17,22,24–28 Since the promising results of the translational sub-study20 to the multicenter EORTC trial,1 detection of MGMT promoter methylation has been in the focus of interest. Methylation analysis can be performed using small amounts of tumor-derived DNA, and MGMT promoter methylation is a specific characteristic of tumor cells never observed in normal cell compartments.23 Thus, contaminations with nonmalignant cell-derived DNA should not lead to false-positive results. Based on these benefits, the use of promoter methylation as a parameter for patient stratification for TMZ therapy was recently suggested.28 Although the data by Hegi et al.20 are in accordance with some other studies,31 a lack of a significant association between MGMT promoter methylation and TMZ response and/or OS in primary or recurrent GBM was reported.17,32–34 In case of astrocytoma grade II, MGMT promoter methylation was even suggested as a negative prognostic marker for progression-free survival.35 Interestingly, the majority of studies using chloroethylating nitrosoureas demonstrated a significant association between MGMT promoter methylation and therapy response.15,36,37 Accordingly, MGMT promoter methylation was predictive for response to Carmustine (BCNU), but not to TMZ + cisplatin, in GBM patients.38
The reasons for the inconsistent results with regard to the predictive quality of MGMT promoter methylation might be based on the higher patient number analyzed by Hegi et al.20 However, the impressive correlation between MGMT protein detection by Western blot and patient survival in the TMZ-treated patient group in our study suggests that it is not the patient number but the parameter per se that determines the predictive value. Unfortunately, no MGMT protein detection method was used by Hegi et al.,20 making it impossible to establish a comparable analysis as done in this study. Additionally, it has to be mentioned that promoter methylation analysis by MSP, especially from paraffin-embedded tissues, is technically demanding. This is also obvious from the distinct center dependency of methylation detection in the translational sub-study leading to successful determination of this parameter in 36% of patients only.20 Technical problems with MSP can be ruled out in our study for two reasons: (i) for detection of MGMT promoter methylation, we used tumor cell culture–derived DNA, thus avoiding problems resulting from tissue fixation and paraffin embedding; and (ii) in a subgroup of tumors, MSP was done comparably from snap-frozen tissue and tumor cells, confirming in all cases the presence of methylated MGMT promoter sequences. The latter observation also excludes possible changes in the MGMT promoter methylation status during short-term in vitro cultivation of tumor cells.
In the present study, the two MGMT parameters showed strong correlation, confirming that promoter methylation is a major factor regulating protein expression. Nevertheless, as obvious from the scattergrams (Fig. 1C) and depicted by selected cases (Fig. 2), there are several tumor cell samples not following this correlation. This indicates that in a subgroup of tumors with methylated MGMT promoter, the protein is still expressed and vice versa. Comparable observations have been reported consistently in repeated biopsies from inoperable GBM.26 High percentages of MGMT-negative tumor cells have also been detected by immunohistochemistry in tumors harboring only unmethylated DNA sequences.27,39 This might explain why the few studies addressing the correlation between MGMT promoter status and gene expression so far delivered contradictory results. Although some earlier reports found a good correlation,16,40,41 others described a rather loose association or no correlation at all.17,26,27,39,42 In agreement with our data, most of the studies suggested that these parameters cannot be used interchangeably for each other. Such observations might also explain why in the study of Hegi et al.20 TMZ treatment led to a borderline OS and significant progression-free survival benefit, even in the patient subgroup with unmethylated MGMT promoter.
The reasons for the discrepancy between promoter methylation and MGMT expression in a patient subgroup are unclear so far. Generally, epigenetic silencing via promoter methylation is an important but obviously not always decisive factor regulating MGMT expression in GBM cells. This assumption is supported by the data of Sasai et al.43 demonstrating activation of MGMT expression by demethylating agents at the mRNA but not protein level in transformed malignant astrocytes and GBM cells, suggesting posttranscriptional repression of MGMT expression. Additionally, a transcriptional activation of MGMT expression by wild-type p53 was reported in murine astrocytes and human gliomas also when harboring a methylated MGMT promoter.44,45 Accordingly, immunohistochemical staining of p53 and MGMT were mutually exclusive in astrocytoma sections.46
With regard to therapy response and OS, MGMT protein expression turned out to be of distinctly higher predictive power when compared with promoter methylation status in our patient samples. Accordingly, MGMT immunostaining, but not promoter methylation, was an independent prognostic factor with regard to OS of patients with anaplastic astrocytoma treated with TMZ.17 Predictive quality of MGMT protein expression with respect to TMZ response and/or disease progression was also reported in mixed gliomas,18 inoperable GBM,47 and recently in a small series of recurrent GBM by Western blot from tumor extracts.48 Accordingly, in a recent comparative analysis, MGMT immunostaining correlated with promoter methylation. However, only the protein levels correlated with MGMT activity in GBM tissues.27 Nevertheless, not all studies using MGMT immunostaining demonstrated correlations with TMZ response and/or patient survival.42 Additionally, several problems are associated with MGMT immunostaining, including tumor cell heterogeneity, infiltration of MGMT-positive inflammatory cells and microglia as well as induction of MGMT during therapy.28 All these factors restrain standardization and determination of cut-off levels.24,27,45 Accordingly, our attempts to analyze MGMT protein expression by immunohistochemistry in selected patient samples used in this study were inconclusive, and immunostaining neither correlated with MGMT promoter methylation nor MGMT expression detected by Western blot, even when using identical antibodies (data not shown). Nevertheless, our data based on tumor cell explants and Western blot analysis prove the high quality of MGMT protein expression as a predictive marker for TMZ response in GBM patients.
In summary, we used a completely new approach to analyze the impact of MGMT status on TMZ therapy response and survival of GBM patients. Determination of MGMT protein levels in GBM cell primo-cultures explanted from surgical specimens turned out to be a highly predictive factor for TMZ therapy-mediated survival benefit with superior information power as compared to the analysis of promoter methylation by MSP. Consequently, reliable and reproducible MGMT protein detection methods have to be developed and tested with regard to their predictive value for TMZ therapy response in prospective studies involving larger patient cohorts.
Acknowledgments
The authors thank Gabriele Reisinger and Kerstin Schauer for skillful technical assistance. This work was sponsored by the Forschungsförderungsfond der Österreichischen Krebshilfe Oberösterreich, the Initiative Krebsforschung of the Medical University Vienna, and the Herzfelderśche Familienstiftung.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.van den Bent MJ, Hegi ME, Stupp R. Recent developments in the use of chemotherapy in brain tumours. Eur J Cancer. 2006;42(5):582–588. doi: 10.1016/j.ejca.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 3.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4(4):296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 4.Kaina B, Christmann M, Naumann S, et al. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst). 2007;6(8):1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Reardon DA, Rich JN, Friedman HS, et al. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol. 2006;24(8):1253–1265. doi: 10.1200/JCO.2005.04.5302. [DOI] [PubMed] [Google Scholar]
- 6.Srivenugopal KS, Yuan XH, Friedman HS, et al. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chloroethyl)-1-nitrosourea. Biochemistry. 1996;35(4):1328–1334. doi: 10.1021/bi9518205. [DOI] [PubMed] [Google Scholar]
- 7.Glassner BJ, Weeda G, Allan JM, et al. DNA repair methyltransferase (MGMT) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis. 1999;14(3):339–347. doi: 10.1093/mutage/14.3.339. [DOI] [PubMed] [Google Scholar]
- 8.Hansen RJ, Nagasubramanian R, Delaney SM, et al. Role of O6-methylguanine-DNA methyltransferase in protecting from alkylating agent-induced toxicity and mutations in mice. Carcinogenesis. 2007;28(5):1111–1116. doi: 10.1093/carcin/bgl218. [DOI] [PubMed] [Google Scholar]
- 9.Bobola MS, Silber JR, Ellenbogen RG, et al. O6-Methylguanine-DNA methyltransferase, O6-benzylguanine, and resistance to clinical alkylators in pediatric primary brain tumor cell lines. Clin Cancer Res. 2005;11(7):2747–2755. doi: 10.1158/1078-0432.CCR-04-2045. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Gerson SL. Targeted modulation of MGMT: clinical implications. Clin Cancer Res. 2006;12(2):328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- 11.Wedge SR, Newlands ES. O6-Benzylguanine enhances the sensitivity of a glioma xenograft with low O6-alkylguanine-DNA alkyltransferase activity to temozolomide and BCNU. Br J Cancer. 1996;73(9):1049–1052. doi: 10.1038/bjc.1996.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman HS, Keir S, Pegg AE, et al. O6-Benzylguanine-mediated enhancement of chemotherapy. Mol Cancer Ther. 2002;1(11):943–948. [PubMed] [Google Scholar]
- 13.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J Clin Oncol. 2002;20(9):2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 14.Stupp R, Ruegg C. Integrin inhibitors reaching the clinic. J Clin Oncol. 2007;25(13):1637–1638. doi: 10.1200/JCO.2006.09.8376. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59(4):793–797. [PubMed] [Google Scholar]
- 17.Brell M, Tortosa A, Verger E, et al. Prognostic significance of O6-methylguanine-DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res. 2005;11(14):5167–5174. doi: 10.1158/1078-0432.CCR-05-0230. [DOI] [PubMed] [Google Scholar]
- 18.Friedman HS, McLendon RE, Kerby T, et al. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol. 1998;16(12):3851–3857. doi: 10.1200/JCO.1998.16.12.3851. [DOI] [PubMed] [Google Scholar]
- 19.Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 20.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 21.Idbaih A, Omuro A, Ducray F, et al. Molecular genetic markers as predictors of response to chemotherapy in gliomas. Curr Opin Oncol. 2007;19(6):606–611. doi: 10.1097/CCO.0b013e3282f075f3. [DOI] [PubMed] [Google Scholar]
- 22.Stupp R, Hegi ME, Gilbert MR, et al. Chemoradiotherapy in malignant glioma: standard of care and future directions. J Clin Oncol. 2007;25(26):4127–4136. doi: 10.1200/JCO.2007.11.8554. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23(1):1–8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- 24.Capper D, Mittelbronn M, Meyermann R, et al. Pitfalls in the assessment of MGMT expression and in its correlation with survival in diffuse astrocytomas: proposal of a feasible immunohistochemical approach. Acta Neuropathol (Berl) 2008;115(2):249–259. doi: 10.1007/s00401-007-0310-x. [DOI] [PubMed] [Google Scholar]
- 25.Friedman HS, Maxwell J. The fallacy of single-agent chemotherapy for cancer. J Clin Oncol. 2007;25(23):3550. doi: 10.1200/JCO.2007.12.3596. [DOI] [PubMed] [Google Scholar]
- 26.Grasbon-Frodl EM, Kreth FW, Ruiter M, et al. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J Cancer. 2007;121(11):2458–2464. doi: 10.1002/ijc.23020. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell JA, Johnson SP, Quinn JA, et al. Quantitative analysis of O6-alkylguanine-DNA alkyltransferase in malignant glioma. Mol Cancer Ther. 2006;5(10):2531–2539. doi: 10.1158/1535-7163.MCT-06-0106. [DOI] [PubMed] [Google Scholar]
- 28.Stupp R, Hegi ME. Methylguanine methyltransferase testing in glioblastoma: when and how? J Clin Oncol. 2007;25(12):1459–1460. doi: 10.1200/JCO.2006.09.7139. [DOI] [PubMed] [Google Scholar]
- 29.Spiegl-Kreinecker S, Pirker C, Marosi C, et al. Dynamics of chemosensitivity and chromosomal instability in recurrent glioblastoma. Br J Cancer. 2007;96(6):960–969. doi: 10.1038/sj.bjc.6603652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger W, Spiegl-Kreinecker S, Buchroithner J, et al. Overexpression of the human major vault protein in astrocytic brain tumor cells. Int J Cancer. 2001;94(3):377–382. doi: 10.1002/ijc.1486. [DOI] [PubMed] [Google Scholar]
- 31.Paz MF, Yaya-Tur R, Rojas-Marcos I, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10(15):4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 32.Blan JL, Wager M, Guilhot J, et al. Correlation of clinical features and methylation status of MGMT gene promoter in glioblastomas. J Neurooncol. 2004;68(3):275–283. doi: 10.1023/b:neon.0000033385.37098.85. [DOI] [PubMed] [Google Scholar]
- 33.Brandes AA, Tosoni A, Cavallo G, et al. Temozolomide 3 weeks on and 1 week off as first-line therapy for recurrent glioblastoma: phase II study from Gruppo Italiano Cooperativo di Neuro-Oncologia (GICNO) Br J Cancer. 2006;95(9):1155–1160. doi: 10.1038/sj.bjc.6603376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yachi K, Watanabe T, Ohta T, et al. Relevance of MSP assay for the detection of MGMT promoter hypermethylation in glioblastomas. Int J Oncol. 2008;33(3):469–475. [PubMed] [Google Scholar]
- 35.Komine C, Watanabe T, Katayama Y, et al. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression-free survival in patients with low-grade diffuse astrocytomas. Brain Pathol. 2003;13(2):176–184. doi: 10.1111/j.1750-3639.2003.tb00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamiryo T, Tada K, Shiraishi S, et al. Correlation between promoter hypermethylation of the O6-methylguanine-deoxyribonucleic acid methyltransferase gene and prognosis in patients with high-grade astrocytic tumors treated with surgery, radiotherapy, and 1-(4-amino-2-methyl-5-pyrimidinyl)methyl-3-(2-chloroethyl)-3-nitrosourea-based chemotherapy. Neurosurgery. 2004;54(2):349–357. doi: 10.1227/01.neu.0000103422.51382.99. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe T, Katayama Y, Komine C, et al. O6-Methylguanine-DNA methyltransferase methylation and TP53 mutation in malignant astrocytomas and their relationships with clinical course. Int J Cancer. 2005;113(4):581–587. doi: 10.1002/ijc.20625. [DOI] [PubMed] [Google Scholar]
- 38.Balana C, Ramirez JL, Taron M, et al. O6-Methyl-guanine-DNA methyltransferase methylation in serum and tumor DNA predicts response to 1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastoma multiforme. Clin Cancer Res. 2003;9(4):1461–1468. [PubMed] [Google Scholar]
- 39.Rood BR, Zhang H, Cogen PH. Intercellular heterogeneity of expression of the MGMT DNA repair gene in pediatric medulloblastoma. Neuro-Oncology. 2004;6(3):200–207. doi: 10.1215/S1152851703000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costello JF, Futscher BW, Tano K, et al. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269(25):17228–17237. [PubMed] [Google Scholar]
- 41.Pieper RO, Costello JF, Kroes RA, et al. Direct correlation between methylation status and expression of the human O6-methylguanine DNA methyltransferase gene. Cancer Commun. 1991;3(8):241–253. doi: 10.3727/095535491820873092. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez FJ, Thibodeau SN, Jenkins RB, et al. MGMT immunohistochemical expression and promoter methylation in human glioblastoma. Appl Immunohistochem Mol Morphol. 2008;16(1):59–65. doi: 10.1097/PAI.0b013e31802fac2f. [DOI] [PubMed] [Google Scholar]
- 43.Sasai K, Akagi T, Aoyanagi E, et al. O6-Methylguanine-DNA methyltransferase is downregulated in transformed astrocyte cells: implications for anti-glioma therapies. Mol Cancer. 2007;6:36. doi: 10.1186/1476-4598-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blough MD, Zlatescu MC, Cairncross JG. O6-Methylguanine-DNA methyltransferase regulation by p53 in astrocytic cells. Cancer Res. 2007;67(2):580–584. doi: 10.1158/0008-5472.CAN-06-2782. [DOI] [PubMed] [Google Scholar]
- 45.Srivenugopal KS, Shou J, Mullapudi SR, et al. Enforced expression of wild-type p53 curtails the transcription of the O6-methylguanine-DNA methyltransferase gene in human tumor cells and enhances their sensitivity to alkylating agents. Clin Cancer Res. 2001;7(5):1398–1409. [PubMed] [Google Scholar]
- 46.Yuan Q, Matsumoto K, Nakabeppu Y, et al. A comparative immunohistochemistry of O6-methylguanine-DNA methyltransferase and p53 in diffusely infiltrating astrocytomas. Neuropathology. 2003;23(3):203–209. doi: 10.1046/j.1440-1789.2003.00504.x. [DOI] [PubMed] [Google Scholar]
- 47.Chinot OL, Barrie M, Fuentes S, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J Clin Oncol. 2007;25(12):1470–1475. doi: 10.1200/JCO.2006.07.4807. [DOI] [PubMed] [Google Scholar]
- 48.Nagane M, Kobayashi K, Ohnishi A, et al. Prognostic significance of O6-methylguanine-DNA methyltransferase protein expression in patients with recurrent glioblastoma treated with temozolomide. Jpn J Clin Oncol. 2007;37(12):897–906. doi: 10.1093/jjco/hym132. [DOI] [PubMed] [Google Scholar]