Abstract
Hypermethylation of the O6-methylguanine-DNA-methyltransferase (MGMT) gene has been shown to be associated with improved outcome in glioblastoma (GBM) and may be a predictive marker of sensitivity to alkylating agents. However, the predictive utility of this marker has not been rigorously tested with regard to sensitivity to other therapies, namely radiation. To address this issue, we assessed MGMT methylation status in a cohort of patients with GBM who underwent radiation treatment but did not receive chemotherapy as a component of adjuvant treatment. Formalin-fixed, paraffin-embedded tumor samples from 225 patients with newly diagnosed GBM were analyzed via methylation-specific, quantitative real-time polymerase chain reaction following bisulfite treatment on isolated DNA to assess MGMT promoter methylation status. In patients who received radiotherapy alone following resection, methylation of the MGMT promoter correlated with an improved response to radiotherapy. Unmethylated tumors were twice as likely to progress during radiation treatment. The median time interval between resection and tumor progression of unmethylated tumors was also nearly half that of methylated tumors. Promoter methylation was also found to confer improved overall survival in patients who did not receive adjuvant alkylating chemotherapy. Multivariable analysis demonstrated that methylation status was independent of age, Karnofsky performance score, and extent of resection as a predictor of time to progression and overall survival. Our data suggest that MGMT promoter methylation appears to be a predictive biomarker of radiation response. Since this biomarker has also been shown to predict response to alkylating agents, perhaps MGMT promoter methylation represents a general, favorable prognostic factor in GBM.
Keywords: glioblastoma, methylation, MGMT, prognostic marker, radiotherapy
Introduction
The O6-methylguanine-DNA-methyltransferase (MGMT) gene encodes for an important DNA repair protein which acts by removing alkyl products from the O6 position on guanine. A so-called “suicide enzyme,” following removal of the alkyl groups, the newly alkylated MGMT protein, is then marked for degradation by ubiquitinization.1,2 Proper functioning of the gene is important for maintaining cell integrity. Epigenetic silencing of the MGMT gene by methylation of the CpG islands of the promoter region has been shown to correlate with loss of gene transcription and protein expression.1 Loss of expression of the MGMT protein results in decreased DNA repair and retention of alkyl groups, thereby allowing alkylating agents such as carmustine (BCNU), lomustine (CCNU), and temozolomide to have greater efficacy in patients whose tumors exhibit hypermethylation of the MGMT promoter and reducing the MGMT protein concentration.3–7 Although MGMT protein expression is expressed in a wide variety of tumors including colon, head and neck, and lung cancers, infiltrative gliomas remain one of the most intriguing and potentially informative tumor groups to study the impact of MGMT expression.8
Glioblastomas (GBMs) are classified as grade IV/IV gliomas by the World Health Organization (WHO) and portend a dismal prognosis with most patients surviving only 1–2 years despite aggressive treatment.9 Although age, extent of resection, and performance status remain the most reliable prognostic markers in patient survival, the search continues for a marker that will predict outcome and allow for a more effective, tailor-made treatment regimen. MGMT expression has recently gained interest as a potential predictive marker for response to chemotherapy, particularly alkylating agents like temozolomide.
The current standard treatment of newly diagnosed GBM is based on a phase III trial conducted by the European Organisation for Research and Treatment of Cancer (EORTC), which compared radiation alone vs radiation plus concurrent and adjuvant temozolomide. This study showed a significant improvement in survival in the temozolomide arm, with a 2-year survival rate of 26.5%, compared with 10.4% in the radiation-only arm.10 Since then, the use of concurrent radiation and temozolomide followed by adjuvant temozolomide has become standard. Because of the in vitro observations (described above) during the preclinical development of temozolomide, as well as prior clinical data linking MGMT promoter methylation with sensitivity to alkylating agents,3 a subset of the cases from the EORTC trial were tested for MGMT methylation status.4 The authors concluded that MGMT promoter methylation conferred a survival advantage only in patients who received temozolomide. In other words, the methylated promoter was a predictive marker of response to temozolomide. This conclusion was driven by the fact that there was a statistically significant improvement in the survival time of patients who had promoter methylation and received temozolomide, although the benefit observed from the MGMT promoter methylation in the patients of the control arm (radiotherapy only) was not statistically significant (P = .06). It should be noted that this was performed on a subset (n = 206) of the original patient population of the EORTC trial (n = 573). Further, although not statistically significant, inspection of the survival curves reveal a trend toward improved survival in patients with unmethylated tumors who received temozolomide vs those who did not. This is most evident at the 2-year survival point, where those in the unmethylated tumor group who received temozolomide had an approximate 10%–15% actuarial overall survival time compared with 0% in those who did not receive temozolomide. Lastly, the time to progression of patients in the control arm (radiation therapy alone) appeared to be more favorable in the patients whose tumors had MGMT promoter methylation, suggesting that this biomarker is associated with improved radiation response; that has relevance to the interpretation of these data, as radiation response has been shown to be a strong predictor of improved overall survival time in patients with GBM.11
To more rigorously test whether MGMT promoter methylation was a predictive marker of chemo-sensitivity alone, or represented a more general prognostic marker that predicted for responsiveness to different modalities of therapy, namely radiotherapy, we determined the methylation status of 225 tumor specimens from patients treated prior to the establishment of concurrent and adjuvant temozolomide as the standard of care and analyzed for its association with clinical outcomes. Our hypothesis was that if MGMT promoter methylation is only a predictive biomarker of response to akylating agents (namely temozolomide), then it should not have a prognostic effect on patients receiving radiation therapy alone. The null hypothesis then would be that MGMT methylation predicts response to radiation as well as alkylating agents and may represent a general prognostic biomarker of outcome in GBM, regardless of treatment.
Materials and Methods
Patient Population
Patients with tissue confirmed diagnosis of GBM (WHO grade IV) were selected from The University of Texas M. D. Anderson Cancer Center Neuropathology Tissue Bank. All patients with tissue sufficient for MGMT promoter methylation assessment were considered evaluable. All samples were from patients with newly diagnosed GBM who had not received prior treatment. A retrospective analysis of the patient charts was performed to collect patient data such as demographics, extent of surgical resection, treatment modalities, time to progression, and overall survival. We chose to focus our evaluation on patients who were treated prior to the adaptation of concurrent/adjuvant temozolomide as standard therapy. Two hundred and twenty-five cases with sufficient tissue for molecular analysis were identified. In addition to external beam radiation therapy, 53 patients received adjuvant chemotherapy and the remaining 172 patients did not receive any chemotherapeutic agents until after the first tumor recurrence, permitting the determination of time to progression and overall survival in the absence of concurrent/adjuvant alkylating chemotherapy in this latter group. For the analysis of radiation response, 183 cases were identified as having (1) no adjuvant therapy prior to the assessment of radiation response, and (2) pre- and postradiotherapy magnetic resonance imaging (MRI) studies available for assessment and comparison.
DNA Extraction/Bisulfite Treatment
Routinely processed formalin-fixed, paraffin-embedded GBM samples were selected from the 225 cases. The hematoxylin and eosin-stained slides were reviewed by a neuropathologist, and appropriate blocks were selected for tumor. Following deparaffinization, DNA extraction was performed using the Epicentre MasterPure Complete DNA Purification Kit (Epicentre Biotechnologies, Madison, WI). Bisulfite treatment was then performed on the extracted DNA via the Zymo Research EZ DNA Methylation-Gold Kit (Cat. #D5005/D5006) to convert unmethylated cytosine to uracil. Up to 2000 ng of DNA per sample were bisulfite treated to obtain adequate converted DNA for quantitative real-time polymerase chain reaction (qRT-PCR).
Methylation-specific qRT-PCR and Determination of MGMT Promoter Methylation
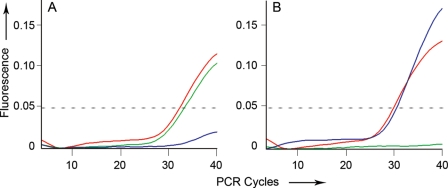
qRT-PCR was performed using the eluted bisulfite-treated DNA. PCR reactions were set at 20 µL volumes using up to 5 µL of bisulfite treated DNA, methylation-specific primers and probes, and 1× TaqMan Universal PCR Master Mix without AmpErase. Primers and probes used to detect methylated and unmethylated MGMT sequences are shown in Table 1. The qRT-PCR was performed using a Chromo4TM Real-Time PCR Detector from Bio-Rad (Hercules, CA) under the following conditions: 10 minutes at 95°C, then 40 cycles of 95°C for 15 seconds, 55°C for 15 seconds, and 60°C for 45 seconds. Following methylation-specific qRT-PCR, the resultant curves were reviewed to assess for MGMT promoter methylation by analyzing the amplification of both the methylated and unmethylated sequences. Amplification of the methylated MGMT promoter sequence was considered to represent MGMT promoter methylation. In cases with concurrent amplification of both the methylated and unmethylated sequences, the samples were interpreted as methylated allowing for DNA extraction from adjacent nontumoral tissue or tumor heterogeneity. This was systematically achieved by setting the threshold level of fluorescence detection of FAM at 0.05 or higher after at least 40 PCR cycles, based on qRT-PCR curves. Examples of the quantitative methylation curves (with this cut-off line) are shown in Fig. 1.
Table 1.
Sequences used in the qRT-PCR analysis
| MGMT-methylated |
| Forward 5′-GCGTTTCGACGTTCGTAGGT-3′ |
| Reverse 5′-CACTCTTCCGAAAACGAAACG-3′ |
| Probe 5′-CGCAAACGATACGCACCGCGA-3′ |
| MGMT-unmethylated |
| Forward 5′-TGTGTTTTGGATATGTTGGGATAGT-3′ |
| Reverse 5′-AACTCCACACTCTTCCAAAAACAA-3′ |
| Probe 5′-TTTTTGTGGTGTGTATTGTT-3′ |
| COL2A1 |
| Forward 5′-TCTAACAATTATAAACTCCAACCACCAA-3′ |
| Reverse 5′-GGGAAGATGGGATAGAAGGGAATAT-3′ |
| Probe 5′-CCTTCATTCTAACCCAATACCTATCCCACCTCTAAA-3′ |
Fig. 1.
MGMT qRT-PCR curves. qRT-PCR shows amplification of methylated MGMT promoter (green curve), unmethylated MGMT promoter (blue curve), and a COL2A1 control (red curve). (A) Example of a tumor with MGMT promoter methylation; (B) Example of a tumor without MGMT promoter methylation.
Endpoint Definitions and Statistical Analysis
To radiographically assess the response of tumors to radiotherapy, the preirradiation MRI was compared with the post-treatment MRI. Postirradiation MRIs were obtained between 4 and 10 weeks after the completion of radiotherapy. Patients with a ≥25% increase in the cross-sectional area of the enhancing component of a subtotally resected tumor were considered to have progressed as described previously.12 This radiation response scoring system has been shown to be highly correlated with overall survival in an independent patient series.13 Patients who underwent a gross total resection and had re-appearance of frank enhancing tumor (not the typical postradiation rim enhancement of the cavity) were considered to have progressed. The time to progression is defined as from the date of the diagnostic procedure (biopsy or tumor resection) until the date of first evidence of radiographic progression. Patients were censored for time to progression if there was no documented progression at the last imaging study. The overall survival time was defined as from the date of diagnosis to the date of death. Patients who were alive at last follow-up were censored. The univariable time to progression and survival time comparisons between groups were determined using the log-rank method. Multivariable analyses were performed using Cox regression models. Comparisons of characteristic distributions between groups were performed using a Fisher's exact test and all P values were 2-sided.
Results
Patient Characteristics
The demographic information of this patient set is summarized on Table 2. The median age was 58.1 years. The distribution of the RTOG recursive partition analysis classes14,15 were 16%, 68%, and 16% for classes III, IV, and V + VI, respectively. Fifty-four patients (24%) had methylation of the MGMT promoter. The distribution of the RPA classes among the unmethylated and methylated tumors was essentially identical to that of the general population (with 19%, 62%, and 18% for classes III, IV, and V + VI in the unmethylated group and 13%, 68%, and 21% for classes III, IV, and V + VI in the methylated group). The median survival for the whole group was 50.4 weeks.
Table 2.
Patient demographic information
| Characteristic | N (%) |
|---|---|
| Age (yr) | |
| < 50 | 61 (27) |
| ≥ 50 | 164 (73) |
| KPSa | |
| 90–100 | 120 (53) |
| 70–80 | 94 (42) |
| < 70 | 11 (5) |
| Extent of resection | |
| GTRb | 111 (49) |
| STR/bxc | 114 (51) |
| RPAd | |
| III | 35 (16) |
| IV | 154 (68) |
| V | 36 (16) |
| MGMT promoter status | |
| Unmethylated | 171 (76) |
| Methylated | 54 (24) |
aKarnofsky performance score.
bGross-total resection.
cSubtotal resection/biopsy.
dRTOG recursive partitioning analysis.
Impact of MGMT Promoter Methylation on Radiation Response
Of the 225 cases, 183 had sufficient clinical and radiographic information to assess the response to radiotherapy. There were 138 unmethylated and 45 methylated cases in this cohort. The proportion of patients with a GBM that harbored an unmethylated MGMT promoter had a tumor progression rate that was twice that seen in unmethylated cases (58% vs 29%, respectively, P < .001, Fisher's exact test [Table 3]).
Table 3.
Radiation response of 183 patients receiving radiation alone after resection of GBM stratified by MGMT promoter methylation status
| MGMT status | Progression on post-XRT scan | Stable/response on post-XRT scan | Total |
|---|---|---|---|
| Unmethylated MGMT promoter | 80 (58%)a | 58 (42%) | 138 |
| Methylated MGMT promoter | 13 (29%)a | 32 (71%) | 45 |
a P = .001, Fisher's exact.
The Impact of MGMT Promoter Methylation on Time to Progression and Overall Survival
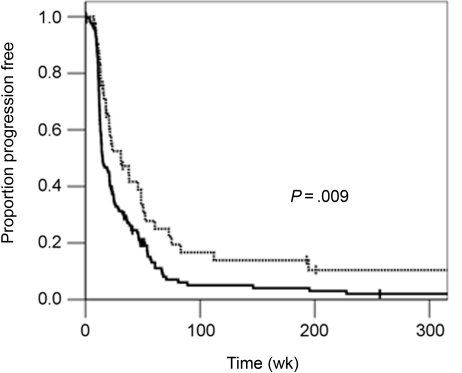
There were 172 cases that did not receive any adjuvant alkylating agents. However, they may have received these agents for salvage after tumor recurrence. This allowed for the MGMT promoter methylation status to be evaluated with respect to time to progression without the confounding effect of alkylating agent administration. Figure 2 demonstrates an improved time to progression when the MGMT promoter is methylated. The median progression time of the methylated group is twice that of the unmethylated group (31 vs 15 weeks, P = .009, log-rank). Multivariable analysis reveals that the MGMT promoter methylation status is an independent factor in determining time to progression when accounting for known risk factors such as age, performance, and extent of tumor resection (Table 4).
Fig. 2.
Progression-free survival and MGMT promoter methylation status. Kaplan–Meier curves showing progression-free survival for patients with methylated (dotted line) vs unmethylated (solid line) tumors.
Table 4.
Cox regression model for time to progression and overall survival time in 172 GBM patients not receiving adjuvant/concurrent alkylating chemotherapy
| Time to progression |
Overall survival |
|||
|---|---|---|---|---|
| Variable | P-value | HR | P-value | HR |
| Agea | .040 | 1.2 | <.001 | 1.3 |
| MGMT | .017 | 1.6 | .023 | 1.6 |
| KPSb | .012 | 0.3 | .001 | 0.7 |
| STR/bx vs GTRc | .006 | 1.6 | Not significant | |
aCoded as decade.
bKarnofsky performance score (score divided by 10 for this analysis).
cSubtotal resection/biopsy vs gross-total resection.
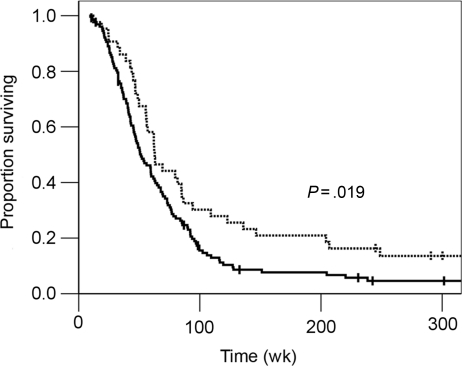
Figure 3 shows that there is a survival benefit in patients with tumors with MGMT promoter methylation. The median overall survival time was 63 weeks in the MGMT methylated vs 51 weeks in the unmethylated tumors (P = .019, log-rank). The actuarial 2-year overall survival rate was nearly double when the tumor MGMT promoter was methylated compared with the MGMT unmethylated group (30% vs 16%). Multivariable analysis reveals the MGMT promoter methylation status to be an independent prognostic factor when accounting for age, performance status, and extent of resection (Table 4).
Fig. 3.
Overall survival and MGMT promoter methylation status. Kaplan–Meier curves showing overall survival for patients with methylated (dotted line) vs unmethylated (solid line) tumors.
Discussion
Our data suggest that MGMT promoter methylation is a prognostic biomarker in GBM even in the absence of chemotherapy with alkylating agents. MGMT promoter methylation, therefore, may not only predict an improved response to temozolomide, but may represent a surrogate marker of a more treatment-responsive tumor in general. We have shown that there was an approximately 50% reduction in the rate of tumor progression during radiation therapy in methylated tumors vs those that were unmethylated. We also demonstrated that methylation of the MGMT promoter resulted in a nearly 2-fold delay in tumor progression compared with tumors with unmethylated MGMT promoter, even in the absence of the use alkylating agent chemotherapy.
A prior report examined a cohort of 219 GBM patients and explored the prognostic impact of MGMT promoter methylation in a variety of treatment settings.16 This study concluded that MGMT promoter methylation was not prognostic in patients who received radiation only; that the benefit was seen only in patients who received adjuvant temozolomide; and that this advantage was most pronounced when temozolomide was given concurrently with radiation. Although these data are somewhat at odds with ours, we note that 85 cases in this study met the same criteria of the 172 cases that were used in our primary progression time and overall survival analyses. Our greater numbers may have increased the likelihood of finding a significant relationship. Further, there are no radiotherapy-alone response data in this study. Analysis of clinical radiation response provides the clearest measurement of the impact of MGMT promoter methylation status on response to other modalities, as the clinical courses (such as salvage therapies) are highly variable after definitive local therapy. Lastly, the manner in which the methylation status was determined is different between these two studies. The differences in techniques may also account for the differences between the two studies.
MGMT promoter methylation was found to be an independent prognostic factor for progression and overall survival after accounting for other known risk factors. Promoter methylation may predict a better response to any form of therapy, including radiation therapy, as we have shown in this study. Although the mechanism for temozolomide resistance in unmethylated tumors is appealing and intuitive, the mechanism of radiation resistance is less clear. It is possible that MGMT promoter methylation is a surrogate marker for other, yet to be delineated processes that contribute to the overall aggressive biology of these tumors. Nonetheless, because of our observations, and the results of the subset analysis of the EORTC trial, it is reasonable to conclude that the MGMT promoter methylation status is an important prognostic factor in GBM that is on par with age and performance status. Though this requires validation in a prospective setting, it should also be considered, like age and performance status, part of patient stratification and randomization in future GBM trials.
Funding
Supported by a K-12 grant (KL2 RR024149) and an Institutional Research Grant to C.E.P. and grants from the V Foundation, Rose Foundation, Brain Tumor Funders' Collaborative and NIH/NCI (P50CA127001) to K.D.A.
Acknowledgments
These data were presented at the 50th Annual Meeting of the American Society for Therapeutic Radiology and Oncology (ASTRO) and the 13th Annual Scientific Meeting of the Society for Neuro-Oncology.
Conflict of interest statement. K.D.A. and H.C. are consultants for Castle Biosciences. This work was not supported by Castle Biosciences. None of the other authors have a conflict of interest to declare.
References
- 1.Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1–8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- 2.Srivenugopal KS, Yuan XH, Friedman HS, Ali-Osman F. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chloroethyl)-1-nitrosourea. Biochemistry. 1996;35:1328–1334. doi: 10.1021/bi9518205. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Paz MF, Yaya-Tur R, Rojas-Marcos I, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 6.Belanich M, Pastor M, Randall T, et al. Retrospective study of the correlation between the DNA repair protein alkyltransferase and survival of brain tumor patients treated with carmustine. Cancer Res. 1996;56:783–788. [PubMed] [Google Scholar]
- 7.Idbaih A, Omuro A, Ducray F, Hoang-Xuan K. Molecular genetic markers as predictors of response to chemotherapy in gliomas. Curr Opin Oncol. 2007;19:606–611. doi: 10.1097/CCO.0b013e3282f075f3. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 9.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. Lyon, France: WHO Press (IARC); 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 11.Barker FG, 2nd, Prados MD, Chang SM, et al. Radiation response and survival time in patients with glioblastoma multiforme. J Neurosurg. 1996;84:442–448. doi: 10.3171/jns.1996.84.3.0442. [DOI] [PubMed] [Google Scholar]
- 12.Barker FG, 2nd, Chang SM, Larson DA, et al. Age and radiation response in glioblastoma multiforme. Neurosurgery. 2001;49:1288–1297. doi: 10.1097/00006123-200112000-00002. discussion 1297–1288. [DOI] [PubMed] [Google Scholar]
- 13.Pelloski CE, Lin E, Zhang L, et al. Prognostic associations of activated mitogen-activated protein kinase and Akt pathways in glioblastoma. Clin Cancer Res. 2006;12:3935–3941. doi: 10.1158/1078-0432.CCR-05-2202. [DOI] [PubMed] [Google Scholar]
- 14.Shaw EG, Seiferheld W, Scott C, et al. Reexamining the radiation therapy oncology group (RTOG) recursive partitioning analysis (RPA) for glioblastoma multiforme (GBM) patients. Int J Radiat Oncol Biol Phys. 2003;57:S135–S136. [Google Scholar]
- 15.Scott CB, Scarantino C, Urtasun R, et al. Validation and predictive power of Radiation Therapy Oncology Group (RTOG) recursive partitioning analysis classes for malignant glioma patients: a report using RTOG 90-06. Int J Radiat Oncol Biol Phys. 1998;40:51–55. doi: 10.1016/s0360-3016(97)00485-9. [DOI] [PubMed] [Google Scholar]
- 16.Criniere E, Kaloshi G, Laigle-Donadey F, et al. MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol. 2007;83:173–179. doi: 10.1007/s11060-006-9320-0. [DOI] [PubMed] [Google Scholar]