Abstract
The purpose of this study was to determine whether chromosome 10q loss is a predictor of tumor aggressiveness and poor clinical outcome in patients with oligodendroglial tumors alone or together with loss of heterozygosity (LOH) on chromosomes 1p and 19q. A microsatellite analysis was performed on sections from 130 patients with grade II and grade III oligodendroglial tumors to assess the allelic status of chromosomes 1p, 19q, and 10q, plus detailed clinical and radiological information was taken prospectively. Median age at diagnosis was 45.5 years. Seventy-eight patients had disease progression after initial therapy; median progression-free survival (PFS) was 27.5 months. Age <47 years, postoperative Karnofsky performance score >65, no contrast enhancement on MRI, grade II, and complete removal on surgery were significantly correlated with a better PFS. Median overall survival (OS) was 40.5 months. Pure oligodendroglioma and temozolomide chemotherapy were correlated with better OS. 10q LOH was correlated with anaplastic grade and 1p19q LOH correlated with pure oligodendroglioma. There was a significant association between LOH status and the tumors' response to chemotherapy: 92.3% with 1p19q LOH, 83.3% without allelic losses, 50% with 1p19q10q LOH, and 14.5% with 10q LOH. Patients with 10q LOH alone had PFS of 6 months and a 3-year survival rate of 1%, when compared with 36 months and 85%, respectively, in patients with 1p19q LOH but without 10q LOH. 1p loss was correlated with better PFS (P < .005) and OS (P = .0007), whereas 10q loss was correlated with decreased PFS (P < .0001) and OS (P < .0001). 10q LOH predicted a survival disadvantage in patients with oligodendroglial tumors irrespective of 1p/19q LOH status.
Keywords: chemosensitivity, gliomas, loss of heterozygosity, prognosis, prospective study.
Gliomas are a part of a wide and heterogeneous tumor type that accounts for approximately half of the new cases of primary brain tumors diagnosed annually in adults. At the present time, a histological classification system is the standard for determining glioma prognosis and is still essential before administering therapies.1 The most frequently used classification for pathological diagnosis is the WHO classification.2 However, there are significant interobserver disparities in these classifications and gradings.3
Gliomas are a heterogeneous entity. The clinical course and response to treatment are highly variable. The five-year survival rates for gliomas are reported to range from 2% to 75%.4,5 On average, prognosis is largely influenced by the histological classification, but patients' variability within each grade and type remains clinically significant. It is also likely that variability in neuropathologists' assessments accounts for some of this within-grade and within-type variability.
This within-group variability has prompted searches for additional prognostic factors, such as other clinical and molecular prognostic markers.6 Even for the same histological type, gliomas show a variety of molecular genetic alterations.7,8 It is reported that genetic profiles may help classify gliomas according to their response to chemotherapy and their risk of recurrence.9, 10 This new knowledge adds to the complexity of gliomas and prompts the need for developing novel treatment strategies. However, these genetic alterations are not found in all gliomas and are not strictly correlated with tumor type. Hence, despite its importance being questioned, the relatively easy to perform and inexpensive histopathological diagnosis remains widely used.
This clinical, histopathological, biomolecular, and genetic variability is challenging. The decision making and prognosis for an individual patient is difficult in the absence of a sensitive and specific predictive model. Therefore, further research is needed. The discovery of new predictors, even if applicable only to a subset of patients, will help decision making. It is more likely that they will be part of more complex multivariate models, combining and investigating the correlation between clinical, histopathological, biomolecular, and genetic variables. The hope is that these multivariate models will lead ultimately to better classifications, decision trees and patient care.
Various molecular techniques, including mutation analysis, allelotyping, in situ hybridization, comparative genomic hybridization, and gene-expression profiling have been used to study gliomas.11 These studies have shown that gliomas result from the accumulation of several distinct chromosomal alterations such as loss of heterozygosity (LOH) on the short arm of chromosome 1 (1p), the long arm of chromosome 19 (19q), and the long arm of chromosome 10 (10q). These acquired genomic alterations in tumor cells have an important role in formulating the prognosis of patients with gliomas, in addition to the histological classification, because some correlate with the clinical outcome. Established indicators of the favorable outcome of oligodendroglial tumors include LOH on chromosomes 1p and 19q, which may indicate a loss of function of as yet unknown tumor-suppressor genes contained in those regions. Such alterations are strongly associated with better response to nonsurgical treatments and longer survival times, and continue to provide important prognostic information.12–15 Conversely, deletion of chromosome 10q has been shown to correlate with an aggressive behavior in gliomas, although this chromosomal alteration is less frequently found in oligodendroglial tumors than in astrocytomas.16,17 The presence of LOH on 1p and 19q does not ensure a completely accurate prognosis grouping, because all patients reported with this codeletion do not have better response to nonsurgical treatments and longer survival times. This observation is critical for the precise prognosis of an individual patient. The refinement of prognosis with the use of the analysis of LOH on 10q in patients with or without 1p19q codeletion provides the opportunity for a clinical trial that would evaluate the benefit of the identification of a subgroup of patients whose gliomas are estimated to be at high risk for recurrence.
Thus, in this study, we assessed the allelic status of chromosomes 1p, 19q, and 10q in a cohort of patients with oligodendrogliomas and mixed oligoastrocytomas by microsatellite analysis on formalin-fixed paraffin-embedded sections. We correlated the molecular results with the clinical data, especially to determine the impact of each genotype on survival and the response to chemotherapy and/or radiotherapy. To this end, we wished to investigate whether chromosome 10q loss is a predictor of tumor aggressiveness and poor clinical outcome in patients with oligodendroglial tumors; to use it to predict the individual risk of disease progression; and to improve the selection of appropriate patients individually, for managing new strategies and choices of treatments. Provision of optimal care for an individual patient requires prudence to achieve the best overall survival (OS) and quality of life.
Patients and Methods
Clinical Data
This work is based on the prospective analysis of clinical and molecular information on patients treated in the Neurosurgery Department of Lille University Medical Center for primary brain glioma, from January 2003 to December 2005. Data were collected from patients aged 18 or older who presented for initial consultation or routine follow-up. The selection of the patients was limited by the pathological diagnosis of a low-grade or an anaplastic cerebral supratentorial glioma—grade II and grade III, according to the WHO classification—including oligodendroglioma and oligoastrocytoma, and the availability of paired blood and tumor tissue for molecular analysis. Patients were excluded from the analysis if they had associated cancer, familial history of gliomas, or had received preoperative radiation therapy or chemotherapy.
Patients had detailed clinical information taken at diagnosis and during follow-up. The following parameters were determined: gender, first symptom, time interval between the time of appearance of the first symptoms and the date of diagnosis, initial tumor location, tumor volume, MRI contrast enhancement, time interval between the time of diagnosis and the date of the initial surgery, type of surgery treatment (biopsies, partial resection, complete resection), pre- and postoperative Karnofsky performance score (KPS), type of adjuvant treatment (radiotherapy, type of chemotherapy), radiological response to adjuvant treatment, time of the recurrent tumor growth (progression-free survival [PFS]), and patient status at the last follow-up (dead or alive). Postoperative imaging studies, all done within the 3 days after surgery, were compared with MRIs that were available during the follow-up of the patient. The radiological response to radiotherapy or chemotherapy was evaluated over at least 3 cycles. A 25% change in volume was necessary for the recognition of progression or regression. Follow-up findings were confirmed in all patients as of January 2008.
There were no missing data. No patient was lost to follow-up.
Neuropathology
Immediately after surgical removal, tumor samples were placed in tissue-culture media and shipped to a central reference laboratory for studies of tumor biology. Surgical specimens were saved as formalin-fixed and paraffin-embedded for histopathological examination. Samples of tumors were diagnosed by a neuropathologist and were graded according to WHO classification. Paraffin-embedded sections from tumors that were composed of at least 90% tumor cells were selected for the molecular analysis.
Molecular Analysis
Tumor DNA was isolated from 30-µm paraffin-embedded sections by overnight proteinase K digestion following xylene/ethanol deparaffinization and extracted using the QIAamp DNA Mini® kit (Qiagen) or NucleoSpin Tissue kit (Macherey-Nagel) according to the manufacturer's instructions. Corresponding constitutional DNA was extracted from peripheral blood leukocyte with the use of the QIAmp DNA Blood Maxi kit (Qiagen).
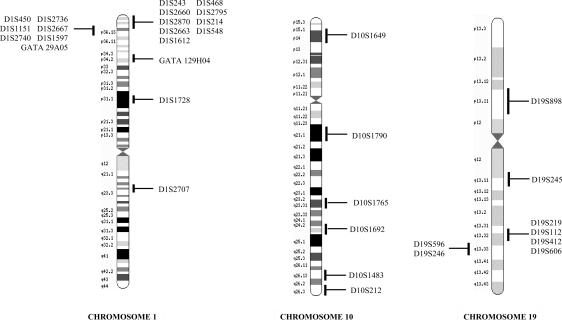
LOH on chromosomes 1p, 19q, and 10q was detected by microsatellite analysis on blood and tumor DNA, respectively, using the following 20 polymorphic markers: D1S468, D1S214, D1S1612, D1S2736, D1S1597 (located on 1p36.32–36.21); GATA129H04 (located on 1p34.2); D1S1728 (located on 1p31.1); D19S245, D19S178, D19S112, D19S412, D19S606, D19S596, D19S246 (located on 19q13.11–13.33); D10S1649 (located on 10p14); D10S1790 (located on 10q21.1); D10S1765 (located on 10q23.31); D10S1692 (located on 10q24.32); and D10S1483, D10S212 (located on 10q26.13–26.3). These microsatellite markers span the regions of chromosomes 1p and 19q that are commonly lost in oligodendroglial tumors—between 1p36.32 and 1p31.1 on chromosome 1p and between 19q13.11 and 19q13.13 on chromosome 19q—or on the entire long arm of chromosome 10q near the genes of interest (Fig. 1 and Table 1). The primer sequences of all markers were obtained from the Genome Database (http://www.gdb.org). The order of microsatellite markers on the chromosomes was according to relevant data on the Web sites at Ensemble (http://ensembl.org) and at GeneLoc (http://genecards.org). Microsatellite markers were selected based on amplicon size and heterozygosity score. One of each specific primer pairs was labelled using 3 different fluorochromes: 6-FAM (blue), NED (yellow), and HEX (green) (Applied Biosystems) for use in a single-run analysis. Six multiplex PCR amplifications were performed using Qiagen Multiplex PCR kit (Qiagen). The reaction mixture (50 µL) contained 25 µL of 2× Multiplex Master Mix, 5 µL of each sense and antisense primer (2 pmol/μL), and 1 µL genomic DNA (100 ng/μL) or 5 µL tumor DNA. Multiplex PCR A consisted of the microsatellite markers D1S468, D1S214, D1S1612, D1S2736; multiplex PCR B consisted of GATA129H04, D1S1728, D19S245, D19S112; multiplex PCR C consisted of D19S606, D19S596, D19S246; multiplex PCR D consisted of D19S178, D19S412, D1S1597; multiplex PCR E consisted of D10S212, D10S1692, D10S1765; and multiplex PCR F consisted of D10S1649, D10S1790, and D10S1483. PCR cycles were performed using a GeneAmp® PCR system 9700 (Applied Biosystems) as follows: initial denaturation step at 95°C for 10 minutes, followed by 5 cycles with denaturation at 95°C for 20 seconds, annealing at 60°C for 1 minute 30 seconds with −1°C at each cycle, and extension at 72°C for 1 minute and by 30 cycles with denaturation at 95°C for 20 seconds, annealing at 55°C for 1 minute 30 seconds, and extension at 72°C for 1 minute. The final extension step was for 30 minutes at 60°C. PCR products were run on an automatic sequencer ABI prism Model 377 XL® or 3100-Avent Genetic Analyzer (Applied Biosystems). Data were collected automatically during electrophoresis and then analysed with the Gene Scan software (Applied Biosystems).
Fig. 1.
Chromosomal location of the 33 polymorphic markers used.
Table 1.
Data on polymorphic microsatellite markers used
| Markers | Chromosomal location | Chromosomal location (base pair) | Percentage of heterozygosity | Allele size |
|---|---|---|---|---|
| Chromosome 1 | ||||
| D1S243 | p36.33 | 2129134–2129295 | 86 | 142–170 |
| D1S468 | p36.32 | 350819–3708342 | 75 | 173–191 |
| D1S2660 | p36.32 | 4704510–4704770 | 78 | 253–261 |
| D1S2795 | p36.31 | 5499155–5499378 | 75 | 214–224 |
| D1S2870 | p36.31 | 6212351–6212560 | 75 | 190–212 |
| D1S214 | p36.31 | 6884605–6884800 | 79 | 120–142 |
| D1S2663 | p36.31 | 7180181–7180375 | 85 | 183–205 |
| D1S548 | p36.23 | 7365426–7365588 | 67 | 148–172 |
| D1S1612 | p36.23 | 8040572–8040684 | 83 | 94–130 |
| D1S450 | p36.22 | 9508007–9508269 | 81 | 243–367 |
| D1S2736 | p36.22 | 10615664–10615783 | 73 | 122–132 |
| D1S1151 | p36.22 | 11387148–11387420 | 92 | 263–332 |
| D1S2667 | p36.22 | 11409625–11409894 | 82 | 246–272 |
| D1S2740 | p36.22 | 11843587–11843676 | 65 | 80–104 |
| D1S1597 | p36.21 | 13656774–13656943 | 66 | 159–179 |
| GATA29A05 | p36.13 | 17428904–17629222 | 71 | 179–211 |
| GATA129H04 | p34.2 | 41499079–41499293 | 88 | 204–256 |
| D1S1728 | p31.1 | 81610619–81810778 | 68 | 158–174 |
| D1S2707 | q23.2 | 158339075–158339223 | 82 | 137–159 |
| Chromosome 19 | ||||
| D19S898 | p13.11 | 18334781–18334960 | 82 | 178–200 |
| D19S245 | q13.11 | 38789997–38790201 | 68 | 195–211 |
| D19S219 | q13.32 | 50685577–50685750 | 77 | 160–190 |
| D19S112 | q13.32 | 51070821–51070950 | 86 | 120 |
| D19S412 | q13.32 | 51702813–51702921 | 80 | 89–113 |
| D19S606 | q13.32 | 52665381–52665558 | 81 | 172–190 |
| D19S596 | q13.33 | 53942842–53943054 | 53 | 213–221 |
| D19S246 | q13.33 | 55647457–55647662 | 84 | 185–229 |
| Chromosome 10 | ||||
| D10S1649 | p14 | 9460072–9460211 | 84 | 126–150 |
| D10S1790 | q21.1 | 54875447–54875631 | 83 | 179–201 |
| D10S1765 | q23.31 | 89591521–89591700 | 83 | 166–184 |
| D10S1692 | q24.32 | 104578855–104579053 | 88 | 182–211 |
| D10S1483 | q26.13 | 123273584–123273727 | 82 | 130–158 |
| D10S212 | q26.3 | 134299591–134299779 | 70 | 189–201 |
For heterozygous samples, a reduction of at least 50% (allelic imbalance factor, IF, of 0.50) in the peak area of one allele in the tumor (normalized against the retained allele) was used to score LOH. In each case, the IF was determined by calculating the ratio of alleles for both the constitutional (C) and the tumor (T) sample, and then the tumor ratio was divided by the constitutional ratio:
If the value obtained was greater than 1.00, the reciprocal 1/R was used. Thirteen additional markers located on chromosome 1p or 19q were also analyzed when the LOH status need to be confirmed: D1S243 (located on 1p36.33), D1S2845, and D1S2660 (located on 1p36.32); D1S2795, D1S2870, and D1S2663 (located on 1p36.31); D1S548 (located on 1p36.23); D1S450, D1S1151, D1S2667, and D1S2740 (located on 1p36.22); GATA29A05 (located on 1p36.13); and D19S219 (located on 19p13.32) (Fig. 1 and Table 1).
We determined chromosome losses as the complete or partial loss of the 1p, 19q, and 10q. During the assessment of allelic status, investigators were blinded to the characteristics of the patients and to follow-up data.
Statistical Methods
The statistical endpoints in our analyses were PFS, OS, and response to chemotherapy. Events for the PFS analysis were defined as relapse or disease progression. The time to an event for the relapse was calculated as the date of surgery to the time of the first relapse or the time of last contact with the patient if no event occurred. The time to an event for the OS analysis was calculated as the date of surgery until the time of death or the time of last contact if the patient was alive. Data on survival were censored if patients had died from potentially other causes. Note that no patient was lost to follow-up and that all patients progressed before dying. Also note that these definitions of PFS and OS are taken from the time of surgery (ie the time of the therapeutic intervention) and not the time of diagnosis as in conventional definitions of PFS and OS, as we are interested in understanding the interplay of LOH with the consequences of surgery for the patient.
Two analysis approaches were taken: First, univariate analyses to evaluate the importance of each factor on its own, then a multivariate analysis in order to allow for any codependencies between factors. We calculated univariate hazard ratios with the proportional-hazards model. Tests of association were performed with the use of Fisher's exact test for categorical data and Wilcoxon rank-sum test for continuous data. Survival curves were constructed according to the methods of Kaplan and Meier and comparisons of the survival curves were performed with a two-sided log-rank test. Multivariate analyses were performed with the use of a Cox proportional-hazards regression model to identify variables that were independently predictive of outcome. All covariates were retained in the model to illustrate the lack of effect in the presence of other significant factors.
Results
Characteristics of the Patients and Correlations with Prognosis
One hundred and forty-five patients were included prospectively at the time of their surgery into the molecular study. We removed 15 patients presenting with partial LOH on 1p, 19q, or 10q due to the possible different prognosis reported in the literature. Table 2 lists the relevant demographic and clinical characteristics of the 130 patients whose gliomas were analyzed by microsatellite analysis.
Table 2.
Clinical and molecular features in oligodendroglial tumors (130 patients)
| Descriptive data | Results | |
|---|---|---|
| Clinical features | ||
| Median age at diagnosis, y (range) | 45.5 (19–73.5) | |
| Number: male/female | 68/62 | |
| Median preoperative KPS (range) | 90% (30%–100%) | |
| Median postoperative KPS (range) | 90% (50%–100%) | |
| Symptoms at diagnosis | ||
| Seizure | 76 (58.5%) | |
| Combination of neurological deficit and epilepsy | 31 (24%) | |
| Unusual headache | 11 (8.5%) | |
| Raised intracranial pressure | 9 (7%) | |
| No symptoms | 3 (2%) | |
| MRI contrast enhancement (%) | 76 (58%) | |
| Surgery | ||
| Biopsy | 57 (44%) | |
| Partial removal | 41 (31%) | |
| Total removal | 32 (25%) | |
| Histopathology | ||
| Grade 2 oligodendroglioma | 28 (22%) | 64 |
| Grade 3 oligodendroglioma | 36 (28%) | |
| Grade 2 oligoastrocytoma | 24 (18%) | 66 |
| Grade 3 oligoastrocytoma | 42 (32%) | |
| Genomic features | ||
| No deletion | 46 (25.5%) | |
| 1p and/or 19q LOH | 33 (35.5%) | |
| 10q LOH | 40 (30.5%) | |
| 1p, 19q, and 10q LOH | 11 (8.5%) | |
Abbreviations: KPS, Karnofsky performance status; LOH, loss of heterozygosity.
Overall, 116 patients received adjuvant treatment: (i) postoperative radiotherapy followed by chemotherapy in 25 patients, (ii) postoperative chemotherapy alone in 18 patients, (iii) postoperative radiotherapy alone in 73 patients, and (iv) chemotherapy at tumor progression in 26 patients. Thirty patients received temozolomide and 39 patients received PCV (procarbazine, CCNU, vincristine) regimen as chemotherapy. Thirty-six of the 69 chemotherapy-treated patients showed positive or stable radiological tumor responses.
Relevant data concerning the ability to predict response to chemotherapy are listed in Table 3. There was no difference in the frequency of the response to chemotherapy in each histological subgroup of gliomas (P = .2), and between appearance or not for contrast enhancement on MRI (P = .06). The time to start chemotherapy, immediately following radiotherapy, or when gliomas relapsed, was not associated with the response to chemotherapy (P = .8). The duration of survival was independent of the timing of the introduction of chemotherapy; it was not that because the chemotherapy was introduced earlier, patients would survive longer. Of the tumors treated by chemotherapy, 41% presented a positive response to the PCV regimen and 66.7% were clinically and radiologically improved by temozolomide. The result was not significant (P = .51). In univariate analysis, the only significant clinical factor relating to the response to chemotherapy was the postoperative KPS (P = .03).
Table 3.
Correlations between analyzed data and response to chemotherapy
| Data | Categories compared | Positive response, % | P value |
|---|---|---|---|
| Clinical | |||
| Sex | Male vs female | 55% vs 48% | .6 |
| Age (y) | ≤ 47 vs > 47 | 52% vs 49% | .4 |
| Postoperative KPS (%) | ≤ 65 vs > 65 | 35% vs 65% | .03 |
| Radiological | |||
| Contrast enhancement | No vs yes | 70% vs 45% | .06 |
| Neuropathology | |||
| Tumor type | O vs OA | 56% vs 48.5% | .6 |
| Grade | 2 vs 3 | 68.5% vs 46% | .1 |
| Treatment | |||
| Delay of chemotherapy | Postoperative vs relapse | 48% vs 54% | .78 |
| Type of chemotherapy | PCV vs TMZ | 41% vs 66.7% | .051 |
| Allelic status | |||
| 1p and 19q LOH | 92.3% | <.0001 | |
| vs no deletion | vs 83.3% | ||
| vs 1p, 19q, and 10q LOH | vs 50% | ||
| vs 10q LOH | vs 14.5% | ||
Abbreviations: PFS, progression-free survival; KPS, Karnofsky performance status; O, oligodendroglioma; OA, oligoastrocytoma; B, biopsy; PR, partial removal; CR, complete removal; PCV, procarbazine, CCNU, vincristine; TMZ, temozolomide; LOH, loss of heterozygosity.
Data regarding the ability of the various clinical, radiological, and neuropathology factors to predict clinical outcome are reported in Tables 4 (PFS) and 5 (OS). The mean PFS for all patients was 27.5 months. Fifty-two patients relapsed and 26 patients had a continuous tumor progression despite various therapy strategies. At the time of analysis, mean survival was 40.5 months, and 60 patients (46%) had died. As expected, in univariate analysis, 5 variables studied had significant prognostic value for OS and PFS. Age >47, postoperative KPS <65, positive contrast enhancement, and grade 3 tumors were associated with a significantly higher risk of death or progression, whereas complete removal on surgery—confirmed on postoperative MRI—was associated with a better prognosis. Tumor histology and type of chemotherapy used were only significant in the OS analysis.
Table 4.
Univariate analysis: correlation between analyzed data and progression-free survival
| Data | Categories compared | Mean PFS (m) | P value |
|---|---|---|---|
| Mean PFS period | 27.5 | ||
| Clinical | |||
| Sex | Male vs female | 28.6 vs 23.1 | .6 |
| Age (y) | ≤ 47 vs > 47 | 27.7 vs 22.1 | .001 |
| Postoperative KPS (%) | ≤ 65 vs > 65 | 4.7 vs 29.5 | < .0001 |
| Radiological | |||
| Contrast enhancement | No vs yes | 32.8 vs 22.3 | .0007 |
| Neuropathology | |||
| Tumor type | O vs OA | 24 vs 25.5 | .19 |
| Grade | 2 vs 3 | 32.1 vs 23.1 | .0037 |
| Treatment | |||
| Quality of surgery | B vs PR vs CR | 22.3 vs 24 vs 32 | .015 |
| PR vs CR | 24 vs 32 | .2 | |
| Genomic data | |||
| LOH 1p | No LOH vs LOH | 22.6 vs 32.5 | < .0001 |
| LOH 19q | No LOH vs LOH | 24.1 vs 30.4 | .0035 |
| LOH 10q | No LOH vs LOH | 40.5 vs 7.9 | < .0001 |
Abbreviations: PFS, progression-free-survival; KPS, Karnofsky performance status; O, oligodendroglioma; OA, oligoastrocytoma; B, biopsy; PR, partial removal; CR, complete removal; LOH, loss of heterozygosity.
Table 5.
Univariate analysis: correlation between analyzed data and overall survival
| Data | Categories compared | Overall 3-y survival rate (%) | P value |
|---|---|---|---|
| Clinical | |||
| Sex | Male vs female | 58 vs 51 | .56 |
| Age (y) | ≤ 47 vs > 47 | 65 vs 40 | .002 |
| Postoperative KPS (%) | ≤ 65 vs > 65 | 0 vs 60 | < .0001 |
| Radiological | |||
| Contrast enhancement | No vs yes | 70 vs 42.5 | .0002 |
| Neuropathology | |||
| Tumor type | O vs OA | 60 vs 48 | .033 |
| Grade | 2 vs 3 | 66 vs 45 | .0009 |
| Treatment | |||
| Quality of surgery | B vs PR vs CR | 39 vs 58 vs 72 | .01 |
| PR vs CR | 58 vs 72 | .15 | |
| Delay for chemotherapy | Post-op vs relapse | 42 vs 25 | .5 |
| Type of chemotherapy | PCV vs TMZ | 26.5 vs 46 | .02 |
| Genomic data | |||
| LOH 1p | No LOH vs LOH | 45.5 vs 73 | .0002 |
| LOH 19q | No LOH vs LOH | 48 vs 68 | .003 |
| LOH 10q | No LOH vs LOH | 86.5 vs 3 | < .0001 |
Abbreviations: OS, overall survival; KPS, Karnofsky performance status; O, oligodendroglioma; OA, oligoastrocytoma; B, biopsy; PR, partial removal; CR, complete removal; PCV, procarbazine, CCNU, vincristine; TMZ, temozolomide; LOH, loss of heterozygosity.
Assessment of the Allelic Status of Chromosomes 1p, 19q, and 10q
Data regarding the genomic alterations analyzed in cohort of 130 patients affected by oligodendroglial tumors are reported in Table 6. Thirty-three tumors (25.5%) showed combined losses of 1p and 19q. LOH on 10q, without deletions on chromosomes 1p19q, was identified in 40 tumors (30.5%). Only 11 tumors (8.5%) had LOH on 1p, 19q, and 10q. Forty-six tumors (35.5%) had no LOH.
Table 6.
Genomic alterations by histology and grade of the tumors
| Genetic alterations | O II | O III | OA II | OA III | Number of patients (%) |
|---|---|---|---|---|---|
| 1p and/or 19q LOH | 9 (7%) | 15 (11.5%) | 3 (2.5%) | 6 (4.5%) | 33 (25.5%) |
| No deletion | 12 (9%) | 5 (4%) | 16 (12.5%) | 13 (10%) | 46 (35.5%) |
| 1p, 19q, and 10q LOH | 1 (0.75%) | 3 (2.5%) | 1 (0.75%) | 6 (4.5%) | 11 (8.5%) |
| 10q LOH | 6 (4.5%) | 13 (10%) | 4 (3%) | 17 (13%) | 40 (30.5%) |
| Total | 28 | 36 | 24 | 42 | 130 |
Abbreviations: O II, oligodendroglioma grade II; O III, oligodendroglioma grade III; OA II, oligoastrocytoma grade II; OA III, oligoastrocytoma grade III; LOH, loss of heterozygosity.
Correlations of Allelic Status with Clinical Characteristics and Evaluation of the Prognostic Value of 10q Loss
LOH status was independent of the patient's sex (P = .5) but correlated significantly with patient's age (P = .0009). At the tumor's diagnosis, mean age was 39 in the absence of LOH (median: 36.5), 45 in 1p and 19q LOH (median: 46), 50 in 10q LOH (median: 52.5), and 54.5 years old in 1p, 19q, and 10q LOH (median: 56.5). In our cohort of patients, 10q LOH was associated with anaplastic grade (P = .0023), whereas 1p and 19q LOH was mainly correlated with pure oligodendroglial tumors whatever the grade (P = 0.0015) (data not shown).
As expected, the LOH status was an important factor for the tumor response to chemotherapy (P < .0001) (Table 3). Sixty-nine patients received chemotherapy during the study. The clinical and radiological response to chemotherapy was 92.3% for tumors with 1p and 19q LOH, 83.3% for tumors without allelic losses, 50% for tumors with 1p, 19q, and 10q LOH, and only 14.5% for those with 10q LOH alone.
A univariate analysis of patients' outcomes showed that both 1p LOH and 19q LOH were highly associated with increases in both PFS (P < .0001 and P = .0035, respectively) and 3-year survival rate (P = .0002 and P = .003, respectively). Analysis of the subgroup of cases showed that LOH on 10q was significantly associated with a decreased probability of PFS (P < .0001) and 3-year survival rate (P < .0001). Patients in whom tumors showed 10q LOH had PFS of 7.9 months and a 3-year survival rate of 3%, when compared with 40.5 months and 86.5%, respectively, in patients in whom tumors did not have 10q LOH (Tables 4 and 5). Respectively, the 3-year survival rate and PFS was 85% and 36 months for patients with tumors having 1p and 19q LOH, 50% and 21 months for patients with tumors without allelic losses, 5% and 9 months for patients with tumors having 1p, 19q, and 10q LOH, and only 1% and 6 months for those with 10q LOH alone.
Multivariate Analysis
Several genetic and clinical factors with significant prognostic value were interrelated in their ability to predict clinical outcome. To identify the most powerful prognostic factors, we performed multivariate analyses with the Cox proportional-hazards model.
The results were roughly the same between PFS and OS. Among the genetic variables, LOH on 1p had a favorable prognosis (relative risk of relapse associated with 1p LOH, 0.57; P < .005, and relative risk of death associated with 1p LOH, 0.54; P = .0007), whereas LOH on 10q was significantly associated with decreased PFS and OS (relative risk of relapse associated with 10q LOH, 3.5; P < .0001, and relative risk of death associated with 10q LOH, 4.6; P < .0001).
Chemotherapy given immediately after radiotherapy in grade III tumors was also found to be independently associated with increased PFS, but not with OS (P = .004 and P = .3, respectively). The assessment was not correct for OS because the patients received chemotherapy at tumor progression.
Discussion
The aim of this prospective study was to investigate the impact of chromosome 1p, 19q, and/or 10q losses of heterozygosity in patients with oligodendroglial tumors, in terms of survival (PFS and OS) and sensitivity of the tumors to chemotherapy. Our analysis of PFS and OS yielded similar results. There is a very small probability of a favorable outcome in patients with recurrent glioma.
Univariate testing confirmed that 3 clinical variables (age, KPS, and extent of surgical treatment) were significant predictors of a favorable outcome18,19 (Tables 3 and 4). All chromosomal alterations analyzed were correlated with outcome: LOH on 1p and 19q was associated with longer survival; whereas LOH on 10q was associated with shorter survival. After the multivariate model was restricted to each group, LOH on 1p was independently associated with better OS and PFS, confirming previous reports.20 However, aggressive disease developed in a subgroup of patients despite the LOH on 1p. LOH on 10q was present in a subgroup of patients whose tumors had LOH on 1p and/or 19q. In this study, allelic loss of chromosome 10q was found to be a better prognosis indicator than allelic loss of chromosomes 1p and 19q.
Advances in the molecular genetics of gliomas have attempted to use molecular markers for assessing prognosis in patients with brain tumors. Cairncross (1998)18 was the first to report that LOH involving 1p and 19q is a stronger predictor of chemosensitivity and longer PFS for patients with anaplastic oligodendrogliomas than are the traditional prognosis factors of age and KPS. However, it is difficult to know whether these markers reflect sensitivity to treatments—radiotherapy and chemotherapy—or are inherent differences in the biological characteristics of the tumors, because of the uncertainty about the mechanism of this effect and the accumulation of genetic defects. To investigate possible biological characteristics, we propose distinguishing patients who had no adjuvant treatment during a study from those in which at least 1 treatment was done, allowing for the surgical procedure. Unfortunately, the scope of our study was limited—more patients and a longer follow-up period might have allowed demonstrating this. Little is known about the molecular correlates of the LOH 1p19q, but they are still among the most important reported factors in predicting survival.20,21 The ability to detect risk factors for the prognosis and the response to treatment could make the therapeutic strategy more effective and less toxic among some patients. Our findings regarding 10q LOH in patients with oligodendroglial tumors, presenting with 1p19q LOH or not, suggest that this abnormality should be searched for in clinical trials to detect patients at high risk.22
Our study found that patients with 10q deletion as the unique abnormality had by far the worst prognosis, followed by patients with 1p and/or19q and 10q deletions, then those without any deletion, whereas patients with 1p19q codeletion had the better estimated survival times and response to treatment. Patients with oligodendroglial tumors who had 10q loss alone (31% of the patients) were at a higher risk for recurrence and death than those who did not have such loss (3-year event-free survival, 1% vs 69%). In patients such as the former, more aggressive or innovative therapeutic approaches at diagnosis may be considered. Patients with LOH on 1p alone (26% of the patients) had an excellent prognosis (3-year event-free survival, 65%). In this group of patients, therapy should be as minimal as possible and should be viewed in the light of possible late effects.
The association between LOH on 1p, 19q, or 10q and other genomic alterations, histological or clinical data, has been described in other studies.14,17,21,23,24 Some of them reported a mutually exclusive deletion in patients; those who had gliomas with a 1p19q codeletion versus those who had gliomas with LOH on 10q.16 In our study, 11 patients had LOH on 1p and/or 19q associated with LOH on 10q. This association was not so common and not completely exclusive. The explanation for these discrepant results in our study is not clear. Other authors found LOH on 10q more often in pure astrocytomas25 or mainly in grade III or IV gliomas.26 We found this tendency, with 30% grade III and 9% grade II gliomas presenting with 10q LOH. All of the authors described poor prognosis for patients who have gliomas with LOH on 10q. We found a 3-year event-free survival of 35% in patients with concomitant allelic loss of chromosome 1p, 19q, and 10q. Taken together, these data indicate that LOH on 10q identifies more patients who are likely to have an unfavorable outcome than does analysis of 1p19q losses alone. We showed that LOH on 10q was a significant predictor of unfavorable outcome. Our results demonstrate that the molecular profile assessment of LOH on 1p, 19q, and 10q in oligodendroglial tumors provides information about prognosis in patients with grade II and III oligodendroglial tumors.
The principal advantage of using allelic LOH 10q as a prognostic factor is in identifying high-risk patients among those with grade II and grade III gliomas in order to adapt the therapeutic strategy. The clinical conclusion of this study is to recommend analysis of the chromosome 10q at the same time as chromosomes 1p and 19q to customize treatment. Predicting which tumors will be more aggressive has clinical implications and may lead to earlier and stronger treatment, and also to suggest several therapies for clinical studies. We propose neuropathology should be supplemented by the molecular analysis of chromosomes 1p, 10q, and 19q; and, with clinical and radiological data, one should consider therapeutic choices, meanwhile waiting for new strategies and choices of treatments.
Prospective studies are needed to determine whether more recent drugs would benefit patients with gliomas whose malignant cells have molecular markers associated with a reduced efficacy of standard chemotherapy regimens. These would move us toward the goal of individualized cancer treatment based on the molecular characteristics of the tumor. The implications of our findings for clinical management follow the increasing tendency to tailor therapy on an individual basis according to risk factors detected at the time of diagnosis. Rapid and better identification of such genetic imbalances and the precise molecular mechanisms underlying tumor development could provide better guidelines for glioma diagnosis and help decision making for treatments and the accurate interpretation of data from clinical trials of new therapies. The objective is to lead to more personalized cancer treatment, potentially to direct the most intensive treatments to patients with the most aggressive tumors, whereas sparing other patients from the adverse effects of unnecessarily intensive therapy.
Funding
This work supported in part by grants from the Journées de Neurologie de Langue Française and by the Comité du Nord de la Ligue Nationale contre le Cancer.
Acknowledgments
We thank Marie-Paule Delescaut and Annette Leclercq for their technical assistance, Sophie Bardin for her helpful secretary assistance, and Olivier Delrieu for all his advice. All authors read and approved the final version of the article.
Conflict of interest statement. None declared.
References
- 1.Vallat AV, Poirier J, Gray F, Chatel M. Tumeurs du système nerveux central. Classifications histologiques et topographiques, épidémiologie. EMC Neurologie. 1997 17-205-A-10. [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumors of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coons SW, Johnson PC, Scheithauer BW, Yates AJ, Pearl DK. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79(7):1381–1393. doi: 10.1002/(sici)1097-0142(19970401)79:7<1381::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 4.Surawicz TS, Davis F, Freels S, Laws ER, Jr, Menck HR. Brain tumor survival: results from the National Cancer Data Base. J Neurooncol. 1998;40(2):151–160. doi: 10.1023/a:1006091608586. [DOI] [PubMed] [Google Scholar]
- 5.Davis FG, Freels S, Grutsch J, Barlas S, Brem S. Survival rates in patients with primary malignant brain tumors stratified by patient age and tumor histological type: an analysis based on Surveillance, Epidemiology, and End Results (SEER) data, 1973–1991. J Neurosurg. 1998;88(1):1–10. doi: 10.3171/jns.1998.88.1.0001. [DOI] [PubMed] [Google Scholar]
- 6.Behin A, Hoang-Xuan K, Carpentier AF, Delattre JY. Primary brain tumors in adults. Lancet. 2003;361(9354):323–331. doi: 10.1016/S0140-6736(03)12328-8. [DOI] [PubMed] [Google Scholar]
- 7.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64(6):479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura M, Shimada K, Ishida E, Nakase H, Konishi N. Genetic analysis to complement histopathological diagnosis of brain tumors. Histol Histopathol. 2007;22(3):327–335. doi: 10.14670/HH-22.327. [DOI] [PubMed] [Google Scholar]
- 9.Fuller GN, Hess KR, Rhee CH, et al. Molecular classification of human diffuse gliomas by multidimensional scaling analysis of gene expression profiles parallels morphology-based classification, correlates with survival, and reveals clinically-relevant novel glioma subsets. Brain Pathol. 2002;12(1):108–116. doi: 10.1111/j.1750-3639.2002.tb00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. [PubMed] [Google Scholar]
- 11.Caskey LS, Fuller GN, Bruner JM, et al. Toward a molecular classification of the gliomas: histopathology, molecular genetics, and gene expression profiling. Histol Histopathol. 2000;15(3):971–981. doi: 10.14670/HH-15.971. [DOI] [PubMed] [Google Scholar]
- 12.Cairncross G, Berkey B, Shaw E, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol. 2006;24(18):2707–2714. doi: 10.1200/JCO.2005.04.3414. [DOI] [PubMed] [Google Scholar]
- 13.Smith JS, Perry A, Borell TJ, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18(3):636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 14.Bauman GS, Ino Y, Ueki K, et al. Allelic loss of chromosome 1p and radiotherapy plus chemotherapy in patients with oligodendrogliomas. Int J Radiat Oncol Biol Phys. 2000;48(3):825–830. doi: 10.1016/s0360-3016(00)00703-3. [DOI] [PubMed] [Google Scholar]
- 15.Ino Y, Betensky RA, Zlatescu MC, et al. Molecular subtypes of anaplastic oligodendroglioma: implications for patient management at diagnosis. Clin Cancer Res. 2001;7(4):839–845. [PubMed] [Google Scholar]
- 16.Hoang-Xuan K, He J, Huguet S, et al. Molecular heterogeneity of oligodendrogliomas suggests alternative pathways in tumor progression. Neurology. 2001;57(7):1278–1281. doi: 10.1212/wnl.57.7.1278. [DOI] [PubMed] [Google Scholar]
- 17.Bissola L, Eoli M, Pollo B, et al. Association of chromosome 10 losses and negative prognosis in oligoastrocytomas. Ann Neurol. 2002;52(6):842–845. doi: 10.1002/ana.10405. [DOI] [PubMed] [Google Scholar]
- 18.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90(19):1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 19.Van den Bent MJ, Carpentier AF, Brandes AA, et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not OS in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol. 2006;24(18):2715. doi: 10.1200/JCO.2005.04.6078. [DOI] [PubMed] [Google Scholar]
- 20.Dehais C, Laigle-Donadey F, Marie Y, et al. Prognostic stratification of patients with anaplastic gliomas according to genetic profile. Cancer. 2006;107(8):1891–1897. doi: 10.1002/cncr.22211. [DOI] [PubMed] [Google Scholar]
- 21.Ino Y, Zlatescu MC, Sasaki H, et al. Long survival and therapeutic responses in patients with histologically disparate high-grade gliomas demonstrating chromosome 1p loss. J Neurosurg. 2000;92(6):983–990. doi: 10.3171/jns.2000.92.6.0983. [DOI] [PubMed] [Google Scholar]
- 22.Thiessen B, Maguire JA, McNeil K, Huntsman D, Martin MA, Horsman D. Loss of heterozygosity for loci on chromosome arms 1p and 10q in oligodendroglial tumors: relationship to outcome and chemosensitivity. J Neurooncol. 2003;64(3):271–278. doi: 10.1023/a:1025689004046. [DOI] [PubMed] [Google Scholar]
- 23.Daumas-Duport C, Beuvon F, Varlet P, Fallet-Bianco C. Gliomas: WHO and Sainte-Anne Hospital classifications. Ann Pathol. 2000;20(5):413–428. [PubMed] [Google Scholar]
- 24.Zlatescu MC, TehraniYazdi A, Sasaki H, et al. Tumor location and growth pattern correlate with genetic signature in oligodendroglial neoplasms. Cancer Res. 2001;61(18):6713–6715. [PubMed] [Google Scholar]
- 25.von Deimling A, Fimmers R, Schmidt MC, et al. Comprehensive allelotype and genetic anaysis of 466 human nervous system tumors. J Neuropathol Exp Neurol. 2000;59(6):544–558. doi: 10.1093/jnen/59.6.544. [DOI] [PubMed] [Google Scholar]
- 26.Sanson M, Leuraud P, Aguirre-Cruz L, et al. Analysis of loss of chromosome 10q, DMBT1 homozygous deletions, and PTEN mutations in oligodendrogliomas. J Neurosurg. 2002;97(6):1397–1401. doi: 10.3171/jns.2002.97.6.1397. [DOI] [PubMed] [Google Scholar]