Abstract
Inteins catalyze protein splicing in a fashion similar to how self-splicing introns catalyze RNA splicing. Split-inteins catalyze precise ligation of two separate polypeptides through trans-splicing in a highly specific manner. Here we report a method of using protein trans-splicing to circumvent the packaging size limit of gene therapy vectors. To demonstrate this method, we chose a large dystrophin gene and an adeno-associated viral (AAV) vector, which has a small packaging size. A highly functional 6.3-kb Becker-form dystrophin cDNA was broken into two pieces and modified by adding appropriate split-intein coding sequences, resulting in split-genes sufficiently small for packaging in AAV vectors. The two split-genes, after codelivery into target cells, produced two polypeptides that spontaneously trans-spliced to form the expected Becker-form dystrophin protein in cell culture in vitro. Delivering the split-genes by AAV1 vectors into the muscle of a mouse model of Duchenne muscular dystrophy rendered therapeutic gene expression and benefits.
Introduction
The protein trans-splicing method was inspired by the discovery of a natural case of protein trans-splicing in the photosynthetic cyanobacterium Synechocystis sp. PCC 6803 (Wu et al., 1998a). The DnaE protein of this organism is encoded in two split-genes, with one gene encoding the N-terminal part of DnaE followed by a 123-amino acid split-intein sequence and another gene encoding the C-terminal part of DnaE preceded by a 36-amino acid split-intein sequence. Two polypeptides expressed from the split-genes undergo spontaneous protein trans-splicing to produce the complete DnaE protein. The splicing reaction is catalyzed by the split-intein sequences, removes precisely the split-intein sequences, and joins specifically the N- and C-terminal parts of DnaE with a peptide bond. Artificial split-inteins have also been engineered from contiguous inteins that are naturally embedded in host proteins and catalyze cis-splicing reactions. For example, the split-intein used in this study was engineered from a contiguous DnaB intein by using only its N-terminal 106-amino acid sequence (N-intein) and C-terminal 48-amino acid sequence (C-intein) (Wu et al., 1998b). Split-intein and contiguous inteins have been found in many microorganisms but not in humans. Because protein splicing is catalyzed by the intein itself, in general without host specificity and assistance from other cellular factors, it was thought that a microbial intein could work in human cells in a human protein, provided that the intein was placed immediately before a nucleophilic residue (serine, cysteine, or threonine) in the human protein.
Dystrophin gene therapy for Duchenne muscular dystrophy (DMD) presented an ideal test case for the protein trans-splicing method. DMD is caused by recessive mutations in the dystrophin gene and is the most common and lethal childhood muscle disorder, affecting 1 in every 3500 male births (Koenig et al., 1987). Gene therapy offers a promising treatment for DMD, and the adeno-associated viral (AAV) vector is the most efficient and nontoxic vehicle for delivering genes to muscle tissues, as has been demonstrated with a number of muscle genes and reporter genes (Xiao et al., 1996, 2000; Fisher et al., 1997; Snyder et al., 1997; Hartigan-O'Connor and Chamberlain, 2000; Wang et al., 2000; Watchko et al., 2002). Because AAV can optimally package approximately 4.7 kb of DNA, an AAV vector has less than 4 kb of room to accommodate a transgene in addition to the required accessory cis-acting sequences such as the promoter, polyadenylation signal, flanking inverted terminal repeats (ITRs), and so on. The room available for the transgene becomes even smaller when large regulatory elements or tissue-specific enhancers are used. Although the full-length dystrophin gene has a coding sequence of 11 kb, a 6.3-kb Becker-form dystrophin cDNA has proven highly functional (England et al., 1990; Phelps et al., 1995). This cDNA was isolated from a Becker muscular dystrophy patient more than 60 years of age with mild symptoms (England et al., 1990). However, this 6.3-kb dystrophin cDNA is still too large for the AAV vector. We hypothesized that this problem could be resolved by a protein trans-splicing strategy. In this report, we have split the 6.3-kb dystrophin cDNA into two pieces and linked the intein catalytic elements at the junctions of the split site. These two separate coding sequences were packaged in AAV1 vectors under the control of a cytomegalovirus (CMV) promoter. Simultaneous delivery of the dual vectors resulted in efficient production and splicing of the two polypeptides to form one full-sized Becker-form dystrophin both in vitro and in vivo in mdx mice.
Materials and Methods
Construction of split dystrophin genes
DNA fragments from the dystrophin gene and the split-inteins were amplified by polymerase chain reaction (PCR), using high-fidelity Pyrococcus furiosus (Pfu) DNA polymerase, and inserted into the rAAV plasmid pXXUF1 (Li et al., 1999) between the NotI and SaII cutting sites. The N-dys gene contains the first 1066 codons and a 10-bp 5′ untranslated leader sequence of the dystrophin gene. The C-dys gene contains the last 917 codons and an 8-bp 5′ untranslated leader sequence of the dystrophin gene. Split-intein coding sequences were from the Synechocystis sp. PCC 6803 (Ssp) DnaB intein or the Rhodothermus marinus (Rma) DnaB intein, with the first 106 codons placed in the N-dys gene and the last 51 codons in the C-dys gene (Liu and Hu, 1997). DNA sequences were confirmed by sequencing all of the intein-containing dystrophin split-genes.
Transfection and Western blot analysis
A total of 25 μg of plasmid DNA was dissolved in 1 ml of 0.25 M CaCl2 and quickly mixed with 1 ml of HBS buffer (50 mM HEPES, 280 mM NaCl, and 1.5 mM Na2HPO4, pH 7.12) before adding it to human 293 cells for transfection. Eight to 12 hr after transfection, the medium was replaced with fresh Dulbecco's modified Eagle's medium (GIBCO DMEM; In-vitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS) and antibiotics. The cells were harvested 48 hr post-transfection and lysed in sodium dodecyl sulfate (SDS)- and dithiothreitol (DTT)-containing gel-loading buffer in a boiling water bath. Total cellular proteins were resolved on an SDS–8% polyacrylamide gel and blotted onto nitrocellulose membrane. Western blot analysis was done with either monoclonal antibody NCL-DYS2 (Novocastra brand, distributed by Vector Laboratories, Burlingame, CA) against the C-terminal part of dystrophin or the monoclonal antibody NCL-DYS3 (Novocastra) against the N-terminal part of dystrophin.
Experiments with mdx mice
Production and CsCl density gradient purification of AAV1 vectors were carried out according to previously published methods (Snyder et al., 1996). Muscle cryosections (thickness, 8 μm) were immunofluorescently stained with a Mouse on Mouse (M.O.M.) kit (Vector Laboratories) to block the endogenous immunoglobulin background on the sections, according to the manufacturer's protocol. The cryosections were immediately treated with the blocking buffer without the fixation step. The immunofluorescence staining was done with either the NCL-DYS2 or NCL-DYS3 antibody (described previously), followed by a secondary rabbit anti-mouse antibody labeled with either the green fluorescent dye fluorescein isothiocyanate (FITC) or the red fluorescent dye cyanine 3 (Cy3; Novocastra), respectively. Alternatively, rabbit polyclonal antibodies against human dystrophin rod domain repeats R1R2 or R22R23 were used for the immuno-fluorescence staining of mouse muscle.
Results and Discussion
As illustrated in Fig. 1, a large gene can be split into two pieces, and each piece is linked with an appropriate coding sequence of a split-intein. Each of the resulting split-genes, containing the intein coding sequence, is sufficiently small to be packaged into an AAV vector. The two split-genes are both delivered into a target cell to produce the corresponding intein-containing split-proteins that spontaneously undergo protein trans-splicing to produce the mature protein, in this case the Becker-form dystrophin. Simultaneous delivery of the two split-genes into the same target cells, especially in muscle cells, is readily achievable, because a target cell typically receives several viral particles via a standard AAV-based gene delivery method.
FIG. 1.
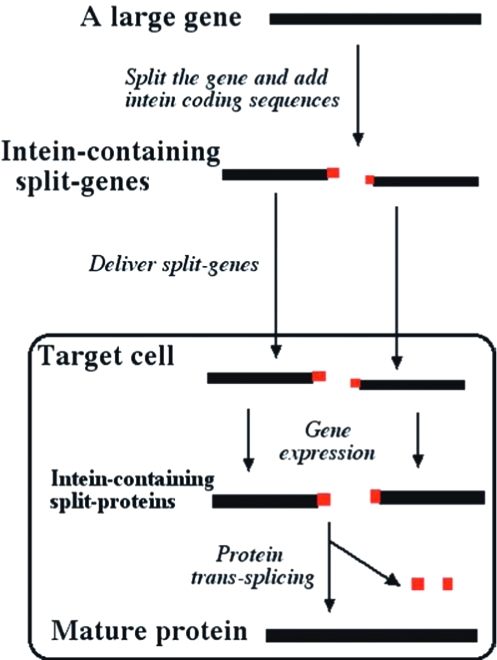
An illustration of the protein trans-splicing method for overcoming the size limitation of gene delivery vectors. A large gene is converted into two smaller, intein-containing split-genes and delivered into a target cell by means of two vectors. Expression of the two split-genes produces two intein-containing split-proteins that spontaneously undergo protein trans-splicing to produce the mature protein.
To accomplish the above-described goal, we first wished to identify an optimal split site in the dystrophin gene, so that the resulting dystrophin fragments would be suitable for efficient protein trans-splicing. Dystrophin is a large, rod-like protein having an N-terminal globular domain, a C-terminal globular domain, and a long central rod domain consisting of 24 coiled-coil repeats (rods) (Koenig and Kunkel, 1990). The central rod domain of the Becker-form dystrophin consists of approximately 8.5 rods, and each rod is thought to fold independently. On the basis of this structure, we selected four alternative split sites near the middle of the 6.3-kb dystrophin gene. In the corresponding 1983-amino acid Becker dystrophin protein sequence, the four split sites are in front of amino acid residues S950, T985, S1067, and S1101, respectively, so that an intein will be placed in front of a nucleophilic amino acid (cysteine, threonine, or serine) that is required for protein splicing. We initially tested these split sites for protein trans-splicing in Escherichia coli cells. A 420-amino acid middle portion of the dystrophin protein (amino acid residues 855–1274 containing the four split sites) was used in this test, because the complete dystrophin protein was difficult to produce in E. coli. We also chose a previously engineered Ssp DnaB split-intein that showed efficient trans-splicing in a model protein in E. coli (Wu et al., 1998b). Adding the coding sequences of this split-intein to each of the four split sites produced a pair of intein-containing dystrophin split-genes. Each pair of the split-genes (as a two-gene operon on a plasmid) was expressed in E. coli cells to produce a corresponding pair of intein-containing dystrophin polypeptides and to detect protein trans-splicing. Only one of the four different pairs of split-genes, which corresponded to the split site at S1067, showed a detectable level of protein trans-splicing, and the splicing efficiency was estimated to be ~50% (data not shown).
The preceding test results allowed us to select the S1067 split site as the optimal site for subsequent studies. The S1067 split site is located inside coiled-coil repeat number 6 (rod 6) of the Becker-form dystrophin, which corresponds to rod 22 of the full-length dystrophin (Koenig et al., 1987; Koenig and Kunkel, 1990). More specifically, the split site is inside an unstructured turn between helix 2 and helix 3 of the spectrin-like triple-helical coiled-coil structure of rod 6 (Fig. 2B). The known close packing between helix 2 and helix 3 may have helped bringing together the two polypeptides for the trans-splicing, although the split-intein parts (intein-N and intein-C) can associate tightly without help from the flanking exteins (Wu et al., 1998b). Because the split site is located in the central rod domain of dystrophin, the resulting dystrophin polypeptides are thought to fold independently before splicing.
FIG. 2.
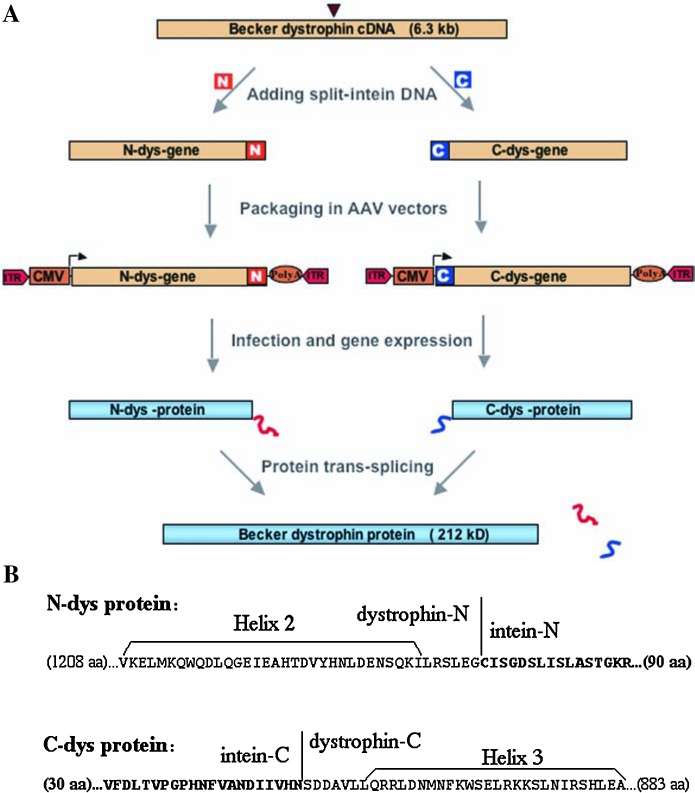
Application of the protein trans-splicing method to dystrophin and AAV vectors. (A) Schematic illustration of the experimental procedure. The 6.3-kb Becker dystrophin cDNA is split at the site marked with an arrowhead, and the split-intein coding sequences (N and C) are added as illustrated. The recombinant split-genes (N-dys gene and C-dys gene) are each packaged in an AAV vector, which contains inverted terminal repeats (ITRs), a poly(A) site, and the CMV promoter. After infection of a target cell with both split-genes and after gene expression, the resulting split-proteins (N-dys protein and C-dys protein) spontaneously undergo protein trans-splicing to produce the Becker dystrophin protein. (B) Amino acid sequences of the N-dys protein and the C-dys protein, which were generated with the S1067 split site of Becker dystrophin and the Ssp DnaB split-intein. Only sequences near the dystrophin–intein junctions are shown; the lengths of omitted sequences are indicated with numbers in parentheses, and the intein sequences are indicated with boldface letters. Sequences of helix 2 and helix 3 inside the sixth of the 8.5 spectrin-like coiled-coil repeats (rods) of the Becker dystrophin are also marked.
After finding the optimal S1067 split site, we constructed a pair of intein-containing split-genes, using the complete Becker dystrophin gene (Fig. 2). These split-genes are named the N-dys gene and C-dys gene, respectively. The N-dys gene encodes the N-dys protein consisting of the N-terminal half (residues 1–1066) of Becker dystrophin followed by the N part (106 amino acids) of the Ssp DnaB split-intein, whereas the C-dys gene encodes the C-dys protein consisting of the C part (51 amino acids) of the Ssp DnaB split-intein followed by the C-terminal half (residues 1067–1983) of Becker dystrophin. The two split-genes were inserted in two separate AAV vector plasmids under the control of a strong CMV promoter for expression in mammalian cells.
To test for dystrophin synthesis through protein trans-splicing, the two AAV vector plasmids containing the intein-containing dystrophin split-genes were introduced into tissue-cultured human cells (the 293 cell line) through DNA cotransfection, and the resulting protein products were analyzed on Western blots (Fig. 3A). In cells transfected with both the N-dys gene and the C-dys gene, protein trans-splicing generated the mature dystrophin protein. As a negative control, cells transfected with either the N-dys gene or the C-dys gene alone produced only the corresponding precursor protein, as expected. The mature dystrophin protein was first identified on the basis of the fact that its apparent size closely matched its predicted size, which was determined by polyacrylamide gel electrophoresis under denaturing and reducing conditions. It was further identified by the fact that it was recognized both with an antibody against the N-terminal part of dystrophin and with an antibody against the C-terminal part of dystrophin. Small amounts of the N-dys and C-dys proteins remained unspliced, indicating that the protein trans-splicing did not go to completion. The remaining N- and C-dys proteins were heterogeneous in size, suggesting partial degradation of these proteins.
FIG. 3.
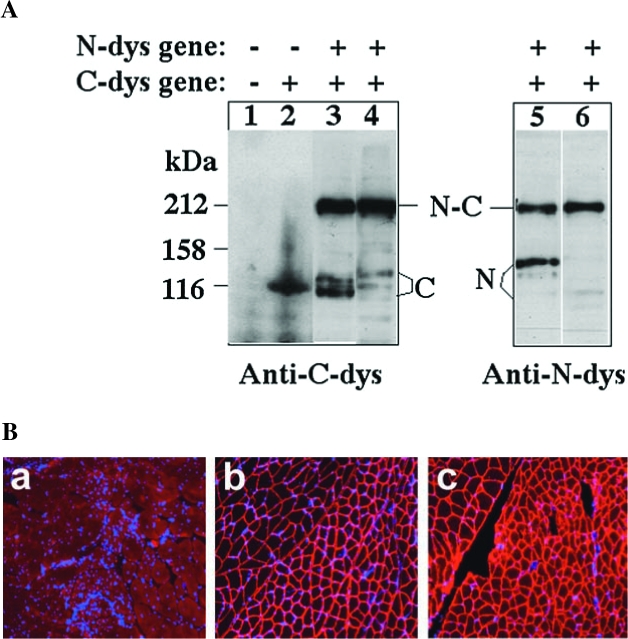
Observation of dystrophin production and function, using the protein trans-splicing method. (A) Western blot analysis of dystrophin production. The human 293 cell line was transfected with either the N-dys gene or C-dys gene, or both, as indicated (−, absent; +, present), using a total of 25 μg of plasmid DNA per transfection. Cells were harvested 48 hr after transfection, and ~50 μg of total cellular protein was resolved by polyacrylamide gel electrophoresis under denaturing (SDS) and reducing (DTT) conditions. Western blotting was done to visualize dystrophin, using the monoclonal antibody NCL-DYS3 against the N-terminal part of dystrophin (Anti-N-dys) or the monoclonal antibody NCL-DYS2 against the C-terminal part of dystrophin (Anti-C-dys). The N-dys protein (marked with the letter N), the C-dys protein (marked with the letter C), and mature dystrophin (marked with the letters N-C) have predicted sizes of 135, 111, and 212 kDa, respectively. In lanes 2, 3, and 5, the N-dys and C-dys proteins contain the Ssp DnaB split-intein. In lanes 4 and 6, the N-dys and C-dys proteins contain the Rma DnaB split-intein. (B) Immunofluorescence staining of functional dystrophin. The gastrocnemius muscles of 3-week-old male mdx mice were coinjected with AAV1-N-dys and AAV1-C-dys vectors and examined after 6 months. Each muscle was injected with 40 μl of AAV1-N-dys (1.5 × 1011 vector genomes [VG]) and AAV1-C-dys (0.5 × 1011 VG) at a 3:1 ratio. Cryosections of mdx muscle were stained immunofluorescently either with a rabbit anti-R1R2 antibody against the N-terminal part of dystrophin (panel b) or a rabbit anti-R22R23 antibody against the C-terminal part of dystrophin (panel c). As a negative control, a cryosection of untreated mdx muscle was stained immunofluorescently with the anti-R1R2 antibody (panel a). All panels were then counterstained for cell nuclei with DAPI (blue). Note the lack of centrally localized nuclei in the AAV-treated muscle, indicating the protective effect of the gene vectors.
To improve the efficiency of protein trans-splicing in dystrophin synthesis, we replaced the Ssp DnaB split-intein with an Rma DnaB split-intein in the preceding experiment. The Rma DnaB intein shares 74% sequence similarity with the Ssp DnaB intein but is from a thermophilic organism (Liu and Hu, 1997). Higher efficiency protein trans-splicing was observed with the Rma DnaB split-intein, with nearly all of the N-dys protein and C-dys protein converted into mature dystrophin protein (Fig. 3A). To achieve this efficient trans-splicing, a 3:1 ratio of the N-dys gene relative to the C-dys gene was used in the cotransfection experiment, as using equal amounts of the N-dys gene and the C-dys gene resulted in less mature dystrophin and excess amounts of unspliced C-dys protein (data not shown). This may be explained by a more rapid degradation of the N-dys protein, as seen in the mdx mouse, where a premature stop codon in the dystrophin gene does not lead to an accumulation of the expected N-terminal partial dystrophin.
To test the protein trans-splicing method in an animal model, the two intein-containing dystrophin split-genes (N-dys gene and C-dys gene) were packaged into AAV1 vectors and coinjected into the leg muscle (gastrocnemius) of mdx mice, a commonly used animal model of Duchenne muscular dystrophy. The production of functional dystrophin through protein trans-splicing was revealed by immunofluorescence staining of muscle thin cryosections, separately with an anti-N-dys antibody recognizing the N-terminal part of dystrophin and with an anti-C-dys antibody recognizing the C-terminal part of dystrophin. As shown in Fig. 3B, the anti-N-dys and anti-C-dys antibodies revealed overlapping and submembrane staining patterns in the muscle cross-sections. The submembrane localization and recognition by the anti-N-dys antibody are strong indicators of a mature dystrophin, because it is known that the N-terminal part of dystrophin alone is unable to localize to the submembrane region and is rapidly degraded in mdx mice. The submembrane localization of dystrophin, as revealed with the anti-N-dys antibody, was observed in nearly 100% of the myofibers of treated mdx muscle at the site of the local injection, indicating dystrophin production through trans-splicing in these cells. The trans-spliced dystrophin protein rendered therapeutic benefits to the dystrophic muscle by improving muscle morphology and histology. For example, the dystrophin-positive myofibers showed more uniform sizes and few centrally localized nuclei (Fig. 3B, panels b and c), whereas the untreated mdx muscle showed the extensive presence of centrally localized nuclei, a sign of muscle degeneration and regeneration (Fig. 3B, panel a). All these indicate that mature and functional dystrophin was produced in the muscle cells of mdx mice through efficient protein trans-splicing, which opens up a new avenue of dystrophin gene therapy for Duchenne muscular dystrophy.
Our findings demonstrated that the protein trans-splicing method could be used to overcome the size limitation of gene therapy vectors, particularly the AAV vectors. In the current study, this method nearly doubled the size range of genes that can be delivered by AAV vectors. This method can potentially expand the size range even further, if one splits a gene into three or more parts and trans-splice the resulting protein parts, using two or more non-cross-reacting split-inteins. The protein trans-splicing method should be generally applicable to other large therapeutic genes besides the dystrophin gene, with notable examples including the cystic fibrosis transmembrane conductance regulator (CFTR) gene for cystic fibrosis, the factor VIII gene for hemophilia A (Chao et al., 2002; Mah et al., 2003; Chen et al., 2007), and other large muscle genes such as the laminin-2 and dysferlin genes implicated in congenital muscular dystrophy and limb girdle muscular dystrophy 2B. The protein trans-splicing method should also be generally applicable to other viral vectors. For example, the 11-kb complete dystrophin gene, in the form of two intein-containing pieces, may then be packaged in two classic retroviral or lentiviral vectors to produce a full-length dystrophin protein.
The protein trans-splicing method shows certain unique advantages as well as disadvantages, when compared with other, similar methods such as the dual AAV vectors (Duan et al., 2000; Nakai et al., 2000; Sun et al., 2000; Yan et al., 2000). A major advantage is its precise and orientation-specific ligation of the two split gene products. Splitting a large gene into two pieces and packaging them in separate vectors is a well-studied strategy by which to overcome the size limitation of AAV vectors. The dual AAV vector methods relied on heterodimerization of two separate AAV vector DNA molecules and, thus, the fusion of the two split half-genes inside a target cell. Because the fusion of vectors A and B appears random in orientation whereas single AAV vector DNA tends to self-circularize, the theoretical probability of the correct tail-to-head fusion of vector A DNA and vector B DNA is significantly less than 25% of the introduced dual vectors. Another dual vector method relied on homologous DNA recombination to fuse the split-genes in a target cell (Duan et al., 2001), but homologous recombination in mammalian cells is notoriously inefficient. In contrast, the protein trans-splicing method does not require vector DNA dimerization or homologous recombination. Each split-gene vector functions on its own to produce the precursor split-proteins. The protein molecules A and B are spontaneously ligated effectively and specifically in a precise tail-to-head orientation by the intein sequences. Other methods of handling the packaging limits of AAV include the use of extremely small promoters (Flotte et al., 1993; Sarkar et al., 2003). But small promoters often result in lower gene expression and tissue specificity. Miniaturized genes have also been constructed by deleting putatively nonessential regions of large genes (Zhang et al., 1998; Wang et al., 2000; Harper et al., 2002; Qiao et al., 2005), but many genes may not tolerate large deletions without losing at least some known or unknown functions.
A potential drawback of the protein trans-splicing method is the use of intein sequences that may be immunogenic, although this problem was not observed in our studies of mdx mice. Should this problem occur, immunosuppression could be used to curb it. Alternatively, tissues with immune privilege such as the CNS and eye could be used as the preferred targets. In addition, one might be able to select the least immunogenic intein sequences from the large number of available and different split-inteins. Finally, the efficiency of trans-splicing may still improve. We have observed different splicing efficiencies with different split-inteins (Ssp DnaB split-intein vs. Rma DnaB split-intein) and with different split sites in the dystrophin protein. A future improvement of the protein trans-splicing method will be to achieve near 100% splicing efficiency through better choices of the split-intein and optimization of the split site in the host proteins. In summary, the protein trans-splicing method provides a novel alternative approach to overcome the packaging limit of gene vectors.
Acknowledgments
The authors thank Zhuma Hu and Jianan Gao for technical assistance. This work was supported by a grant from the Muscular Dystrophy Association (USA) and by a grant from the Canadian Institutes of Health Research to X.-Q.L, and by NIH grants to X.X.
Author Disclosure Statement
No competing financial interests exist.
References
- Chao H. Sun L. Bruce A. Xiao X. Walsh C.E. Expression of human factor VIII by splicing between dimerized AAV vectors. Mol. Ther. 2002;5:716–722. doi: 10.1006/mthe.2002.0607. [DOI] [PubMed] [Google Scholar]
- Chen L. Zhu F. Li J. Lu H. Jiang H. Sarkar R. Arruda V.R. Wang J. Zhao J. Pierce G.F. Ding Q. Wang X. Wang H. Pipe S.W. Liu X.Q. Xiao X. Camire R.M. Xiao W. The enhancing effects of the light chain on heavy chain secretion in split delivery of factor VIII gene. Mol. Ther. 2007;15:1856–1862. doi: 10.1038/sj.mt.6300268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D. Yue Y. Yan Z. Engelhardt J.F. A new dual-vector approach to enhance recombinant adeno-associated virus-mediated gene expression through intermolecular cis activation. Nat. Med. 2000;6:595–598. doi: 10.1038/75080. [DOI] [PubMed] [Google Scholar]
- Duan D. Yue Y. Engelhardt J.F. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: A quantitative comparison. Mol. Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- England S.B. Nicholson L.V. Johnson M.A. Forrest S.M. Love D.R. Zubrzycka-Gaarn E.E. Bulman D.E. Harris J.B. Davies K.E. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- Fisher K.J. Jooss K. Alston J. Yang Y. Haecker S.E. High K. Pathak R. Raper S.E. Wilson J.M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat. Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- Flotte T.R. Afione S.A. Solow R. Drumm M.L. Markakis D. Guggino W.B. Zeitlin P.L. Carter B.J. Expression of the cystic fibrosis transmembrane conductance regulator from a novel adeno-associated virus promoter. J. Biol. Chem. 1993;268:3781–3790. [PubMed] [Google Scholar]
- Harper S.Q. Hauser M.A. Dellorusso C. Duan D. Crawford R.W. Phelps S.F. Harper H.A. Robinson A.S. Engelhardt J.F. Brooks S.V. Chamberlain J.S. Modular flexibility of dystrophin: Implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- Hartigan-O'Connor D. Chamberlain J.S. Developments in gene therapy for muscular dystrophy. Microsc. Res. Tech. 2000;48:223–238. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<223::AID-JEMT10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Koenig M. Kunkel L.M. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J. Biol. Chem. 1990;265:4560–4566. [PubMed] [Google Scholar]
- Koenig M. Hoffman E.P. Bertelson C.J. Monaco A.P. Feener C. Kunkel L.M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Li J. Dressman D. Tsao Y.P. Sakamoto A. Hoffman E.P. Xiao X. rAAV vector-mediated sarcoglycan gene transfer in a hamster model for limb girdle muscular dystrophy. Gene Ther. 1999;6:74–82. doi: 10.1038/sj.gt.3300830. [DOI] [PubMed] [Google Scholar]
- Liu X.Q. Hu Z. A DnaB intein in Rhodothermus marinus: Indication of recent intein homing across remotely related organisms. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7851–7856. doi: 10.1073/pnas.94.15.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah C. Sarkar R. Zolotukhin I. Schleissing M. Xiao X. Kazazian H.H. Byrne B.J. Dual vectors expressing murine factor VIII result in sustained correction of hemophilia A mice. Hum. Gene Ther. 2003;14:143–152. doi: 10.1089/104303403321070838. [DOI] [PubMed] [Google Scholar]
- Nakai H. Storm T.A. Kay M.A. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nat. Biotechnol. 2000;18:527–532. doi: 10.1038/75390. [DOI] [PubMed] [Google Scholar]
- Phelps S.F. Hauser M.A. Cole N.M. Rafael J.A. Hinkle R.T. Faulkner J.A. Chamberlain J.S. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum. Mol. Genet. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- Qiao C. Li J. Zhu T. Draviam R. Watkins S. Ye X. Chen C. Li J. Xiao X. Amelioration of laminin-α2-deficient congenital muscular dystrophy by somatic gene transfer of miniagrin. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11999–12004. doi: 10.1073/pnas.0502137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar R. Xiao W. Kazazian H.H., Jr. A single adeno-associated virus (AAV)-murine factor VIII vector partially corrects the hemophilia A phenotype. J. Thromb. Haemost. 2003;1:220–226. doi: 10.1046/j.1538-7836.2003.00096.x. [DOI] [PubMed] [Google Scholar]
- Snyder R.O. Xiao X. Samulski R. Production of recombinant adeno-associated viral vectors. In: Dracopoli N., editor; Haines J., editor; Krof B., editor; Moir D., editor; Seidman C., editor; Seidman J., editor; Smith D., editor. Current Protocols in Human Genetics. John Wiley & Sons; New York: 1996. pp. 12.11.11–12.12.23. [Google Scholar]
- Snyder R.O. Spratt S.K. Lagarde C. Bohl D. Kaspar B. Sloan B. Cohen L.K. Danos O. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum. Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- Sun L. Li J. Xiao X. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nat. Med. 2000;6:599–602. doi: 10.1038/75087. [DOI] [PubMed] [Google Scholar]
- Wang B. Li J. Xiao X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watchko J. O'Day T. Wang B. Zhou L. Tang Y. Li J. Xiao X. Adeno-associated virus vector-mediated minidystrophin gene therapy improves dystrophic muscle contractile function in mdx mice. Hum. Gene Ther. 2002;13:1451–1460. doi: 10.1089/10430340260185085. [DOI] [PubMed] [Google Scholar]
- Wu H. Hu Z. Liu X.Q. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 1998a:959226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Xu M.Q. Liu X.Q. Protein trans-splicing and functional mini-inteins of a cyanobacterial dnaB intein. Biochim. Biophys. Acta. 1998b;1387:422–432. doi: 10.1016/s0167-4838(98)00157-5. [DOI] [PubMed] [Google Scholar]
- Xiao X. Li J. Samulski R.J. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J. Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X. Li J. Tsao Y.P. Dressman D. Hoffman E.P. Watchko J.F. Full functional rescue of a complete muscle (TA) in dystrophic hamsters by adeno-associated virus vector-directed gene therapy. J. Virol. 2000;74:1436–1442. doi: 10.1128/jvi.74.3.1436-1442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z. Zhang Y. Duan D. Engelhardt J.F. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Wang D. Fischer H. Fan P.D. Widdicombe J.H. Kan Y.W. Dong J.Y. Efficient expression of CFTR function with adeno-associated virus vectors that carry shortened CFTR genes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10158–10163. doi: 10.1073/pnas.95.17.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]


