Abstract
Aberrant promoter hypermethylation of several known or putative tumor suppressor genes occurs frequently during the malignant transformation in gliomas. We hypothesized that quantitative analysis of methylated genes will provide prognostic values in malignant glioma patients. We used an immunocapturing approach followed by real-time polymerase chain reaction analysis to detect altered patterns of promoter methylation in O-6-methylguanine-DNA methyltransferase (MGMT), p16INK4a, tissue inhibitor of metalloproteinase-3 (TIMP-3), and thrombospondin 1 (THBS1). The tumor tissue and paired serum as well as cerebrospinal fluid (CSF) from 66 patients with malignant gliomas were studied. Serum and CSF from 20 age-matched noncancer individuals were used as control. Promoter hypermethylation in MGMT, p16INK4a, TIMP-3, and THBS1 was detected at high frequencies in tumor tissue, serum, and CSF. None of the control serum or CSF showed aberrant methylation. Hypermethylation in serum and CSF DNA was all accompanied with methylation in the corresponding tumor tissues with 100% specificity. Highly elevated MGMT, p16INK4a, and THBS1 methylation levels in gliomas serum were the sole independent factors predicting inferior overall survival in this cohort. For progression-free survival, hypermethylation of MGMT and THBS1 in CSF were the independent prognostic factors. Multiple gene promoter hypermethylation analysis appears to be promising as a prognostic factor in glioma and as a mini-invasive tumor marker in serum and/or CSF DNA. Evaluation of these changes may help in selecting glioma patients for optimal adjuvant treatments and modifying chemotherapy.
Keywords: body fluids, epigenetic biomarker, glioma, prognosis, promoter methylation
Approximately 22 500 new cases of malignant primary brain tumors are diagnosed in the United States each year, among which nearly 70% are malignant gliomas.1,2 Although relatively uncommon, malignant gliomas are associated with disproportionately high morbidity and mortality. A grading scheme proposed by the WHO distinguishes 4 different grades of gliomas; of which, glioblastoma (GBM) WHO grade 4 is the most common (accounting for 60%–70% of all malignant gliomas) and the most malignant variant, with a median survival of only 12–15 months despite optimal treatment; while anaplastic gliomas WHO grade 3 including anaplastic astrocytomas (for 10%–15%), anaplastic oligodendrogliomas, and anaplastic oligoastrocytomas (AOA) (together for 10%) indicate a median survival of 2–5 years.1–3
The global hypomethylation of repetitive genomic sequences is a common alteration in cancer cells, and tissue-specific and imprinted genes can also show loss of DNA methylation. At the same time, the CpG islands of tumor-suppressor genes of the cancer undergo hypermethylation, which leads to the transcriptional inactivation of these genes and the loss of their normal cellular functions. This altered pattern of epigenetic modifications contributes to many of the hallmarks of cancer cells.
Promoter hypermethylation as a potential biomarker for early diagnosis as well as prognosis has been successfully used to detect neoplastic DNA in body fluids from several types of cancer, including sputum,4 bronchoalveolar lavage,5 serum, and plasma6 from lung cancer patients; urine sediment,7 blood, and ejaculates8 from prostate cancer patients; and ductal fluid9 and plasma10 from breast cancer patients.
To our knowledge, this approach has not been developed in body fluids from glioma patients. Therefore, we examined 4 tumor-related genes, O-6-methylguanine-DNA methyltransferase (MGMT), p16INK4a, tissue inhibitor of metalloproteinase-3 (TIMP-3), and thrombospondin-1 (THBS1), the expression of which is frequently silenced by aberrant methylation, not only in tumor tissues, but also in corresponding serum and cerebrospinal fluid (CSF) from glioma patients and noncancer controls. We used high-throughput methylated DNA immunoprecipitation (MeDIP) to immonocapture methylated genomic DNA and combined it with quantitative detection using promoter-specific real-time polymerase chain reaction (PCR).11 We then correlated our findings with disease states and clinicopathologic features in glioma patients, with overall survival (OS) as the primary end point.
Patients and Methods
Sample Collection and DNA Preparation
Tumor samples of 66 consented patients with pathologically confirmed malignant gliomas (WHO grades 3 and 4) were included in the present study. Patients underwent radical neurosurgery of primary tumors at the Department of Neurosurgery of Xijing Hospital (Xi'an, P.R. China), between January 2004 and December 2007. Patients aged 18–72 years treated with either gross total (57) or subtotal (9) removal of the tumor were eligible for the study. All of the patients had received stereotactic radiosurgery and a median of 3 courses of intravenous carmustine (100 mg/m2) given at 4-week intervals postoperatively. Major clinical and biologic characteristics of these patients are summarized in Table 1. Corresponding serum and CSF samples were obtained at the time of photographic diagnosis before surgery or other forms of treatment, and stored at −80°C before DNA extraction. Seum and CSF samples from 20 age-matched noncancer individuals were used as the control. Local institutional review board approval was obtained to use archived materials for research purposes.
Table 1.
Clinical characteristics of study sample
| WHO grade | n | Sex (M/F) | Histology/WHO grade | n | Median age (y) | Median PFS (m) | Number of tumor regrowth | Median OS (m) | Alive at LO |
|---|---|---|---|---|---|---|---|---|---|
| 3 | 23 | 16/7 | AA/3 | 18 | 45 (18–71) | 9.3 (1.0–28.7) | 5/18 | 12.1 (4.4–28.7) | 2/18 |
| AOA/3 | 3 | 56 (21–68) | 25.1 (23.5–41.0) | 2/3 | 25.7 (25.1–41.0) | 2/3 | |||
| AO/3 | 2 | 34 (32–36) | 10.3 (10.3–28.2) | 2/2 | 10.3 (10.3–28.2) | 1/2 | |||
| 4 | 43 | 24/19 | pGBM/4 | 35 | 59 (18–72) | 9.8 (2.0–22.8) | 17/35 | 10.6 (2.4–22.8) | 3/35 |
| sGBM/4 | 8 | 47 (22–61) | 6.9 (1.1–11.9) | 4/8 | 7.0 (1.1–17.5) | 1/8a |
Abbreviations: n, case number; M, male; F, female; y, years; m, months; LO, last observation; AA, anaplastic astrocytoma; AOA, anaplastic oligoastrocytoma; AO, anaplastic oligodendroglioma; pGBM, primary glioblastoma; sGBM, secondary glioblastoma.
aDeath of 1 patient not tumor related.
We prepared genomic DNA from tissue and serum/CSF samples by overnight proteinase K treatment, phenol–chloroform extraction, ethanol precipitation, and RNase digestion.11
Methylated DNA Immunoprecipitation
MeDIP was performed as previously described.11 Briefly, we used 2 µg of sonicated (300–1000 bp) genomic DNA fragments as a starting material, then denatured the DNA for 10 minutes at 95°C, and immunoprecipitated it overnight at 4°C with 4 µg antibody against 5-methyl-cytidine (Eurogentec). After incubation with 60 µL of rabbit anti-IgG magnetic beads (BioLabs S1430S) for 2 hours at 4°C, the mixture was washed with 1 mL of immunoprecipitation buffer (10 mM sodium phosphate [pH 7.0], 140 mM NaCl, 0.05% Triton X-100). The beads were treated with proteinase K for 3 hours at 50°C and the methylated DNA was recovered by phenol–chloroform extraction followed by ethanol precipitation.
Real-time PCR on MeDIP Samples
We carried out real-time PCR12 with 20 ng immunoprecipitated methylated DNA and one-fifth of input DNA. We used the SYBR Green PCR master mix (Applied Biosystems) and an ABI PriSM 7700 Sequence Detection System to perform real-time PCR. Primers used were as follows: MGMT-F: 5′-CAGGACCGGGATTCTCACTA-3′, MGMT-R: 5′-CCAAATGGCCCGTACCTT-3′; p16INK4a-F: 5′-TCTTCCACATCACCGATCCTT-3′, p16INK4a-R: 5′-TCCTTTCCTTGCCCTGCTT-3′; TIMP-3-F: 5′-TTGGGGCGGAGTGGAGAA-3′, TIMP-3-R: 5′- CCGTTAGTAGTGAATGGGGACA-3′; THBS1-F: 5′- GCGCTGAGGCTTCAGTCC-3′; and THBS1-R: 5′- GGTGTCCTGATGAGTTGGTTTG-3′. Reactions were done in duplicates and standard curves were calculated on serial l0-fold dilutions (1 × 10−1–1 × 10−8) of input genomic DNA. The ratios of the signals in the immunoprecipitated DNA vs input DNA were calculated as a measure for representing the relative enrichment of methylation in the particular sample. The resulting values were standardized against the unmethylated control CpGenome Universal Unmethylated DNA (Milipore).
Statistical Analysis
Follow-up data were updated in February 2009. The OS was defined as the time from the date of surgery to date of death as a result of any cause. Progression-free survival (PFS) was calculated from the date of surgery to date of documented tumor recurrence or further growth of residual tumor or death from any cause. Patients who were alive at the date of the last follow-up were censored on that date plus 1 day. Probability of survival was estimated using the Kaplan–Meier method and compared using the log-rank test. To test whether variables differed across groups, the χ2 test or Fisher's exact test was used according to the test condition. A Cox proportional hazards model was performed to establish independent factor(s) for survival. The maximal χ2 method was adapted to determine which methylation value best segregated patients into poor and good prognosis subgroups (in terms of likelihood of surviving).13 Statistical significance was defined as P < .05. All of the tests are 2-sided. Statistical analysis was performed using the SPSS software package, version 16.0 (SPSS, Inc.).
Results
Gene Promoter Hypermethylation in Tumor Tissue, Serum, and CSF DNA
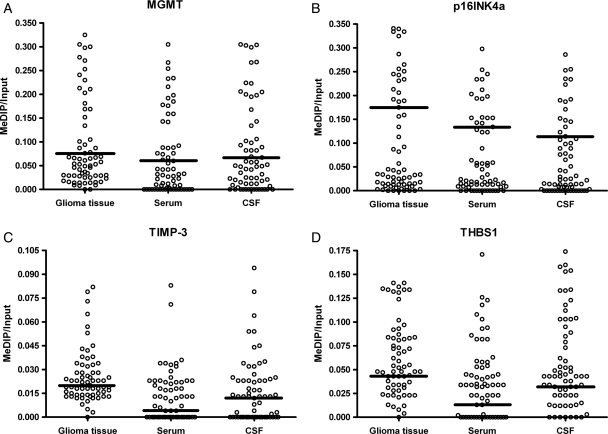
Initially, 85 patients were included according to the inclusion criteria, but patients lost to follow-up were excluded before the current experiment of hypermethylation detection. The survival analysis showed no statistically significant difference between 66 finally included patients and all 85 patients. We collected tumor, serum, and CSF samples of malignant gliomas from 66 patients who underwent curative resection (Table 1), as well as control serum and CSF from 20 age-matched noncancer individuals. Methylation levels of 4 tumor-related genes in tumor, serum, and CSF of glioma patients are shown in Fig. 1.
Fig. 1.
Methylation levels of MGMT, p16INK4a, TIMP-3, and THBS1 in glioma tumor tissue as well as paired serum and CSF DNA of malignant glioma patients. Calculation of the MeDIP gene of interest/input gene of interest ratios was based on the fluorescence emission intensity values for both the immunoprecipitated methylated DNA and input genomic DNA of each individual gene and sample obtained by quantitative real-time PCR analysis. Black bars denote the calculated cutoff values that best segregated patients into good and poor prognostic groups.
Hypermethylation in serum or CSF DNA was never detected if this alteration in the corresponding genes was not present in the primary tumor tissue. Moreover, methylated DNA was not detected in 40 control serum and CSF samples from noncancer individuals. None of the control serum or CSF samples had hypermethylated the DNA detected, whereas robust amplification of the interest genes from input DNA was documented. All serum and CSF samples from glioma patients showed hypermethylation in at least 1 of the 4 genes with 100% specificity. The frequency and median methylation values for each gene in tumor tissue, serum, and CSF DNA are listed in Table 2.
Table 2.
Methylation levels in tumor tissue, serum and CSF correlates with survival
| Genes | Number of patients with positive methylation (%) | Median | Range | Cutoff | Good prognostic group |
Poor prognostic group |
P value | ||
|---|---|---|---|---|---|---|---|---|---|
| n | Median OS (95% CI) (m) | n | Median OS (95% CI) (m) | ||||||
| MGMT | |||||||||
| Tumor tissue | 64 (97.0) | 0.054 | 0–0.325 | 0.076 | 42 | 13.5 (9.2–17.8) | 24 | 7.0 (5.2–8.8) | <0.0001 |
| Serum | 47 (71.2) | 0.031 | 0–0.305 | 0.060 | 42 | 14.6 (9.9–19.3) | 24 | 6.5 (4.6–8.4) | <0.0001 |
| CSF | 52 (78.8) | 0.048 | 0–0.304 | 0.067 | 40 | 14.3 (8.6–20.0) | 26 | 6.7 (4.8–8.6) | <0.0001 |
| p16INK4a | |||||||||
| Tumor tissue | 60 (90.9) | 0.034 | 0–0.341 | 0.175 | 47 | 13.5 (10.1–16.9) | 19 | 6.7 (5.0–8.4) | <0.0001 |
| Serum | 52 (78.8) | 0.022 | 0–0.298 | 0.134 | 49 | 12.5 (9.2–15.8) | 17 | 6.5 (4.8–8.2) | <0.0001 |
| CSF | 49 (74.2) | 0.024 | 0–0.286 | 0.114 | 50 | 12.4 (9.3–15.5) | 16 | 6.5 (4.0–9.0) | <0.0001 |
| TIMP3 | |||||||||
| Tumor tissue | 65 (98.5) | 0.021 | 0–0.082 | 0.020 | 32 | 12.1 (8.7–15.5) | 34 | 9.3 (7.3–11.3) | 0.025 |
| Serum | 39 (59.1) | 0.009 | 0–0.083 | 0.004 | 26 | 11.3 (4.3–18.3) | 40 | 10.7 (9.6–11.8) | 0.022 |
| CSF | 41 (62.1) | 0.013 | 0–0.094 | 0.012 | 31 | 11.3 (9.7–12.9) | 35 | 10.8 (9.4–12.2) | 0.303 |
| THBS1 | |||||||||
| Tumor tissue | 65 (98.5) | 0.050 | 0–0.141 | 0.043 | 22 | 16.2 (8.4–24.0) | 44 | 9.6 (8.1–11.1) | 0.001 |
| Serum | 48 (72.7) | 0.034 | 0–0.171 | 0.013 | 20 | 20.5 (14.1–26.9) | 46 | 9.6 (8.0–11.2) | <0.0001 |
| CSF | 59 (89.4) | 0.043 | 0–0.174 | 0.032 | 22 | 16.2 (11.3–21.1) | 44 | 10.1 (8.7–11.5) | 0.002 |
At least one of the genes with methylation was detected in the tumor, serum and CSF of all the 66 glioma patients with 100% specificity.
Abbreviations: n, patients number; m, months.
Overall, identical methylation patterns were found in the serum, CSF, and corresponding tumor DNA. Additionally, hypermethylation levels of each single promoter in serum, CSF, and tumor were all associated within the same patient, respectively (Pearson's correlation coefficient, r ranged from 0.349 to 0.963; P < .0001). Hypermethylation of MGMT in tumor (Spearman's correlation coefficient, r = 0.422; P < .0001) and CSF (r = 0.373; P = .002), p16INK4a in tumor (r = 0.369; P = .002), serum (r = 0.297; P = .015), and CSF (r = 0.311; P = .011), TIMP-3 in tumor (r = 0.338; P = .006), THBS1 in tumor (r = 0.355; P = .003) were positively correlated with increasing WHO grades. No additional significant relationship was shown between the methylation status and patient clinical characteristics.
Epigenetic Alterations and Survival
The median follow-up was 11.3 months. A total of 57 patients were dead at the time of this analysis, and 9 were alive, including 2 patients alive with tumor recurrence. The median OS and PFS of the whole population were 11.2 months (95% CI, 10.4–12.0 months) and 11.0 months (95%CI, 8.0–14.0 months), respectively. The survival data of different WHO grades and histologies are summarized in Table 1.
Cutoff values representing methylation ratios indicated in Table 2 best segregated patients into good and poor prognostic groups. Except for TIMP-3 in CSF, all the low promoter methylation levels in tumor tissue, serum, and CSF correlated with a better survival (Table 2). Furthermore, for each single gene, there was no statistically significant difference between the classification results when applying different cutoff values in tumor, serum, and CSF.
Cox proportional hazards models including age, sex, WHO grade, histology, and methylation status-defined patient subgroups were analyzed to evaluate independent factors for OS and PFS. A very strong trend for age as an independent prognostic factor was observed, but only methylation status of MGMT, p16INK4a, and THBS1 in serum was independent prognostic factor of importance for OS. Intriguingly, methylation status of MGMT and THBS1 in CSF was found to influence PFS significantly and independently (Table 3).
Table 3.
Cox proportional hazards regressions for PFS and OS
| Variable | PFS |
OS |
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age ≥50 y | 1.53 (0.60–3.93) | .372 | 2.02 (0.99–4.09) | .053 |
| Sex: male vs female | 0.87 (0.40–1.87) | .713 | 0.83 (0.44–1.57) | .569 |
| WHO grade: grade 4 vs grade 3 | 0.90 (0.35–2.28) | .821 | 1.87 (0.84–4.17) | .126 |
| Histology: astrocytic vs oligodendrocytic/oligoastrocytic | 5.42 (0.55–53.59) | .148 | 3.18 (0.56–18.17) | .193 |
| Methylation in tumor tissue (high vs low) | ||||
| MGMT | 1.31 (0.45–3.83) | .627 | 2.04 (0.94–4.41) | .070 |
| p16INK4a | 0.90 (0.30–2.71) | .852 | 3.25 (0.65–16.22) | .151 |
| TIMP-3 | 1.49 (0.72–3.07) | .285 | 1.12 (0.64–1.96) | .702 |
| THBS1 | 2.28 (0.57–2.87) | .553 | 1.27 (0.63–2.57) | .499 |
| Methylation in serum (high vs low) | ||||
| MGMT | 1.20 (0.41–3.46) | .739 | 13.46 (5.12–35.40) | <.0001 |
| P16INK4a | 1.17 (0.36–3.79) | .800 | 5.92 (2.31–15.12) | <.0001 |
| TIMP-3 | 1.97 (0.82–4.73) | .129 | 1.94 (1.00–3.78) | .052 |
| THBS1 | 1.73 (0.68–4.41) | .250 | 2.58 (1.16–5.75) | .020 |
| Methylation in CSF (high vs low) | ||||
| MGMT | 8.10 (1.44–45.53) | .018 | 1.62 (0.92–2.88) | .098 |
| p16INK4a | 1.00 (0.17–5.73) | .570 | 1.93 (0.96–3.84) | .064 |
| TIMP-3 | 0.67 (0.30–1.48) | .320 | 0.67 (0.35–1.29) | .234 |
| THBS1 | 1.52 (0.60–3.81) | .049 | 1.56 (0.79–3.12) | .203 |
Since patients with anaplastic gliomas (WHO grade 3) have an apparently different prognosis compared with those with GBM (WHO grade 4), we further analyzed each subgroup separately. Similar results have been reached comparing with the whole data set that all the low promoter methylation levels in tumor tissue, serum, and CSF correlate with a better survival (Table 4). When applying Cox models, similar results compared with the whole data set were gained again in the GBM group that only hypermethylation of MGMT and THBS1 in serum were independent prognostic factors for inferior OS, and that hypermethylation of MGMT and THBS1 in CSF were independent prognostic factors for inferior PFS (Table 5); while in the anaplastic gliomas group, a very strong trend for hypermethylation of p16INK4a in serum as a prognostic factor for OS (P = .052) was observed, but no factor in the models were found to influence survival significantly and independently.
Table 4.
Methylation levels in tumor tissue, serum, and CSF correlates with survival in WHO grades 3 and 4 glioma separately
| Genes | Number of patients with positive methylation (%) | Median | Range | Cutoff | Good prognostic group |
Poor prognostic group |
P value | ||
|---|---|---|---|---|---|---|---|---|---|
| n | Median OS (95% CI) (m) | n | Median OS (95% CI) (m) | ||||||
| MGMT | |||||||||
| Tumor tissue/3 | 21 (91.3) | 0.029 | 0–0.213 | 0.064 | 17 | 19.3 (14.9–23.7) | 6 | 10.3 (6.9–13.7) | <.0001 |
| 4 | 43 (100) | 0.069 | 0.008–0.325 | 0.069 | 21 | 12.4 (10.6–14.2) | 22 | 7.0 (5.3–8.7) | .001 |
| Serum/3 | 16 (70.0) | 0.012 | 0–0.159 | 0.067 | 18 | 17.8 (13.4–22.2) | 5 | 10.3 (6.2–14.4) | <.0001 |
| 4 | 31 (72.1) | 0.042 | 0–0.305 | 0.060 | 23 | 12.4 (10.5–14.3) | 20 | 6.5 (4.1–8.9) | <.0001 |
| CSF/3 | 15 (65.2) | 0.015 | 0–0.200 | 0.058 | 17 | 19.3 (14.9–23.7) | 6 | 10.3 (6.9–13.7) | <.0001 |
| 4 | 37 (86.0) | 0.067 | 0–0.304 | 0.069 | 22 | 11.6 (9.6–13.6) | 21 | 6.5 (4.6–8.4) | <.0001 |
| p16INK4a | |||||||||
| Tumor tissue /3 | 18 (78.3) | 0.015 | 0–0.287 | 0.156 | 19 | 17.8 (14.7–20.9) | 4 | 7.2 (3.3–11.1) | <.0001 |
| 4 | 42 (97.7) | 0.057 | 0–0.341 | 0.206 | 30 | 11.3 (10.1–12.5) | 13 | 6.5 (5.1–7.9) | <.0001 |
| Serum/3 | 15 (65.2) | 0.013 | 0–0.193 | 0.128 | 19 | 17.8 (14.7–20.9) | 4 | 7.2 (3.3–11.1) | <.0001 |
| 4 | 37 (86.0) | 0.053 | 0–0.298 | 0.142 | 30 | 11.3 (10.1–12.5) | 13 | 6.5 (5.1–7.9) | <.0001 |
| CSF/3 | 14 (60.9) | 0.008 | 0–0.223 | 0.114 | 20 | 17.2 (15.7–18.7) | 3 | 7.2 (2.7–11.7) | <.0001 |
| 4 | 35 (81.4) | 0.051 | 0–0.286 | 0.134 | 31 | 11.3 (10.2–12.4) | 12 | 5.4 (3.4–7.4) | <.0001 |
| TIMP3 | |||||||||
| Tumor tissue /3 | 22 (95.7) | 0.018 | 0–0.079 | 0.023 | 17 | 17.2 (16.0–18.4) | 6 | 8.4 (2.8–14.0) | .359 |
| 4 | 43 (100) | 0.023 | 0.008–0.082 | 0.015 | 9 | 13.5 (5.6–21.4) | 34 | 9.3(7.9–10.7) | .088 |
| Serum/3 | 12 (52.2) | 0.007 | 0–0.083 | 0.007 | 10 | 20.5 (0–41.9) | 13 | 16.9 (10.8–23.0) | .100 |
| 4 | 27 (62.8) | 0.012 | 0–0.071 | 0.004 | 16 | 10.6 (5.1–16.1) | 27 | 9.7 (8.3–11.1) | .079 |
| CSF/3 | 11 (47.8) | 0 | 0–0.079 | 0.013 | 15 | 17.1 (9.8–24.4) | 8 | 16.9 (8.7–25.1) | .656 |
| 4 | 30 (70.0) | 0.013 | 0–0.094 | 0.004 | 15 | 10.6 (9.4–11.8) | 28 | 9.3 (6.8–11.8) | .586 |
| THBS1 | |||||||||
| Tumor tissue/3 | 23 (100) | 0.034 | 0.004–0.137 | 0.024 | 8 | 20.5 (11.7–26.5) | 15 | 11.3 (10.4–12.2) | .014 |
| 4 | 42 (97.7) | 0.069 | 0–0.141 | 0.043 | 10 | 12.5 (7.9–17.1) | 33 | 9.3 (7.5–11.1) | .025 |
| Serum/3 | 14 (60.9) | 0.013 | 0–0.171 | 0.013 | 11 | 25.1 (17.5–32.7) | 12 | 11.3 (10.1–12.5) | .001 |
| 4 | 34 (79.1) | 0.035 | 0–0.126 | 0.020 | 10 | 16.2 (12.6–19.8) | 33 | 9.3 (6.7–11.9) | .002 |
| CSF/3 | 19 (82.6) | 0.029 | 0–0.174 | 0.029 | 11 | 20.5 (12.2–28.8) | 12 | 11.3 (10.1–12.5) | .046 |
| 4 | 40 (93.0) | 0.043 | 0–0.160 | 0.034 | 12 | 11.3 (7.0–15.6) | 31 | 9.3 (7.0–11.6) | .109 |
Abbreviations: n, patients number; m, months; 3, WHO grade 3 glioma, including anaplastic astrocytoma, anaplastic oligoastrocytoma, and anaplastic oligodendroglioma; 4, WHO grade 4, glioblastoma.
Table 5.
Cox proportional hazards regressions for PFS and OS in patients with glioblastomas
| Variable | PFS |
OS |
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age ≥50 y | 1.50 (0.73–2.56) | .553 | 1.48 (0.57–3.85) | .417 |
| Sex: male vs female | 0.51 (0.13–1.98) | .331 | 0.57 (0.24–1.37) | .208 |
| Methylation in tumor tissue (high vs low) | ||||
| MGMT | 1.43 (0.19–10.70) | .058 | 2.22 (0.67–3.06) | .415 |
| p16INK4a | 1.87 (0.73–2.59) | .946 | 6.07 (0.85–43.43) | .408 |
| TIMP-3 | 2.95 (0.45–19.46) | .261 | 1.29 (0.22–2.72) | .694 |
| THBS1 | 1.35 (0.28–6.59) | .708 | 0.94 (0.14–1.44) | .179 |
| Methylation in serum (high vs low) | ||||
| MGMT | 2.36 (0.19–29.43) | .505 | 8.25 (1.69–40.41) | .009 |
| p16INK4a | 1.00 (0.20–3.04) | .904 | 1.04 (0.05–17.25) | .989 |
| TIMP-3 | 2.17 (0.51–9.32) | .298 | 1.53 (0.50–4.65) | .453 |
| THBS1 | 1.60 (0.41–6.21) | .498 | 6.52 (1.86–22.86) | .003 |
| Methylation in CSF (high vs low) | ||||
| MGMT | 19.12 (1.58–231.47) | .020 | 2.77 (0.37–20.81) | .321 |
| p16INK4a | 1.68 (0.10–12.85) | .920 | 1.07 (0.07–12.40) | .956 |
| TIMP-3 | 1.09 (0.14–19.34) | .334 | 1.12 (0.35–2.28) | .810 |
| THBS1 | 1.23 (0.61–3.88) | .032 | 1.06 (0.37–2.44) | .905 |
Discussion
Malignant gliomas typically contain both neoplastic and stromal tissues, which lead to their histologic heterogeneity and thus the discrepancies between pathological grades and clinical outcomes. Molecular studies such as quantitative gene promoter hypermethylation analyses potentially allow for better classification of these tumors and separation of the patients into different prognostic groups. Since malignant transformation in gliomas results from the sequential accumulation of genetic aberrations that are partly modulated by different gene promoter hypermethylation at different stages of tumor initiation and progression,3,14,15 here we investigated aberrant methylation of 4 tumor-related genes that affect several steps of tumor cell biology at different stages in cancer development.
The DNA repair gene MGMT is located on chromosome 10q26 and encodes a protein that removes alkyl groups from the O6 position of guanine, an important site of DNA alkylation, and thus protects the cell from alkylating agents.16 Previous reports showed 30%–80% methylation in primary tumor tissues and glioma cell lines,17–22 while our finding is a bit higher. These discrepancies may be a result of differences in the methylation assays used and the inclusion of tumors with different stages and grades.
From a clinical standpoint, MGMT promoter hypermethylation might be a useful predictor of the chemosensitivity of tumors to alkylating agents. According to previous studies, MGMT promoter hypermethylation could silence the gene, thus decreasing DNA repair activity and increasing the susceptibility of the tumor cells to carmustine17 or temozolomide,20 which were associated with prolonged OS and PFS.17,20 This is of particular importance since carmustine/temozolomide is currently used in the treatment of malignant gliomas regardless of MGMT promoter methylation status. If the significance of MGMT promoter methylation could be further confirmed, patients with unfavorable MGMT methylation status might be selected for other regimens or combinations of carmustine/temozolomide with MGMT inhibitors. But our results failed to confirm MGMT hypermethylation as a favorable prognostic factor.
With the perception that acquisition of MGMT methylation is associated with a grim prognosis from our findings, we searched for a possible explanation for this disparity. Somewhat contradictorily, however, MGMT hypermethylation alone is not a good prognostic factor per se. In fact, it signifies poor prognosis, probably owing to the fact that tumors with epigenetic silencing of MGMT accumulate more mutations.15 Our observation fits the concept that gene silencing by promoter hypermethylation is, in general, a poor prognostic factor, and it occurs more frequently in tumors with increased stages.15,23–25 Although a subset of patients in our cohort have not received standard carmustine/temozolomide treatment, which may partly account for the poor prognosis, we speculate that the idea that MGMT promoter methylation is the only relevant molecular factor in the tumor's chemosensitivity is probably an oversimplification given that the metabolism of chemotherapeutic drugs may be a complicated “network” in which MGMT is only one of the key points.
Cell cycle regulator p16INK4a promoter methylation was frequently found in tumor tissue, serum, and CSF of malignant gliomas (90.9%, 78.8%, and 74.2%, respectively), a rate much higher than that found in previous studies.18,26
Metastatic suppressor gene TIMP-3 encodes the third member of the TIMP family of proteins and is believed to play a significant role in suppressing extracellular matrix remodeling during tumor growth, angiogenesis, invasion, and metastasis.27 Our finding of TIMP-3 methylation is higher than that from other studies based on conventional methylation-specific PCR in primary brain tumors.19,27
Angiogenesis inhibitor THBS1 is an extracellular matrix glycoprotein that influences cell adhesion, motility, and growth, and whose altered expression is associated with neovascularization in human cancers.28 THBS1 was found to be hypermethylated in 17%–61% of primary gliomas,19,29 compared with 72.7%–89.4% methylation in serum and CSF DNA reported in the present study.
The detection of tumor molecular signatures in body fluids has implications for the identification of poor prognosis patients and high-risk patients with preinvasive or early-stage lesions and for monitoring residual disease.4–10 In our study, multiple gene promoter hypermethylation were analyzed, for the first time, in body fluids of glioma patients who had undergone resection. By applying a maximal χ2 method, we divided the patients into good and poor prognostic subgroups with different cutoff values of the 4 genes detected in tumor tissue, serum, and CSF, respectively. The results suggested that for each single gene, the classification results were of no statistically significant difference. Besides, it is noteworthy that detection of promoter methylation in serum/CSF is a specific event in that (i) hypermethylation was not detected in any of the 20 control serum and (ii) the same methylation profiles were found in the serum, CSF, and the corresponding tumor, while hypermethylation was not detected in the serum or CSF of glioma patients without methyaltion in the corresponding tumor. More interestingly, when we further used Cox models to determine independent prognostic factors in this cohort, the methylation status of MGMT, p16INK4a, and THBS1 in serum were retained as independent prognostic factors for OS, and methylation status of MGMT and THBS1 in CSF predicts PFS independently.
Furthermore, our observations have 3 potential clinical applications: (i) identifying aberrant gene promoter methylation in the serum and CSF may help make decisions about optimal therapy. Though WHO grade is the most important prognostic factor for glioma patients, the broad differences of treatment response and survival within grades suggest that the aggressiveness of treatment should not be decided simply by grade. In our cohort, the prognosis of patients was predicted by epigenetic alterations other than WHO grade. Therefore, for those with worse prognosis as indicated by epigenetic profiles, more aggressive adjuvant therapy could be considered. In addition, to date, the gripping evidence that MGMT hypermethylation is the best independent predictor of glioma patients' response to alkylating agents17 and temozolomide20 underlies the importance of improving our understanding of the methylation status correlates of responses to treatments. Compared with previous studies using tumor tissue, body fluids used in our study are easily available through noninvasive or mini-invasive way and for patients unable or unwilling to receive surgery, the purpose to modify chemotherapeutic treatment can still be achieved; and (ii) the detection of aberrant methylation in serum and CSF DNA may offer a promising approach for the mini-invasive diagnosis of glioma. This is of particular importance to patients with early-stage gliomas that are hard to diagnose by neuroimaging. However, as GBM can be separated into 2 main subtypes on the basis of biologic and genetic differences,3,14 primary GBM, which typically occurs in patients older than 50 years and without the transformation period from previous low-grade gliomas, still highlights the value of early diagnosis; and (iii) it would hold much promise to investigate whether the detection of aberrant methylation in the serum/CSF can be used in disease monitoring after curative surgery. If methylated DNA disappears in body fluids shortly after curative surgery, the reappearance of these markers may suggest recurrence or progression of disease that may require more intensive screening and aggressive treatment.6 Although our primary results need to be confirmed in larger and longitudinal studies, detection of aberrant promoter hypermethylation of tumor-related genes in serum and CSF of patients with malignant gliomas may be useful for glioma prognosis evaluation, diagnosis as well as the detection of recurrence or progression. We recommend that microarray techniques be applied in future investigations to determine (i) whether MGMT promoter methylation is the only relevant molecular factor in tumor's chemosensitivity; (ii) if any other interesting genes need more attention; and (iii) how much MGMT could function in the probable network of chemotherapeutic drug's metabolism, making it possible to select the most appropriate therapies on the basis of epigenetic profiles of the patients' tumors.
Traditionally, conversion of ummethylated cytosine with bisulfite followed by quantitative methylation-specific PCR provides unbiased and sensitive detection of methylated DNA; however, it is laborious and cannot easily be applied to screening a large set of sequences or samples.30 Here, we used an antibody specific for 5-methyl-cytidine after random fragmentation to immunocapture methylated genomic fragments, which permits highly efficient and specific enrichment of methylated DNA.11 The resulting enrichment in the immunoprecipitated fraction is determined by standard DNA detection including real-time PCR, which does not require particular bisulfite treatment or 2 pairs of PCR primers. Our results showed that this approach has an increased sensitivity as well as good specificity. Furthermore, the fragmented DNA used in our study has a size range similar to DNA after isolation from formalin-fixed tissues. Thus, this approach enables screening of stored clinical samples and holds more promise for large-scale and high-throughput analysis, especially in clinical application.
In conclusion, our results highlight the role of epigenetic changes involving multiple gene promoter hypermethylation in glioma, and that the detection of methylation status in serum and CSF of patients with malignant gliomas has a prognostic value. Moreover, this approach has many potential clinical applications including primary diagnosis, selecting treatment strategy, monitoring for disease progression and evolution, and measurement of therapeutic response, which need further investigation.
Conflict of interest statement. None declared.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. World Health Organization Classification of Tumours of the Central Nervous System. Lyon: IARC Press; 2007. [Google Scholar]
- 2.Central Brain Tumor Registry US. Central Brain Tumor Registry of the United States (2008) Statistical report: Primary Brain Tumors in the United States 2000–2004 (Years Data Collected) 2008 http://wwwcbtrusorg/reports/2007-2008/2007reportpdf . [Google Scholar]
- 3.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 4.Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topaloglu O, Hoque MO, Tokumaru Y, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–2288. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 6.Usadel H, Brabender J, Danenberg KD, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002;62:371–375. [PubMed] [Google Scholar]
- 7.Hoque MO, Topaloglu O, Begum S, et al. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J Clin Oncol. 2005;23:6569–6575. doi: 10.1200/JCO.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Goessl C, Muller M, Heicappell R, Krause H, Miller K. DNA-based detection of prostate cancer in blood, urine, and ejaculates. Ann N Y Acad Sci. 2001;945:51–58. doi: 10.1111/j.1749-6632.2001.tb03863.x. [DOI] [PubMed] [Google Scholar]
- 9.Fackler MJ, Malone K, Zhang Z, et al. Quantitative multiplex methylation-specific PCR analysis doubles detection of tumor cells in breast ductal fluid. Clin Cancer Res. 2006;12:3306–3310. doi: 10.1158/1078-0432.CCR-05-2733. [DOI] [PubMed] [Google Scholar]
- 10.Silva JM, Dominguez G, Garcia JM, et al. Presence of tumor DNA in plasma of breast cancer patients: clinicopathological correlations. Cancer Res. 1999;59:3251–3256. [PubMed] [Google Scholar]
- 11.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 12.Boehm D, Herold S, Kuechler A, Liehr T, Laccone F. Rapid detection of subtelomeric deletion/duplication by novel real-time quantitative PCR using SYBR-green dye. Hum Mutat. 2004;23:368–378. doi: 10.1002/humu.20011. [DOI] [PubMed] [Google Scholar]
- 13.Hothorn T, Zeileis A. Generalized maximally selected statistics. Biometrics. 2008;64:1263–1269. doi: 10.1111/j.1541-0420.2008.00995.x. [DOI] [PubMed] [Google Scholar]
- 14.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 16.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 17.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 18.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 19.Alonso ME, Bello MJ, Gonzalez-Gomez P, et al. Aberrant promoter methylation of multiple genes in oligodendrogliomas and ependymomas. Cancer Genet Cytogenet. 2003;144:134–142. doi: 10.1016/s0165-4608(02)00928-7. [DOI] [PubMed] [Google Scholar]
- 20.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 21.Lavon I, Zrihan D, Zelikovitch B, et al. Longitudinal assessment of genetic and epigenetic markers in oligodendrogliomas. Clin Cancer Res. 2007;13:1429–1437. doi: 10.1158/1078-0432.CCR-06-2050. [DOI] [PubMed] [Google Scholar]
- 22.Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Gerson SL. Targeted modulation of MGMT: clinical implications. Clin Cancer Res. 2006;12:328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- 24.Soejima H, Zhao W, Mukai T. Epigenetic silencing of the MGMT gene in cancer. Biochem Cell Biol. 2005;83:429–437. doi: 10.1139/o05-140. [DOI] [PubMed] [Google Scholar]
- 25.Oue N, Mitani Y, Motoshita J, et al. Accumulation of DNA methylation is associated with tumor stage in gastric cancer. Cancer. 2006;106:1250–1259. doi: 10.1002/cncr.21754. [DOI] [PubMed] [Google Scholar]
- 26.Costello JF, Berger MS, Huang HS, Cavenee WK. Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res. 1996;56:2405–2410. [PubMed] [Google Scholar]
- 27.Bachman KE, Herman JG, Corn PG, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 28.Roberts DD. Regulation of tumor growth and metastasis by thrombospondin-1. FASEB J. 1996;10:1183–1191. [PubMed] [Google Scholar]
- 29.Li Q, Ahuja N, Burger PC, Issa JP. Methylation and silencing of the thrombospondin-1 promoter in human cancer. Oncogene. 1999;18:3284–3289. doi: 10.1038/sj.onc.1202663. [DOI] [PubMed] [Google Scholar]
- 30.Rakyan VK, Hildmann T, Novik KL, et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]