Abstract
Molecular alterations in glioblastoma have the potential to guide treatment. Here, we explore the relationship between temozolomide (TMZ) response and O6-methylguanine DNA methyltransferase (MGMT) status in brain tumor initiating cells (BTICs). Methylation, expression, and sensitivity were assessed in 20 lines; associations were evaluated by Fisher's exact test. Some BTICs were sensitive. Sensitivity to TMZ was only associated with protein expression (P = .001). There were atypical BTICs including TMZ-resistant lines in which the methylation-specific PCR reaction revealed both methylated and unmethylated bands. BTICs are not uniformly resistant to TMZ; some are sensitive. MGMT status does not predict TMZ response with high precision.
Keywords: brain tumor stem cells, glioblastoma, MGMT methylation, temozolomide
In 2005, silencing of the O6-methylguanine DNA methyltransferase (MGMT) gene by promoter methylation emerged as a potentially useful test to identify patients with glioblastoma (GBM) who would benefit from adding temozolomide (TMZ) chemotherapy to radiotherapy.1,2 Despite the compelling rationale to personalize cancer treatment and data supporting the association between MGMT methylation and benefit from TMZ, MGMT testing is not used clinically. Oncologists have been reluctant to adopt such a test because in the study by Hegi et al.,2 there were several patients with unmethylated tumors who had long survival times after treatment with TMZ and many patients with methylated tumors who had short survival despite TMZ. The failed promise of a predictive test prompted us to explore the relationship between MGMT status and TMZ sensitivity in brain tumor initiating cell (BTIC) lines, which may be a disease reservoir in GBM.3 We used permanent cell lines that were established using the neurosphere culture method developed at the University of Calgary by Reynolds and Weiss.4 These GBM-initiating cell lines, 16 of which express the surface marker CD133, have retained the cardinal features of stem cells, including the ability to self-renew and to differentiate into multiple neural cell lineages.5 Furthermore, they also possess the signature characteristics of transformed cells, including continuous proliferation, growth factor-independent replication, and tumor formation in immune-deficient mice.5 Additionally, they harbor the hallmark molecular alterations that occur in GBM tissues, including p53, PTEN, and epidermal growth factor receptor mutations6 (data not shown). This resource afforded us the opportunity to rigorously evaluate the link between response to TMZ and MGMT methylation in a cell type that may be important to the maintenance of GBM tumors.
Materials and Methods
Cell Culture
All BTIC lines were derived from patients with GBMs. They were established in the laboratory of S.W. (Hotchkiss Brain Institute, University of Calgary) and maintained in basal neural stem cell media plus EGF, FGF, and heparin, as recommended by the manufacturer (Stem Cell Technologies) and described elsewhere.5
Methylation-specific PCR
Methylation-specific PCR was performed as described elsewhere2 and modified from Herman et al.7 Briefly, DNA was isolated using the DNeasy kit (Qiagen): 2 µg of DNA in 20 µL water was denatured in NaOH (0.2 M final concentration) for 15 minutes at 37°C. Following denaturation, 30 µL of 10 mM hydroquinone (Sigma) and 520 µL of 3 M sodium bisulfite (Sigma) were added and incubated overnight at 55°C in the dark. Modified DNA was then purified using the Wizard DNA Purification kit (Promega) and eluted in 20 µL of water. Modification was completed by treatment with NaOH (final concentration of 0.3 M) at 37°C for 15 minutes, followed by ethanol precipitation. The DNA was resuspended in water, and PCR was performed as described.2,6 Each assay was performed with positive and negative controls and repeated to ensure reproducibility.
Real-Time PCR
RNA was isolated from each line, and quantitative RT–PCR was performed for MGMT gene expression. Pelleted cells were washed in cold PBS, and RNA was extracted using the RNEasy kit (Qiagen) following the manufacture's protocol. Sensiscript Reverse Transcriptase (Qiagen) was used for reverse transcription. Real-time PCR was performed on the 7900HT Fast Real-Time PCR system, using an MGMT probe set and GAPDH (all from Applied Biosystems).
Western Blotting
Tumor cell pellets were resuspended in an equal volume of RIPA lysis buffer (50 mM Tris, 10% SDS, 150 mM NaCl, 12 mM sodium deoxycholate, 50 mM NaF, 1% Triton X-100, 0.5% NP40, 1 mM PMSF, 1 mM DTT, 1 µg/mL aprotinin, and 1 µg/mL pepstatin). The suspension was placed on ice for 15 minutes, vortexed for 10 seconds, and spun-down for 10 minutes at 12 000 rpm at 4°C. The supernatant was removed and protein concentration determined using the BioRad Protein Assay kit (specifications from BioRad). Lysates were supplemented with 500 mM DTT and protein sample buffer (Invitrogen) and heated to 70°C for 10 minutes. Samples were loaded and run on precast 4%–12% gradient SDS–PAGE gels (Nupage, Invitrogen) and transferred to nitrocellulose membranes in a semi-dry gel transfer module (Nupage, Invitrogen). Antibody hybridizations were performed, and signals were visualized with a chemiluminescence kit (Amersham). MGMT (FL-207) secondary goat-anti-rabbit (G1305) and anti-actin (C2) antibodies were from Santa Cruz.
BTIC Viability
The sensitivity of BTICs to TMZ was assessed using the alamarBlue viability assay (Medicorp). Briefly, single-cell suspensions of BTICs were plated in a 96-well plate, 5000 cells/well in a final volume of 200 µL/well. Cultures were treated with vehicle (DMSO) or increasing concentrations of TMZ chemotherapy (1–10 µg/mL; Schering Plough). Every second day, alamarBlue was added to the culture media. After 24 hours, absorbance was measured at 560 and 600 nm, and cell viability calculated as directed in the manufacturer's protocol. Final measurements were made 12 days after TMZ treatment began. Each treatment group was repeated in duplicate and each experiment in triplicate. Sensitivity was assessed in 2 ways: relative viability was determined, first, by comparing the absorbance values of TMZ-treated cells with vehicle-treated cells, and second, by the absolute absorbance values for each treatment group. Lines were classified as sensitive if their relative viability decreased compared with controls and their absolute absorbance decreased or remained unchanged. In addition, there was an extended period of observation after treatment to confirm that sensitive lines stopped proliferating (data not shown).
Statistical Considerations
The relationship between TMZ sensitivity and MGMT methylation was analyzed in 2 ways: first, grouping hemi-methylated lines with methylated lines as described in Hegi et al.,2 and then pooling hemi-methylated lines with unmethylated lines. The relationships between promoter methylation, transcript expression, and protein expression were analyzed after excluding hemi-methylated cases; all other comparisons included hemi-methylated lines. Fisher's exact test was used to evaluate the significance of associations (Graphpad Prism Software).
Results and Discussion
TMZ Sensitivity
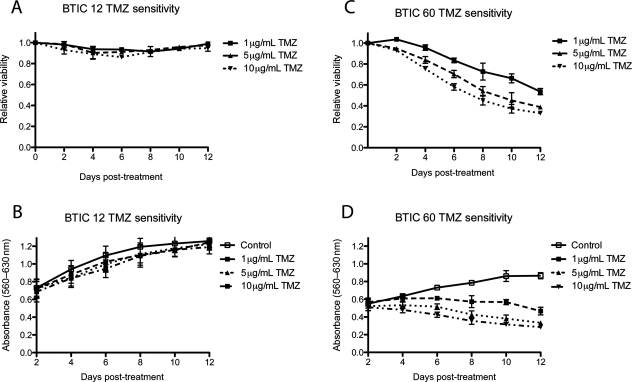
There is a commonly held view that BTICs are resistant to chemotherapy even though the experimental data on this point are conflicting. Some have found that BTICs are resistant to chemotherapy,8 whereas others have reported their sensitivity.9 Here, we performed a comprehensive analysis of the response of BTICs to TMZ chemotherapy using 20 newly established lines from patients with GBM. We assessed sensitivity with a well-characterized cell viability assay (alamarBlue) and chose a dose range (1–10 µg/mL) that spanned the clinically relevant concentrations of TMZ. This range was specifically selected to reflect drug concentrations that have been achieved in the cerebrospinal fluid of patients with GBM undergoing treatment with TMZ.10 Response data for representative sensitive and resistant lines are shown (Fig. 1). Lines varied in their response to TMZ: 9 were sensitive and 11 resistant (Table 1). Some lines were highly sensitive to TMZ and others highly resistant. Response to TMZ was difficult to categorize in 2 lines, BT067 and BT085; both continued to proliferate after TMZ exposure, but more slowly than vehicle-treated cells. For the purpose of subsequent analysis, these lines were scored resistant. There was no association between CD133 expression and response to TMZ (data not shown).
Fig. 1.
Toxicity assays. BTICs were plated (5000 cells/well) and treated with TMZ. Every second day alamarBlue was added and absorbance measured 24 hours later. Two patterns of response were seen. Some lines were resistant to TMZ as exemplified by BT012 for which there was no change in relative viability (A) or absolute absorbance (B) after treatment. Others were sensitive as exemplified by BT060 for which there was a decrease in relative viability (C) and a divergence in absorbance after treatment.
Table 1.
BTIC lines (n = 20) were characterized for MGMT methylation status using MS-PCR and classified as methylated (M), unmethylated (U), or hemi-methylated (U/M)
| Cell line | MGMT methylation | MGMT transcript | MGMT protein | TMZ response |
|---|---|---|---|---|
| BT048 | M | ND | ND | S |
| BT053 | M | ND | ND | S |
| BT084 | M | ND | ND | S |
| BT089 | M | ND | ND | S |
| BT094 | M | ND | ND | S |
| BT041 | U | ND | ND | S |
| BT050 | U | ND | ND | S |
| BT060 | U | Present | ND | S |
| BT069 | U | ND | ND | S |
| BT042 | U | ND | ND | R |
| BT085 | U | ND | ND | R |
| BT012 | U | Present | Present | R |
| BT030 | U | Present | Present | R |
| BT073 | U | Present | Present | R |
| BT074 | U | Present | Present | R |
| BT075 | U | Present | Present | R |
| BT090 | U | Present | Present | R |
| BT025 | U/M | Present | Present | R |
| BT067 | U/M | ND | ND | R |
| BT068 | U/M | Present | Present | R |
Real-time PCR was used to detect MGMT gene transcript (ND, no transcript; Present, transcript detected). Protein expression was examined by Western blot analysis (ND, no protein; Present, protein detected). TMZ response was characterized over time by alamarBlue viability after exposure to increasing concentrations of TMZ. Resistant lines (R) demonstrated minimal or no change in viability in response to clinically relevant doses of TMZ. Sensitive lines (S) demonstrated a significant decrease in viability after drug treatment.
These results derived from a careful survey of 20 BTIC lines from GBMs are noteworthy because they immediately call into question the popular view that BTICs are inherently and invariably resistant to chemotherapy. Perhaps, conflicting data in the literature reflect the fact that conclusions about the behavior of BTICs have been based on small numbers of lines.7,8
MGMT Status
Three patterns of MGMT promoter methylation were observed in BTIC lines—methylated, unmethylated, and hemi-methylated (Fig. 2). Five BTIC lines were methylated, 12 were unmethylated, and 3 displayed a hemi-methylated configuration (Table 1). The MGMT transcript was not detected in methylated lines and was found in only 7 of 12 unmethylated lines; similarly, protein was not detected in methylated lines and was found in only 6 of 12 unmethylated lines (Table 1). When detected, the levels of expression of the MGMT transcript and the MGMT protein were highly variable between the lines (data not shown). A very strong correlation was observed between the transcript and the protein expression (P < .0001). There was a weak association between MGMT promoter methylation and the expression of MGMT transcript (P = .04) and no association between methylation and protein expression (P = .1). These observations are similar to the results of comparable analyses performed in GBM tissues and traditional glioma cell lines derived from GBMs.11
Fig. 2.
Representative methylation-specific PCR (MS-PCR) data. DNA was isolated from each BTIC line and MS-PCR performed for the MGMT promoter. Three methylation patterns were seen: unmethylated (BT012), hemi-methylated (BT025), and methylated (BT048).
Sensitivity vs MGMT
The most widely studied and best characterized mechanism of resistance to TMZ in GBM is the expression of the DNA repair protein, MGMT. MGMT removes methyl adducts from O6-guanine, a site of lethal DNA damage by TMZ.12 GBM tumors that actively express MGMT are more resistant to TMZ than identical looking tumors in which the MGMT gene has been silenced.13 In GBM, the expression of the MGMT is silenced epigenetically via methylation of the MGMT gene promoter. For unknown reasons, methylation of the promoter occurs in up to 50% of GBMs.2 By silencing the MGMT, methylation of the gene promoter renders the tumor more sensitive to TMZ. Given its critical role in governing the response to TMZ, we assessed MGMT promoter methylation in GBM-derived BTICs that had different sensitivities to TMZ. Unlike in Hegi et al.,2 where the benefit from TMZ in GBM was associated with methylation of the MGMT gene promoter, we were unable to demonstrate a statistically significant association between sensitivity to TMZ and MGMT methylation in BTIC lines (P = .3). However, when the hemi-methylated lines were pooled with the unmethylated BTICs instead of methylated BTICs, as occurs clinically,2 we found a significant association between sensitivity to TMZ and methylation status (P = .008; Table 1). Overall, our findings in BTIC lines support the prevailing viewpoint that MGMT methylation status, by itself, should not be used to guide TMZ use in patients with GBM.
Although TMZ sensitivity was not associated with MGMT promoter methylation, except as noted, there were significant associations between response to TMZ and the expression of MGMT transcript (P = .01) and protein (P = .001). These results raise the possibility that transcript and protein expression in GBM tissues may be better indicators of benefit from TMZ than methylation status, although at present both transcript and protein14 are difficult to quantify in tissue sections. Such obstacles to reliable measurement may not be insurmountable, however. Similar challenges were successfully addressed by the breast cancer translational research community, who developed consensus criteria for therapeutic decision-making based on her2-neu expression.15 Her2-neu, like the MGMT protein, is detected by immunohistochemical analysis of tumor tissues that may contain a mixture of normal and neoplastic cells. Of course, no criteria are perfect or apply to all possible situations. Extrapolating from this BTIC study, it would be difficult to personalize the use of TMZ for patients with tumors that behaved like BT042, BT067, and BT085. These lines defy simple interpretation; they express neither the MGMT transcript nor protein, yet are resistant to TMZ.
Hemi-Methylated and Atypical BTICs
One of the unexpected findings in this study was the existence of hemi-methylated lines. The phenomenon of hemi-methylation has been seen before in the analyses of GBM tissues but attributed to the inadvertent contamination of the test sample by normal brain tissue. Normal tissue contamination is clearly an untenable explanation for a hemi-methylated pattern in BTICs, but whether hemi-methylation implies that only 1 of the 2 MGMT alleles is methylated in each cell within the line, or there are 2 subpopulations of tumor initiating cells with different methylation states coexisting in a single line, as suggested by Piccirillo et al.,16 is unknown at this time. In either case, the finding of hemi-methylation in some BTICs raises the intriguing possibility that hemi-methylation might also be a characteristic of some GBM tumors and is not due to normal tissue contamination in all instances. This possibility could have implications for the interpretation of MGMT test results and may have additional significance when coupled with our finding that TMZ sensitivity is significantly associated with MGMT methylation when hemi-methylated lines are assigned to the unmethylated group. Indeed, in a clinical trial, the pooling of TMZ-resistant hemi-methylated cases with sensitive methylated cases could undermine a positive study by shifting poor prognosis patients to a good prognosis subgroup. At the very least, miss-assignment of hemi-methylated tumors could explain short survivors in a series of methylated cases.
Atypically behaving lines also deserve comment. Whereas all methylated lines in this series were sensitive to TMZ, unmethylated lines sometimes had anomalous patterns of MGMT expression or unpredictable responses to TMZ. There were 2 groups of atypical lines. BT041, BT050, BT060, and BT069 had unmethylated promoters, yet were sensitive to TMZ; alternative sites of promoter methylation, not examined in this study, might explain such cases.17,18 BT042 and BT085 did not express MGMT protein, yet were resistant to TMZ. Neither BT042 nor BT085 had a mutation of MSH6; presumably other mechanisms of TMZ resistance were operative in these cases.19 Further characterization of anomalous lines may contribute to our understanding of atypically responding patients and assist in the development of robust predictive tests for TMZ use.
Our data demonstrate that BTICs have varying sensitivities to TMZ. In this study, methylation status did not predict response to TMZ precisely, although the reassignment of hemi-methylated cases to the unmethylated group improved our accuracy. All methylated lines were sensitive to TMZ and lines that expressed MGMT protein were resistant. Although our findings have not yet been confirmed in vivo, it would appear from this and other data that predicting response to TMZ with “clinical grade” precision will require more complex profiling of BTICs and, by inference, GBMs.
Conflict of interest statement. None declared.
Funding
This work was supported by the Alberta Heritage Foundation for Medical Research and the Alberta Cancer Foundation Chair in Brain Tumour Research.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. doi:10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. doi:10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 3.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. doi:10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. doi:10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 5.Kelly JJ, Stechishin O, Chojnacki A, et al. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells. 2009;27:1722–1733. doi: 10.1002/stem.98. doi:10.1002/stem.98. [DOI] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. doi:10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salmaggi A, Boiardi A, Gelati M, et al. Glioblastoma-derived tumorospheres identify a population of tumor stem-like cells with angiogenic potential and enhanced multidrug resistance phenotype. Glia. 2006;54:850–860. doi: 10.1002/glia.20414. doi:10.1002/glia.20414. [DOI] [PubMed] [Google Scholar]
- 9.Beier D, Rohrl S, Pillai DR, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68:5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. doi:10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 10.Ostermann S, Csajka C, Buclin T, et al. Plasma and cerebrospinal fluid population pharmacokinetics of temozolomide in malignant glioma patients. Clin Cancer Res. 2004;10:3728–3736. doi: 10.1158/1078-0432.CCR-03-0807. doi:10.1158/1078-0432.CCR-03-0807. [DOI] [PubMed] [Google Scholar]
- 11.Kitange GJ, Carlson BL, Mladek AC, et al. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol. 2009;92:23–31. doi: 10.1007/s11060-008-9737-8. doi:10.1007/s11060-008-9737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsson M, Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980;255:10569–10571. [PubMed] [Google Scholar]
- 13.Chinot OL, Barrie M, Fuentes S, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J Clin Oncol. 2007;25:1470–1745. doi: 10.1200/JCO.2006.07.4807. doi:10.1200/JCO.2006.07.4807. [DOI] [PubMed] [Google Scholar]
- 14.Preusser M, Charles Janzer R, Felsberg J, et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 2008;18:520–532. doi: 10.1111/j.1750-3639.2008.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. doi:10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 16.Piccirillo SG, Combi R, Cajola L, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807–1811. doi: 10.1038/onc.2009.27. doi:10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 17.Qian XC, Brent TP. Methylation hot spots in the 5′ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997;57:3672–3677. [PubMed] [Google Scholar]
- 18.Everhard S, Tost J, El Abdalaoui H, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neurooncology. 2009;11:348–356. doi: 10.1215/15228517-2009-001. doi:10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip S, Miao J, Cahill DP, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. doi:10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]