Abstract
Angiogenesis inhibitors, such as sunitinib, represent a promising strategy to improve glioblastoma (GBM) tumor response. In this study, we used the O6-methylguanine methyltransferase (MGMT)-negative GBM cell line U87MG stably transfected with MGMT (U87/MGMT) to assess whether MGMT expression affects the response to sunitinib. We showed that the addition of sunitinib to standard therapy (temozolomide [TMZ] and radiation therapy [RT]) significantly improved the response of MGMT-positive but not of MGMT-negative cells. Gene expression profiling revealed alterations in the angiogenic profile, as well as differential expression of several receptor tyrosine kinases targeted by sunitinib. MGMT-positive cells displayed higher levels of vascular endothelial growth factor receptor 1 (VEGFR-1) compared with U87/EV cells, whereas they displayed decreased levels of VEGFR-2. Depleting MGMT using O6-benzylguanine suggested that the expression of these receptors was directly related to the MGMT status. Also, we showed that MGMT expression was associated with a dramatic increase in the soluble VEGFR-1/VEGFA ratio, thereby suggesting a decrease in bioactive VEGFA and a shift towards an antiangiogenic profile. The reduced angiogenic potential of MGMT-positive cells is supported by: (i) the decreased ability of their secreted factors to induce endothelial tube formation in vitro and (ii) their low tumorigenicity in vivo compared with the MGMT-negative cells. Our study is the first to show a direct link between MGMT expression and decreased angiogenicity and tumorigenicity of GBM cells and suggests the combination of sunitinib and standard therapy as an alternative strategy for GBM patients with MGMT-positive tumors.
Keywords: GBM, MGMT, tumor angiogenesis, tyrosine kinase inhibitor
Glioblastoma (GBM) is the most aggressive primary malignant brain tumor in adults. Recently, concomitant temozolomide (TMZ) and radiation therapy (RT) followed by adjuvant TMZ became the standard of care for GBM patients.1 However, correlative studies showed that patients with tumors displaying O6-methylguanine methyltransferase (MGMT) promoter methylation (ie, MGMT(−)) were more likely to benefit from combined RT and TMZ, with a 2-year survival rate of 46% compared with 14% for patients with unmethylated MGMT tumors (ie, MGMT(+)).2 Indeed, the DNA repair protein MGMT is able to counteract the cytotoxic effects of TMZ by removing alkyl groups from the O6-position of guanine.3–5 Thus, alternative strategies for patients with unmethylated MGMT promoters (unresponsive tumors) are required to improve their poor outcome.
Tumoral neovascularization is induced by tumor expression of proangiogenic growth factors,6 and several growth factors and their cognate receptors are known to be overexpressed in GBM.7 Most notably, vascular endothelial growth factor (VEGF) and its primary receptors VEGFR-1/FMS-like tyrosine kinase 1 (FLT-1) and VEGFR-2/KDR/FLK-1 are increased in brain tumors and are widely considered to be the principal mediators of glioma angiogenesis.8–10 Sunitinib malate (Sutent, SU11248) is a multitargeted receptor tyrosine kinase (RTK) inhibitor with antiangiogenic activities. In addition to inhibition of VEGFR-1/-2/-3, sunitinib inhibits several RTKs involved in GBM growth and neovascularization, including platelet-derived growth factor receptors (PDGFRα and β), stem cell growth factor receptor (KIT),11–13 FLT-3, and colony stimulating factor-1 receptor (CSF1-R).14 In preclinical studies, sunitinib alone15 or in combination with RT16 has shown potent antiangiogenic and anti-invasive effects in GBM cell lines. Sunitinib is currently being tested in a phase II study of recurrent GBM. However, the effect of sunitinib in combination with TMZ and RT has not yet been investigated.
Genomic characterization of GBM tumors highlighted the association between MGMT promoter methylation and a hypermutator phenotype that encompasses global changes in DNA methylation and mutations in several genes.17 These alterations would affect functional pathways dictating both tumor behavior and clinical response to TMZ or other drugs. We hypothesized that a combination of antiangiogenic drugs with TMZ and RT must be evaluated in the context of MGMT status, which is so far the only available predictive biomarker of response to TMZ and RT. Thus, we first aimed to investigate cellular effects of sunitinib-based therapy in 3 GBM cell lines with differing MGMT status, including the highly tumorigenic and angiogenic MGMT(−) GBM cell line U87MG and its counterpart stably transfected with MGMT (U87/MGMT). The addition of sunitinib to TMZ and RT significantly improved the antiproliferative effects in these MGMT(+) cells compared with MGMT(−) cells. Additionally, gene expression profiling revealed for the first time that MGMT expression induced gene alterations involved in several functional pathways. Importantly, MGMT expression elicited a switch of the angiogenic balance toward an antiangiogenic profile in a GBM background. We specifically show an association between high MGMT expression and decreased expression of VEGFR-2 and secretion of VEGFA, as opposed to increased levels of VEGFR-1 and its soluble form (sVEGFR-1).
These findings highlight a novel role of MGMT as a critical upstream regulator of genes involved in angiogenesis of GBM tumor cells. Accordingly, we believe that MGMT status should be assessed in future clinical trials testing antiangiogenic therapy with TMZ and RT, which represents a promising strategy for patients with MGMT(+) tumors that are resistant to TMZ.
Materials and Methods
Cell Culture
The T98G GBM cell line was obtained from American Type Culture Collection. U87MG cells transfected with empty vector (U87/EV) and U87MG cells transfected with MGMT (U87/MGMT)18 cells were grown at 37°C/5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; standard medium). The HMEC-1 endothelial cell line was obtained from Dr Edmund Ades19 (Center for Disease Control, Georgia) and was cultured in MCDB medium supplemented with 10% FCS.
In Vitro Drug and RT Treatment
Sunitinib malate (Pfizer) and TMZ (Schering-Plough) were dissolved in dimethylsulfoxide (DMSO). Cells were serum-starved in DMEM containing 0.5% FCS overnight, then exposed to sunitinib (1 µM) for 2 hours, TMZ (100 µM) for 3 hours in serum-starved media, and/or exposed to 4 Gy of 60Co γ-radiation. MGMT was depleted with 20 µM O6-benzylguanine (O6BG) dissolved in DMSO as described previously.20
Proliferation and Clonogenic Survival Assays
Cells growing at 70% confluency were treated as described above, harvested, and seeded in triplicate in a 96-well plate at a density of 5 × 102 cells/well (U87/MGMT, T98G) for 48 hours, or 1 × 103 cells/well (U87/EV) for 72 hours. Cellular proliferation was assessed using the XTT Cell Proliferation Kit (Roche Pharmaceuticals).
Clonogenic survival analysis was performed as described previously.21 Briefly, cells plated at various densities (2 × 102–4.5 × 103) were allowed to adhere overnight and then treated with the drugs and/or RT as described above. After 10–14 days, cells were stained with 1% crystal violet and colonies with >50 cells were counted manually. Surviving fraction was calculated as follows: (colonies formed/total cells plated)/plating efficiency.
ELISA Assay
The conditioned medium of cells growing at 70% confluency for 48 hours was collected and passed through a 0.22-µm filter to remove cell debris. VEGF and sVEGFR-1 ELISA analyses (R&D Systems) were performed according to the manufacturer's instruction. The VEGFA and sVEGFR-1 concentrations were calculated from standard curves generated using recombinant human VEGFA and recombinant human VEGFR-1, respectively.
Western Blot Analysis
Following treatment, cells were washed twice with phosphate-buffered saline (PBS) and lysed with buffer.16 Proteins (30 µg, BCA protein assay kit, Pierce) were electrophoretically separated by 12% SDS-PAGE under reducing conditions and transferred onto polyvinylidine difluoride membranes. Membranes were probed for phospho-Akt (Cell Signaling), Akt1/2/3 (Santa Cruz), phospho-ERK1/2 (Cell Signaling), ERK1/2 (Cell Signaling), β-actin (Sigma-Aldrich), and human MGMT (BD Biosciences) as described previously.22
Gene Expression Microarray Studies
Total RNA was isolated from three independent cell samples of each cell line (U87/EV and U87/MGMT) using Trizol (Sigma-Aldrich) and purified using Qiagen RNeasy columns (Qiagen) according to the manufacturer's instructions. The RNA was quantified using a NanoDrop 1000 Spectrophotometer and its integrity evaluated using a Bioanalyzer 2100 (Agilent) according to the manufacturer's protocols. The RNA was subjected to linear amplification and Cy3 labeling followed by hybridization to Agilent's whole genome arrays in triplicate. Agilent kits were used according to the manufacturer's recommended protocols. Arrays were scanned using an Agilent Scanner, the data were extracted and the quality evaluated using Feature Extraction Software 9.5 (Agilent). The data were normalized and only the entities flagged as being present in the 3 samples were included in the analysis (GeneSpring GX 10, Agilent). Genes that were more than 2-fold up- or down-regulated (P values of <.05, unpaired Student's t-test with Bonferroni multiple testing correction) were identified. Database for annotation, visualization, and integrated discovery23 was used to identify enriched gene ontology (GO) biological themes.24 The GO data mining was conducted at a term specificity level 3.25 The EASE score was set at 0.05 and the minimum number of genes in a category was 5.
Quantitative Real-Time PCR
RNA was extracted from 1 × 106 cells with the RNeasy mini-kit (Qiagen). Briefly, 1 µg of total RNA was reverse transcribed into cDNA using the superscript reverse transcription kit (Invitrogen), and cDNA was quantified by quantitative real-time PCR (QRT-PCR) on an ABI 9700HT system (Applied Biosystems). RT-PCR reactions were done using SYBR green PCR Master Mix (Applied Biosystems) according to the manufacturer's instructions. Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Analysis was performed using the comparative Ct method.26
Primer sequences were as follows: VEGFR-1 sense, 5′-CTCTACTCCTGAAATCTATCAGA-3′, antisense, 5′-TACCATCCTGTTGTACATTTGCT-3′; VEGFR-2 sense, 5′-ACACCAGAAATGTACCAGACCAT-3′, antisense, 5′-TGCCATCCTGCTGAGCATTAG-3′; GAPDH sense, 5′-TCGCCAGCCGAGCCACAT-3′, antisense, 5′-CAATACGACCAAATCCGTTGACT-3′.
Flow Cytometry Analysis
Cells (1 × 106) were harvested, washed twice with PBS, and fixed with 2% paraformaldehyde for 30 minutes. Cells were washed 3 times, and phycoerythrin (PE)-conjugated anti-VEGFR-1 or anti-VEGFR-2 (1:100) was added for 30 minutes. PE-conjugated IgG1 was used as a negative control. Cells were washed 3 times and suspended in 0.5 mL of PBS and 10 000 events were acquired on a BD FACScalibur flow cytometer. Results were analyzed using Cell Quest software (BD Biosciences).
Endothelial Tube Cell Formation Assay
Twenty-four–well plates were coated with 300 µL of matrigel (BD Biosciences) for 30 minutes at 37°C. A total of 2 × 105 HMEC-1 cells in 1 mL of U87/EV or U87/MGMT conditioned media (1:2 dilution with serum-starved DMEM) were added to each well. Conditioned medium was collected following 48 hours of culture in standard medium. After 12 hours, cells were stained with 8 µg/mL calcein AM (Invitrogen) for 30 minutes at 37°C.27 Endothelial tube structures were examined using a Zeiss LSM 510 Axiovert 100 M microscope and a Fluar Zeiss 5× 0.25 NA lens. MetaMorph 7.6 software was used to assess mean tube length, mean tube area, and number of nodes.
Tumor Growth in Mice
A total of 5 × 106 cells (150 µL) were injected subcutaneously into the right flank of Balb/c, NIH III, or CDI/nu nude mice. Tumor growth was monitored twice a week using a digital caliper. Tumor volume was calculated as previously described.28 All experiments were approved by our institutional Animal Care Committee.
Results
Sunitinib-Based Treatment Preferentially Inhibits the Proliferation and Survival of MGMT(+) Cells
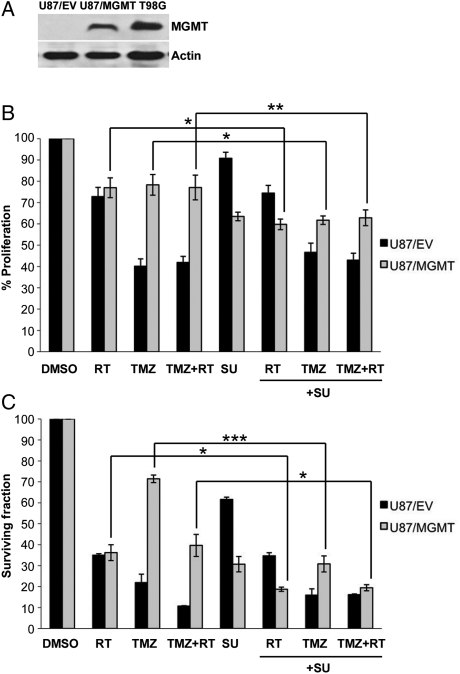
To investigate whether sunitinib in combination with the standard therapy would modulate the cellular response of MGMT(−) and MGMT(+) GBM cell lines, we used the MGMT(−) cell line U87MG (U87/empty vector, U87/EV) and its derived clone stably transfected with MGMT (U87/MGMT).18 As shown by immunoblotting, U87/MGMT and T98G cells, which exhibit constitutive expression of MGMT, had increased levels of MGMT protein compared with U87/EV cells (Fig. 1A).
Fig. 1.
Sunitinib-based treatment decreases proliferation and survival of MGMT(+) cells. (A) MGMT expression determined by immunoblotting, (B) effect of sunitinib (SU, 1 µM), RT (4 Gy), and/or TMZ (100 µM) on proliferation, and (C) clonogenic survival of U87/EV and U87/MGMT cells. Mean values are normalized to a DMSO control. Error bars represent the SEM of at least 3 independent experiments. *P < .05, **P < .01, and ***P < .001.
On the basis of our data,15,16 sunitinib at 1 µM was associated with significant inhibition of cell proliferation and was ultimately selected for subsequent studies. First, we used the XTT assay to test the efficacy of combining sunitinib (1 µM) with TMZ (100 µM) and/or RT (4 Gy) on cellular proliferation (Fig. 1B). As expected, compared with RT alone, TMZ alone and the combination of TMZ and RT significantly decreased the proliferation of U87/EV cells (P = .003 and <.001, respectively), but not of U87/MGMT cells (P = .7 and .7, respectively). Sunitinib alone or in combination with RT, TMZ, or TMZ and RT did not significantly decrease the proliferation of U87/EV cells compared with the same treatments without sunitinib. In contrast, the addition of sunitinib to each treatment decreased the proliferation of U87/MGMT cells to a greater extent than RT alone (P = .02), TMZ alone (P = .03), or TMZ and RT (P = .007).
Next, we assessed the ability of sunitinib-based therapy to inhibit clonogenic survival of MGMT(−) and MGMT(+) cells (Fig. 1C). A drastic loss of colony forming ability occurred when U87/EV cells were treated with the combination of TMZ and RT when compared with RT alone (P = .004), but this effect was not seen with U87/MGMT cells (P = .3). As shown in the proliferation assay, the effect of sunitinib alone was more pronounced on U87/MGMT compared with U87/EV cells (P = .01). Interestingly, the combination of sunitinib with RT, TMZ, or TMZ and RT significantly decreased the surviving fraction of U87/MGMT cells when compared with RT alone (P = .04), TMZ alone (P< .001), or TMZ and RT (P = .02). In contrast, the addition of sunitinib to RT and/or TMZ in MGMT(−) U87/EV cells did not further inhibit cell survival compared with RT and/or TMZ alone. Furthermore, proliferation and clonogenic survival of the MGMT(+) cell line T98G were also significantly inhibited by sunitinib in combination with TMZ and RT (Supplementary Material, Fig. S1). These data suggest that the addition of sunitinib preferentially improved the response of MGMT(+) cells to standard therapy.
Sunitinib Inhibits ERK1/2 and Akt Phosphorylation in MGMT(+) Cells
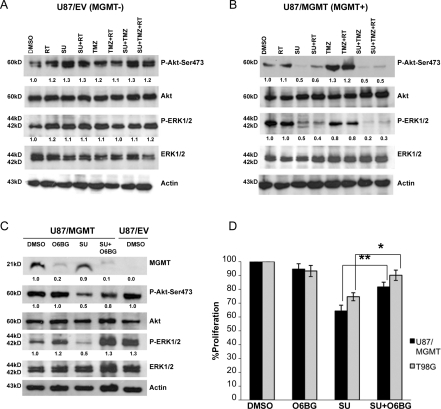
ERK1/2 and Akt, signaling molecules primarily involved in cell proliferation and survival, are downstream of several angiogenic growth factor receptors, including those targeted by sunitinib (such as VEGFRs).8 To assess the effect of sunitinib alone and in combination with standard therapy on these downstream kinases, we analyzed the phosphorylation of ERK1/2 and Akt by immunoblotting 24 hours after RT and/or drug exposure. Compared with the DMSO control, sunitinib alone and sunitinib-based treatment (with RT, TMZ, or TMZ and RT) induced a marked decrease in Akt-Ser473 phosphorylation in U87/MGMT cells but not in U87/EV cells. Similarly, sunitinib-based treatment (RT and/or TMZ) also decreased ERK1/2 phosphorylation in U87/MGMT cells but not in U87/EV cells (Fig. 2A and B).
Fig. 2.
Sunitinib inhibition of ERK1/2 and Akt phosphorylation is dependent on MGMT status. (A) Phosphorylation of ERK1/2 and Akt-Ser473 in response to sunitinib (SU, 1 µM), RT (4 Gy), and/or TMZ (100 µM) in U87/EV cells and (B) in U87/MGMT cells. (C) Phosphorylation of ERK1/2 and Akt-Ser473 in U87/MGMT in response to sunitinib following O6BG (20 µM). DMSO-treated U87/EV cells were used as a negative control. Values below bands represent relative intensities (histogram analysis using Adobe Photoshop) normalized to their respective total forms and the DMSO control conditions. Experiments were repeated twice with similar results. (D) Proliferation of U87/MGMT and T98G cells treated with O6BG and/or sunitinib. Mean values are normalized to the DMSO control. Error bars represent the SEM of 3 independent experiments. *P < .05 and **P < .01.
To investigate whether the inhibitory effect of sunitinib on ERK1/2 and Akt phosphorylation in MGMT(+) cells was related to MGMT status, we depleted MGMT protein levels using O6BG, a substrate analog of MGMT which induces MGMT degradation.29 Compared with the DMSO control, O6BG (20 µM) depleted MGMT expression in U87/MGMT cells by 90%, that is, to a level comparable to U87/EV cells. Sunitinib treatment did not affect MGMT levels, but it decreased the phosphorylation of both Akt and ERK1/2 by 50%. In contrast, depletion of MGMT using O6BG in cells treated with sunitinib completely abrogated the inhibitory effect of sunitinib on the phosphorylation of ERK1/2 and partially blocked the dephosphorylation of Akt (Fig. 2C). Treatment with O6BG did not significantly affect the proliferation of both MGMT(+) cell lines (U87/MGMT and T98G). As expected, compared with either DMSO or O6BG treatment, sunitinib treatment significantly decreased the proliferation of U87/MGMT (P < .001 and <.001, respectively) and T98G cells (P < .001 and P = .006, respectively). Interestingly, the addition of O6BG to sunitinib decreased the antiproliferative effects of sunitinib to levels similar to O6BG treatment alone in both U87/MGMT and T98G cells (P = .163 and =.145, respectively; Fig. 2D). Our data suggest that the antiproliferative effect of sunitinib on U87/MGMT and T98G cells is mediated through decreased phosphorylation of ERK1/2 and Akt and that this inhibitory effect is related to MGMT expression in these cells.
Genes Involved in Angiogenesis Are Differentially Regulated in MGMT(+) Cells
To further investigate the differential response of U87/EV and U87/MGMT cells to sunitinib, we compared the expression of genes in U87/MGMT vs U87/EV cells by cDNA microarray. We identified 3242 genes that were significantly differentially expressed by a minimum fold change of 2 (>99% CI, P value fixed at <.005, Student's t-test). We used the GO Consortium to classify genes into functional groups on the basis of biological process categories.24 GO analysis revealed that several functional pathways not previously related to the known functions of the MGMT protein were affected (Supplementary Material, Fig. S2). These observations will likely set the stage for further investigations to validate the expression of some of these genes and unravel how MGMT affects their expression.
We also investigated genes that could potentially affect GBM angiogenesis and/or tumorigenicity and the response to sunitinib therapy. GO analysis revealed that genes involved in vasculature development and RTK signaling pathways (Table 1) were differentially regulated in U87/MGMT cells compared with U87/EV cells (P = .00014 and .0004, respectively). Interestingly, the expression of known sunitinib targets, such as CSF1-R and VEGFR-2, was decreased in U87/MGMT cells. Additionally, the expression of VEGFA, the most potent stimulator of angiogenic signaling30 and a key determinant of angiogenicity and tumorigenicity of U87MG cells,31 was also decreased in U87/MGMT cells. In contrast, the expression of VEGFR-1, whose encoded protein exhibits 10-fold higher affinity for VEGFA than VEGFR-2 but has weaker tyrosine kinase activity,32 was increased in U87/MGMT cells. PDGFRs, well-known targets of sunitinib,11 were not differentially regulated in U87/MGMT cells. Thus, the gene expression analysis revealed a dramatic switch in the angiogenic profile of U87/MGMT compared with the parental cell line.
Table 1.
Differential expression of genes involved in angiogenesis and sunitinib response in U87/EV and U87/MGMT cells
| Gene | Symbol | U87/MGMT vs U87/EV fold change | GenBank |
|---|---|---|---|
| Biological processes | |||
| Vasculature development | |||
| Forkhead box F2; synonyms | FOXF2 | 296.4 | NM_001452 |
| Angiotensinogen | AGT | 134.5 | NM_000029 |
| Endothelin 1 | EDN1 | 65.38 | NM_001955 |
| EGF-like-domain, multiple 7 | EGFL7 | 55.31 | NM_201446 |
| Forkhead box F1 | FOXF1 | 24.54 | NM_001451 |
| Angiomotin | AMOT | 17.76 | NM_133265 |
| Laminin, α 4 | LAMA4 | 16.68 | NM_002290 |
| Vascular endothelial growth factor receptor 1 | FLT1 | 12.44 | NM_002019 |
| Carcinoembryonic antigen-related cell adhesion molecule 1 | CEACAM1 | 11.9 | NM_001712 |
| 7-Dehydrocholesterol reductase | DHCR7 | 8 | NM_001360 |
| v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 | ERBB2 | 6.798 | NM_001005862 |
| Fibroblast growth factor 18 | FGF18 | 5.937 | NM_033649 |
| Fibroblast growth factor 2 | FGF2 | 3.708 | NM_002006 |
| c-fos induced growth factor (vascular endothelial growth factor D) | FIGF | 3.645 | NM_004469 |
| B-cell translocation gene 1, antiproliferative | BTG1 | 3.54 | NM_001731 |
| Nuclear receptor subfamily 2, group F, member 2 | NR2F2 | −2.35 | NM_021005 |
| RAS p21 protein activator (GTPase activating protein) 1 | RASA1 | −2.63 | NM_002890 |
| Fibroblast growth factor receptor 1 | FGFR1 | −3.062 | NM_023111 |
| Lysyl oxidase | LOX | −3.66 | NM_002317 |
| Vascular endothelial growth factor A | VEGFA | −5.33 | NM_001025366 |
| Alternatively spliced product of the AML1 gene | AML1 | −5.917 | D43967 |
| Homosapiens acid fibroblast growth factor-like protein | GLIO703 | −6.849 | AF211169 |
| Phosphatidic acid phosphatase type 2B | PPAP2B | −9.615 | NM_003713 |
| Neuroplanin 2 | NRP2 | −12.66 | NM_201266 |
| Vascular endothelial growth factor receptor 2 | KDR | −16.73 | NM_002253 |
| SRY (sex determining region Y)-box 17 | SOX17 | −23.641 | NM_022454 |
| Plexin domain containing 1; tumor endothelial marker 7 | PLXDC1 | −32.89 | NM_020405 |
| Serpin peptidase inhibitor, clade E, member 1 | SERPINE1 | −111.48 | NM_000602 |
| Epiregulin | EREG | −173.21 | NM_001432 |
| Integrin, α 7 | ITGA7 | −181.49 | NM_002206 |
| Chondroitin sulfate proteoglycan 4 | CSPG4 | −207.04 | NM_001897 |
| Podoplanin | PDPN | −228.83 | NM_198389 |
| Transmembrane receptor protein tyrosine kinase signaling pathway | |||
| Adrenergic, β-2-, receptor, surface | ADRB2 | 138.7 | NM_000024 |
| Leukocyte tyrosine kinase | LTK | 90.08 | NM_002344 |
| Fibroblast growth factor receptor 3 | FGFR3 | 65.97 | NM_000142 |
| Ephrin-A1 | EFNA1 | 39.62 | NM_004428 |
| Neurturin | NRTN | 36.63 | NM_004558 |
| Vascular endothelial growth factor receptor 1 | FLT1 | 12.44 | NM_002019 |
| Ephrin-A4 | EPHA4 | 7.623 | NM_004438 |
| v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 | ERBB2 | 6.798 | NM_001005862 |
| Fibroblast growth factor 18 | FGF18 | 5.937 | NM_033649 |
| c-fos induced growth factor (vascular endothelial growth factor D) | FIGF | 3.645 | NM_004469 |
| Cas-Br-M (murine) ecotropic retroviral transforming sequence | CBL | 3.572 | NM_005188 |
| SNF1-like kinase 2 | SNF1LK2 | 2.68 | NM_015191 |
| Eukaryotic translation initiation factor 2-α kinase 3 | EIF2AK3 | 2.197 | NM_004836 |
| PTK2 protein tyrosine kinase 2; focal adhesion kinase 1 | PTK2 | −2.246 | NM_153831 |
| met proto-oncogene (hepatocyte growth factor receptor) | MET | −2.397 | NM_000245 |
| Fibroblast growth factor receptor 1 | FGFR1 | −3.062 | NM_023111 |
| Docking protein 1, 62kDa (downstream of tyrosine kinase 1) | DOK1 | −3.56 | NM_001381 |
| Lysyl oxidase | LOX | −3.66 | NM_002317 |
| Eukaryotic translation initiation factor 4A binding protein 2 | EIF4EBP2 | −3.78 | AK001936 |
| Vascular endothelial growth factor A | VEGFA | −5.33 | NM_001025366 |
| Phospholipase C, epsilon 1 | PLCE1 | −8.57 | NM_016341 |
| Fibronectin 1 | FN1 | −12.53 | NM_212482 |
| Ephrin-B3 | EFNB3 | −15.57 | NM_001406 |
| Vascular endothelial growth factor receptor 2 | KDR | −16.73 | NM_002253 |
| Ephrin-B1 | EPHB1 | −19.31 | NM_004441 |
| Fibroblast growth factor 5 | FGF5 | −20.2 | NM_004464 |
| Anaplastic lymphoma kinase (Ki-1) | ALK | −76.26 | NM_004304 |
| SHC (Src homology 2 domain containing) transforming protein 3 | SHC3 | −103.95 | NM_016848 |
| Metastasis suppressor 1 | MTSS1 | −172.2 | NM_014751 |
| Epiregulin | EREG | −173.21 | NM_001432 |
| Necdin homolog (mouse) | NDN | −210.9 | NM_002487 |
| Colony stimulating factor-1 receptor | CSF1-R | −222.46 | NM_005211 |
Differential Expression of VEGFR-1 and -2 Based on MGMT Expression
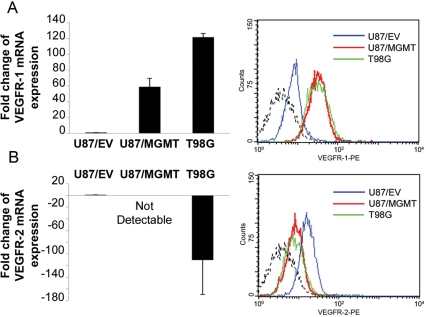
Our gene expression profiling suggested that overexpression of MGMT induced major changes in the expression of several genes involved in angiogenesis, which has great significance for the response to antiangiogenic inhibitors. Thus, we selected VEGFR-1 and -2 for validation of their differential expression using measurements of RNA and/or protein in MGMT(+) and MGMT(−) cell lines. QRT-PCR confirmed that VEGFR-1 mRNA expression was increased in U87/MGMT and T98G cells compared with U87/EV cells. This increase was confirmed at the protein level by flow cytometry (Fig. 3A). The decreased expression of VEGFR-2 in U87/MGMT and T98G cells compared with U87/EV cells was also validated by QRT-PCR and flow cytometry (Fig. 3B).
Fig. 3.
VEGFR-1 and -2 are differentially expressed in MGMT(+) cell lines. Expression of VEGFR-1 (A) and -2 (B) by QRT–PCR (left panel) and flow cytometry (right panel) in U87/EV, U87/MGMT, and T98G. Dashed lines represent the isotype controls. mRNA expression was analyzed using the comparative Ct method, and values are represented as fold change compared with U87/EV cells. Error bars represent the SEM of 3 independent experiments.
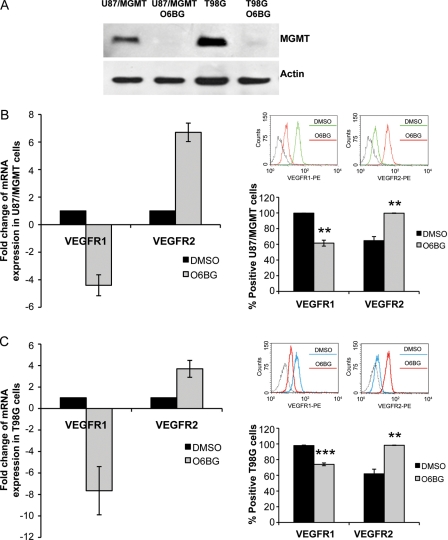
To investigate whether this differential expression was related to MGMT expression, U87/MGMT and T98G cells were treated with O6BG (20 µM, 48 hours) to deplete MGMT, and the expression of VEGFR-1 and -2 was assessed by QRT-PCR and flow cytometry. As shown by immunoblotting, exposure to O6BG completely depleted MGMT protein in U87/MGMT and T98G cells (Fig. 4A). Depletion of MGMT protein was associated with a significant decrease in VEGFR-1 mRNA and protein expression in U87/MGMT (P = .004, Fig. 4B) and T98G cells (P = .0005, Fig. 4C). In contrast, treatment with O6BG significantly increased expression of VEGFR-2 at the mRNA and protein level in U87/MGMT (P = .002, Fig. 4B) and T98G cells (P = .0007, Fig. 4C). Our data support the association between MGMT levels and regulation of VEGFR-1 and -2 expression in GBM cells.
Fig. 4.
Expression of VEGFR-1 and -2 correlates with MGMT expression. (A) Expression of MGMT protein following treatment with O6BG (20 µM, 48 hours) in U87/MGMT and T98G cells. Expression of VEGFR-1 and -2 by QRT-PCR (left panel) and flow cytometry (right panel) in U87/MGMT cells (B) and T98G cells (C) following treatment with O6BG. Dashed lines in the flow cytometry panels represent the isotype controls. mRNA expression was analyzed using the comparative Ct method, and values are represented as fold change compared with the DMSO control condition. Error bars represent the SEM of 3 independent experiments. **P < .01 and ***P < .001.
Decreased Secretion of VEGFA and sVEGFR-1 Was Accompanied by Reduced Angiogenic Potential of MGMT(+) Cell Lines
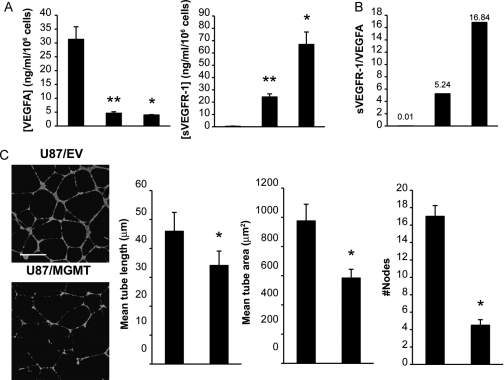
VEGFA, a key angiogenic factor strongly expressed by tumor cells, is involved in the growth and malignant progression of GBM tumors, mostly through VEGFR-1 and VEGFR-2.33,34 According to our differential expression profile, expression of VEGFA was decreased in U87/MGMT cells (Table 2). ELISA analysis showed that U87/EV cells secreted significantly more VEGFA than U87/MGMT or T98G cells (P = .002 and .05, respectively; Fig. 5A), suggesting that the regulation of the endogenous expression of VEGFA is related to MGMT expression in GBM cells.
Table 2.
Tumorigenicity of MGMT-negative and -positive cell lines
| Tumor cell line | Injection vehicle | Mouse strain | # Mice injected | Greatest average tumor volume reached |
||
|---|---|---|---|---|---|---|
| # mice with tumors | Mean ± SEM (mm3) | Days post implantation | ||||
| U87/EV | Serum-free media | Balb/c | 55 | 42 | 2780.75 ± 39.57 | 45 |
| U87/MGMT | Serum-free media | Balb/c | 53 | 1 | 47.5 | 6 |
| U87/MGMT | Full serum media | Balb/c | 6 | 3 | <5 ± 1 | 5 |
| U87/MGMT | 50% matrigel/starved media | Balb/c | 5 | 2 | 54.36 ± 22.95 | 8 |
| U87/MGMT | 50% matrigel/starved media | NIH III | 2 | 2 | 12.65 ± 10.51 | 8 |
| U87/MGMT | 50% matrigel/starved media | CDI/nu | 12 | 2 | 55.85 ± 19.61 | 8 |
| T98G | Serum-free media | Balb/c | 6 | 0 | 0 | >90 |
| T98G | 50% matrigel/starved media | CDI/nu | 3 | 0 | 0 | >90 |
Fig. 5.
Regulation of angiogenic factors in MGMT(+) cells influences angiogenic potential. (A) Secretion of VEGFA and sVEGFR-1 as determined by ELISA assay. Error bars represent the SEM of at least 3 independent experiments. (B) Ratio of sVEGFR-1 to VEGFA; a representation of the relative amount of bioactive VEGFA. (C) Representative photomicrographs (left) of the effect of conditioned medium (24 hours) from U87/EV and U87/MGMT cells on HMEC-1 cell tube formation in matrigel. Cells were fluorescently stained with calcein AM. Scale bar = 500 µm. Tube formation was quantified (right) using MetaMorph 7.6 software, and error bars represent the SEM of 3 independent experiments. *P < .05 and **P < .01.
sVEGFR-1, produced by alternative splicing, inhibits VEGFA signaling by sequestering the VEGF ligand35 and acts as a negative modulator for the bioactivity of VEGFA.36 Quantification of sVEGFR-1 by ELISA showed that this species was undetectable in U87/EV cultures but was secreted by U87/MGMT and T98G cells (Fig. 5A). The marked increase in sVEGFR-1/VEGFA ratio in MGMT(+) cells (Fig. 5B) would be expected to greatly decrease signaling through the VEGFR-1 and -2 receptors.
Next, to investigate the biological significance of differential secretion of VEGFA and sVEGFR-1 between U87/EV and U87/MGMT cells, we used an in vitro angiogenesis assay. We tested how conditioned medium from these two cell lines influenced the ability of HMEC-1 endothelial cells to form tubular structures in matrigel (Fig. 5C). Conditioned medium of U87/EV cells induced a more extensive branching network with tube-like structures displaying multicentric junctions, compared with U87/MGMT-conditioned medium (mean tube length P = .04, mean tube area P = .03, number of nodes P = .03; Fig. 5C). Thus, secretion of angiogenic factors by MGMT(−) cells elicited a significantly greater in vitro angiogenic response than MGMT(+) cells.
Reduced Tumorigenic Potential of MGMT(+) U87/MGMT Cells
Because the inhibition of endogenous expression of VEGFA is known to suppress tumor growth in vivo,31 we compared the tumorigenicity of U87/MGMT to U87/EV cells. Balb/c nu/nu mice were subcutaneously injected with both cell lines, one in each flank. U87/EV cells rapidly generated tumors in this mouse xenograft model, whereas the growth and sustainability of U87/MGMT tumors was drastically suppressed up to 9 weeks post-injection (Supplementary Material, Fig. S3). Because growth factor-reduced matrigel has been shown to enhance the tumorigenicity of GBM cell lines in vivo,37 subcutaneous injection of U87/MGMT cells in a matrigel vehicle was attempted. Although palpable tumors did form initially, they spontaneously regressed within 2 weeks even when other mouse genetic backgrounds (CD1/nu or NIH III) were used as recipients. Orthotopic injection of U87/MGMT cells also did not show evidence of tumor growth in necropsy studies (P. Forsyth, unpublished data). Additionally, among several MGMT-transfected U87 clones, only those expressing lower levels of MGMT were able to form xenografts (M.A., unpublished data). Another MGMT(+) cell line, T98G, previously reported to be poorly tumorigenic,38 also showed decreased tumorigenicity in our model (Table 2). These results reveal for the first time a link between MGMT expression and reduced tumorigenicity of GBM xenografts.
Discussion
Our study highlights for the first time the sensitivity of MGMT(+) vs MGMT(−) GBM cells to sunitinib. To understand how MGMT alters the expression of genes involved in the response to sunitinib, we performed a cDNA microarray study using an MGMT(−) GBM cell line and its MGMT(+) counterpart. Gene expression profiling revealed alterations in the angiogenic profile, as well as differential expression of several RTKs targeted by sunitinib. To our knowledge, our study is the first to suggest a relationship between MGMT expression and the angiogenic profile in human GBM. Notably, a large number of key positive regulators of GBM angiogenesis, such as VEGFA, VEGFR-2, neuropilin 2, CSF3, and acidic fibroblast growth factor,39 were decreased in U87/MGMT cells, whereas other genes known for their antiangiogenic activity, such as semaphorin 3F, endostatin, and COL4A1 (arrestin),39 were increased. For gene validation, we selected genes/proteins that are directly involved in angiogenesis and are targets of sunitinib, namely VEGFR-1 and VEGFR-2.40 MGMT(+) cell lines (U87/MGMT and T98G) displayed higher levels of VEGFR-1 mRNA and protein levels compared with U87/EV cells, whereas they displayed decreased levels of VEGFR-2. More importantly, depleting MGMT using O6BG suggested that the expression of these receptors was directly related to MGMT levels.
The validation in MGMT(+) cells of decreased VEGFA, a primary mediator of angiogenesis, and of increased sVEGFR-1, which sequesters VEGFA and negatively regulates VEGF-mediated angiogenesis,41 has a great significance. A decreased sVEGFR-1/VEGFA ratio was previously shown to correlate with an increased bioavailability of VEGFA and a proangiogenic phenotype in malignant GBM compared with diffuse astrocytoma.42 In our study, we show that MGMT expression was associated with a dramatic increase in the sVEGFR-1/VEGFA ratio, thereby suggesting a decrease in bioactive VEGFA, which shifts the balance in favor of an antiangiogenic phenotype in MGMT(+) GBM cells. Several lines of evidence support the tenet that our MGMT(+) cells display an antiangiogenic phenotype compared with the MGMT(−) GBM cell line: (i) their increased response to sunitinib-based therapy; (ii) the decreased activity of their conditioned media when tested in the in vitro tube formation assay; and (iii) their low tumorigenicity. In this regard, earlier studies showed that inhibition of VEGFA-induced angiogenesis43 and inhibition of endogenous expression of VEGFA suppressed tumor growth in vivo, thereby supporting the role of VEGFA as a major determinant of both angiogenicity and tumorigenicity of U87MG cells.31 Thus, our study is the first to suggest a direct link between MGMT expression and decreased angiogenicity and tumorigenicity of GBM cells.
With respect to the large number of genes that were differentially expressed in U87/MGMT vs U87/EV cells, elucidating the intricate mechanism(s) by which MGMT induced these transcriptional alterations is challenging. We speculate that overexpression of MGMT altered the expression of these genes through indirect mechanisms and at different levels of regulation, including: (i) the regulation of a transcription program that includes the induction of a number of transcription factors, such as zinc finger domain proteins acting as activators or repressors of gene expression (Supplementary Material, Fig. S2), or (ii) regulation through common epigenetic alterations in GBM, such as DNA hypermethylation or hypomethylation at the CpG island promoters affecting genes that control cell growth, apoptosis, and angiogenesis.44 Of interest in this regard is the observation from our gene array data that the expression of DNA methyltransferase 3a was significantly decreased in U87/MGMT cells, and this finding was validated by QRT-PCR (data not shown).
To date, MGMT has been described as a DNA repair protein, which protects DNA from the mutagenic actions of endogenous carcinogens and elicits resistance to alkylating agents.45,46 Our study suggests a novel function(s) for MGMT as a negative upstream regulator of key functional pathways involved in angiogenesis and tumorigenicity. Further work using additional cell lines and archived surgical specimens from GBM patients is needed to decipher how MGMT mediates these functions and to validate the concept that MGMT expression shifts the angiogenic profile. Our in vitro and in vivo studies suggest that MGMT(−) GBM cells did not derive benefit from the addition of sunitinib to standard therapy. The combination of sunitinib with the standard treatment may inhibit tumor growth of MGMT(+) cells by exerting not only direct antiproliferative effects on tumor cells, but also antiangiogenic effects through a concerted action on tumor cells expressing MGMT and the tumor vasculature in vivo. We were unable to test this hypothesis in the current study because of the poor tumorigenicity of the MGMT(+) cell lines. However, as expected, the addition of sunitinib did not significantly improve the in vivo response of U87/EV xenografts to TMZ and RT (data not shown).
The validation of the relationship between MGMT status and an angiogenic profile in GBM tumor samples may ultimately lead to prospective testing of MGMT expression before offering antiangiogenic agents in combination with RT and TMZ in future clinical trials.
Funding
This work was funded by independent research grants from Pfizer and Schering-Plough and by a donation from the St John Bosco Elementary School in Edmonton, AB.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Acknowledgments
We would like to thank Dr Xuejun Sun and the imaging facility manager as well as the vivarium staff for their support and expertise.
Conflict of interest statement. None declared.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Denny BJ, Wheelhouse RT, Stevens MF, et al. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA. Biochemistry. 1994;33:9045–9051. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 5.Kaina B, Christmann M, Naumann S, et al. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst). 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Jain RK, di Tomaso E, Duda DG, et al. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 7.Dunn IF, Heese O, Black PM. Growth factors in glioma angiogenesis: FGFs, PDGF, EGF, and TGFs. J Neurooncol. 2000;50:121–137. doi: 10.1023/a:1006436624862. [DOI] [PubMed] [Google Scholar]
- 8.Byrne AM, Bouchier-Hayes DJ, Harmey JH. Angiogenic and cell survival functions of vascular endothelial growth factor (VEGF) J Cell Mol Med. 2005;9:777–794. doi: 10.1111/j.1582-4934.2005.tb00379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 10.Fischer I, Gagner JP, Law M, et al. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol. 2005;15:297–310. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto KM. Su-11248 Sugen. Curr Opin Investig Drugs. 2004;5:1329–1339. [PubMed] [Google Scholar]
- 12.Joensuu H, Puputti M, Sihto H, et al. Amplification of genes encoding KIT, PDGFRalpha and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme. J Pathol. 2005;207:224–231. doi: 10.1002/path.1823. [DOI] [PubMed] [Google Scholar]
- 13.Anderson JC, McFarland BC, Gladson CL. New molecular targets in angiogenic vessels of glioblastoma tumours. Expert Rev Mol Med. 2008;10:e23. doi: 10.1017/S1462399408000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faivre S, Demetri G, Sargent W, et al. Molecular basis for sunitinib efficacy and future clinical development. Nat Rev Drug Discov. 2007;6:734–745. doi: 10.1038/nrd2380. [DOI] [PubMed] [Google Scholar]
- 15.de Bouard S, Herlin P, Christensen JG, et al. Antiangiogenic and anti-invasive effects of sunitinib on experimental human glioblastoma. Neuro Oncol. 2007;9:412–423. doi: 10.1215/15228517-2007-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schueneman AJ, Himmelfarb E, Geng L, et al. SU11248 maintenance therapy prevents tumor regrowth after fractionated irradiation of murine tumor models. Cancer Res. 2003;63:4009–4016. [PubMed] [Google Scholar]
- 17.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aghi M, Rabkin S, Martuza RL. Oncolytic herpes simplex virus mutants exhibit enhanced replication in glioma cells evading temozolomide chemotherapy through deoxyribonucleic acid repair. Clin Neurosurg. 2006;53:65–76. [PubMed] [Google Scholar]
- 19.Ades EW, Candal FJ, Swerlick RA, et al. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 20.Bobola MS, Silber JR, Ellenbogen RG, et al. O6-methylguanine-DNA methyltransferase, O6-benzylguanine, and resistance to clinical alkylators in pediatric primary brain tumor cell lines. Clin Cancer Res. 2005;11:2747–2755. doi: 10.1158/1078-0432.CCR-04-2045. [DOI] [PubMed] [Google Scholar]
- 21.Abdulkarim B, Sabri S, Zelenika D, et al. Antiviral agent cidofovir decreases Epstein–Barr virus (EBV) oncoproteins and enhances the radiosensitivity in EBV-related malignancies. Oncogene. 2003;22:2260–2271. doi: 10.1038/sj.onc.1206402. [DOI] [PubMed] [Google Scholar]
- 22.Lokker NA, Sullivan CM, Hollenbach SJ, et al. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002;62:3729–3735. [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 27.Di Simone N, De Santis M, Tamburrini E, et al. Effects of antiretroviral therapy on tube-like network formation of human endothelial cells. Biol Pharm Bull. 2007;30:982–984. doi: 10.1248/bpb.30.982. [DOI] [PubMed] [Google Scholar]
- 28.Huang P, Allam A, Taghian A, et al. Growth and metastatic behavior of five human glioblastomas compared with nine other histological types of human tumor xenografts in SCID mice. J Neurosurg. 1995;83:308–315. doi: 10.3171/jns.1995.83.2.0308. [DOI] [PubMed] [Google Scholar]
- 29.Dolan ME, Moschel RC, Pegg AE. Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proc Natl Acad Sci USA. 1990;87:5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park JE, Keller GA, Ferrara N. The vascular endothelial growth factor (VEGF) isoforms: differential deposition into the subepithelial extracellular matrix and bioactivity of extracellular matrix-bound VEGF. Mol Biol Cell. 1993;4:1317–1326. doi: 10.1091/mbc.4.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng SY, Huang HJ, Nagane M, et al. Suppression of glioblastoma angiogenicity and tumorigenicity by inhibition of endogenous expression of vascular endothelial growth factor. Proc Natl Acad Sci USA. 1996;93:8502–8507. doi: 10.1073/pnas.93.16.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JE, Chen HH, Winer J, et al. Placenta growth factor. Potentiation of vascular endothelial growth factor bioactivity, in vitro and in vivo, and high affinity binding to Flt-1 but not to Flk-1/KDR. J Biol Chem. 1994;269:25646–25654. [PubMed] [Google Scholar]
- 33.Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plate KH, Breier G, Weich HA, et al. Vascular endothelial growth factor is a potential tumour angiogenesis factor in human gliomas in vivo. Nature. 1992;359:845–848. doi: 10.1038/359845a0. [DOI] [PubMed] [Google Scholar]
- 35.Kendall RL, Thomas KA. Inhibition of vascular endothelial cell growth factor activity by an endogenously encoded soluble receptor. Proc Natl Acad Sci USA. 1993;90:10705–10709. doi: 10.1073/pnas.90.22.10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornig C, Weich HA. Soluble VEGF receptors. Angiogenesis. 1999;3:33–39. doi: 10.1023/a:1009033017809. [DOI] [PubMed] [Google Scholar]
- 37.Mullen P. The use of Matrigel to facilitate the establishment of human cancer cell lines as xenografts. Methods Mol Med. 2004;88:287–292. doi: 10.1385/1-59259-406-9:287. [DOI] [PubMed] [Google Scholar]
- 38.Rubenstein M, Shaw M, Mirochnik Y, et al. In vivo establishment of T98G human glioblastoma. Methods Find Exp Clin Pharmacol. 1999;21:391–393. doi: 10.1358/mf.1999.21.6.541918. [DOI] [PubMed] [Google Scholar]
- 39.Roskoski R., Jr Vascular endothelial growth factor (VEGF) signaling in tumor progression. Crit Rev Oncol Hematol. 2007;62:179–213. doi: 10.1016/j.critrevonc.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Mendel DB, Laird AD, Xin X, et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res. 2003;9:327–337. [PubMed] [Google Scholar]
- 41.Toi M, Bando H, Ogawa T, et al. Significance of vascular endothelial growth factor (VEGF)/soluble VEGF receptor-1 relationship in breast cancer. Int J Cancer. 2002;98:14–18. doi: 10.1002/ijc.10121. [DOI] [PubMed] [Google Scholar]
- 42.Lamszus K, Ulbricht U, Matschke J, et al. Levels of soluble vascular endothelial growth factor (VEGF) receptor 1 in astrocytic tumors and its relation to malignancy, vascularity, and VEGF-A. Clin Cancer Res. 2003;9:1399–1405. [PubMed] [Google Scholar]
- 43.Kim KJ, Li B, Winer J, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 44.Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Semin Cancer Biol. 2009;19:188–197. doi: 10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Kaina B, Christmann M, Naumann S, et al. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair. 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Pegg AE, Dolan ME, Moschel RC. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog Nucleic Acid Res Mol Biol. 1995;51:167–223. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]