Abstract
We have previously demonstrated that chordomas express activated platelet-derived growth factor receptor (PDGFRB) and that treatment with imatinib, which is capable of switching off the activation of various receptor tyrosine kinases (RTKs) including PDGFRB, benefits a number of patients. The aim of this study was to identify the possible presence of other activated RTKs and their downstream signaling effectors. Cryopreserved material from 22 naïve sporadic chordomas was investigated for the presence of activated RTKs and their cognate ligands and downstream signaling effectors by means of human phospho-RTK antibody arrays, Western blotting, and molecular analysis; immunohistochemistry and fluorescence in situ hybridization were used to analyze the corresponding formalin-fixed and paraffin-embedded samples. We detected activated PDGFRB, FLT3, and colony stimulating factor 1 receptor (CSF1R) of the PDGFR family and highly phosphorylated EGFR, HER2/neu, and (to a lesser extent) HER4 of the EGFR family. The detection of PDGFRB/PDGFB confirmed our previous data. The presence of activated EGFR was paralleled by the finding of high levels of epidermal growth factor (EGF) and transforming growth factor α (TGFα) and PDGFB co-expression and PDGFRB co-immunoprecipitation. Of the downstream effectors, the PI3K/AKT and RAS/MAPK pathways were both activated, thus leading to the phosphorylation of mammalian target of rapamycin (mTOR) and 4E-BP1 among the regulators involved in translational control. Taken together, our results (i) provide a rationale for tailored treatments targeting upstream activated receptors, including the PDGFR and EGFR families; (ii) support the idea that a combination of upstream antagonists and mTOR inhibitors enhances the control of tumor growth; and (iii) indicate that the 4E-BP1/eIF4E pathway is a major regulator of protein synthesis in chordoma.
Keywords: chordomas, mTOR, receptor tyrosine kinases (RTKs), signal transduction.
Chordomas are rare low-grade bone tumors that account for 1%–4% of all malignant bone tumors;1 they arise from the sacrum in 50%–60% of cases, from the skull base in 20%–35%, from the cervical vertebrae in 10%, and from the thoraco-lumbar vertebrae in 5%.2 Their peak incidence is between the fourth and sixth decades of life, although children may also be affected (5%). The M:F ratio is 2–3:1 (more males are affected by sacral chordomas, whereas the frequency of chordomas of the skull base seems to be the same in men and women).3 Chordomas are generally sporadic, but there have also been familial cases4–6 and tumors arising in patients with tuberous sclerosis complex (TSC).7–11
Untreated disease has an indolent but relentless course. The tumors are locally highly aggressive and may also give rise to distant metastases (10%–30% of cases, or more). On a population basis, the reported 5- and 10-year survival rates are about 70% and 40%, which reflects the naturally slow progress of the disease; however, after the development of metastases, median survival is about 1 year.3
Surgery remains the best standard treatment for both localized and metastatic disease; radiotherapy can control tumor growth, particularly cobalt or heavy particle radiation, but the high doses required lead to significant toxicity, and standard cytotoxic chemotherapy is inefficacious.3 Targeted molecular therapy recently been found to be promising as imatinib has induced an anti-tumor response in a number of patients when used alone12 and when combined with cisplatin in patients showing secondary progression on imatinib montherapy.13 Furthermore, combined treatment with imatinib and sirolimus has proved to be effective in patients with imatinib-resistant chordomas.14
We have previously demonstrated that chordomas express activated platelet-derived growth factor receptor (PDGFRB) and that this activation is due to the occurrence of an autocrine/paracrine loop;15 it has also been reported that a subset of chordomas express EGFR and c-MET.16 The presence of activated receptor tyrosine kinases (RTKs) leads to the activation of secondary transducers belonging to the MAPK or PI3K/AKT pathways, which together activate mTOR;17 downstream of mTOR, S6K/S6, and 4E-BP1 harmonize protein synthesis, and promote cell growth and proliferation.
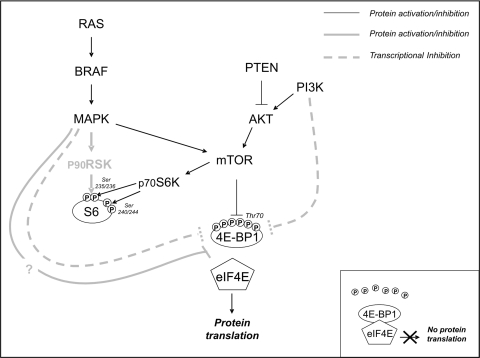
Recent insights into the regulation of translation control have shown that, in addition to the p70 ribosomal S6 kinase (p70S6K)-mediated PI3K/mTOR pathway, S6 may also be regulated by the p90 ribosomal S6 kinase (p90RSK)-mediated RAS/MAPK cascade.18 Furthermore, 4E-BP1 (a repressor of eIF4E) may be inactivated as a result of hyper-phosphorylation by the PI3K/mTOR or RAS/MAPK cascade through a still unidentified mechanism (which abrogates the association between 4E-BP1 and eIF4E),19 or as a result of transcription silencing20,21 (Fig. 1). More importantly, S6 may act as an oncogene or tumor suppressor gene.22,23 Finally, it is likely that the specific functions of the 2 regulators of the translational machinery are closely related to cell type or tumor histotype.24
Fig. 1.
Usual regulation of RTK downstream effectors. The grey lines indicate new pathways related to protein activation/inhibition by means of phosphorylation (thick continuous lines) or transcriptional silencing (dotted lines). In addition to being phosphorylated by p70S6K/mTOR, S6 may be activated by p90RSK. The same is true of 4E-BP1 which, in addition to mTOR, may be phosphorylated by an unknown factor (light-grey thick continuous lines); it may also be regulated by the transcriptional silencing induced by MAPK and/or PI3K (light-grey thick dotted lines). Insert: Non-phosphorylated 4E-BP1 forming a complex with eIF4E that blocks protein translation.
In this study, human phospho-RTK antibody arrays (RTK cards) were used to explore the upstream RTK profile of 7 naïve sporadic chordomas, and detected activated PDGFRB, FLT3, and colony stimulating factor 1 receptor (CSF1R) of the PDGFR family, as well as highly phosphorylated EGFR, HER2/neu, and (to a lesser extent) HER4 of the EGFR family. Taken together, this subsequently validated upstream analysis indicates that both the PDGFR and EGFR families may be activated in chordomas. This upstream analysis was extended to 22 sporadic cases, and downstream effectors were investigated. We found activated PI3K/AKT and RAS/MAPK cascades lead to the phosphorylation of mTOR. The absence of gene deregulation upstream of mTOR in the presence of PDGFRB and EGFR ligands supports the hypothesis that the activation is ligand dependent and that combined RTK and mTOR/MAPK inhibitors25 may lead to better disease control. Our findings also indicate that translational control downstream of mTOR is preferentially driven by the 4E-BP1/eIF4E pathway.
Materials and Methods
Clinical Data and Materials
We analyzed 21 surgical specimens and 1 wide biopsy sample taken from 22 surgically treated patients with a confirmed diagnosis of conventional sporadic chordoma: 16 males and 6 females with a median age of 61 years (range 36–77). Fourteen were primary tumors and 8 were recurrences (Patients 7, 10–12, 14, 19, 20, and 22); 18 were located in the sacrum and 4 in the thoraco-lumbar vertebrae. Freshly frozen material and formalin-fixed paraffin-embedded (FFPE) blocks were available for all but 1 of the patients (Patient 14), for whom there was only frozen material.
All of the analyses involved representative areas of tissue sections carefully selected under microscope control in order to avoid normal tissue or necrosis. The biochemical and molecular analyses were performed using representative cryopreserved tissue and checking 1 hematoxylin-and-eosin-stained frozen section; the immunohistochemical (IHC) and fluorescence in situ hybridization (FISH) analyses were performed using the corresponding FFPE samples.
Written informed consent was obtained from all of the patients.
Analyses of Frozen Material
Biochemical Analysis
RTK Phosphorylation Array. The expression of phosphorylated growth factor RTKs was detected using the Proteome Profiler Array Kit (ARY001, R&D Systems) as laid down in the manufacturer's protocol, and 2 mg of protein lysate per array.
Total Protein Extraction, Immunoprecipitation and Western Blotting: The proteins were extracted from tissue samples stored at −80°C as previously described.26 For the immunoprecipitation (IP) analysis, 1 mg each of protein lysates was precipitated by means of incubation with Protein A Sepharose (Amersham Biosciences) and specific antibodies: 20 µl of rabbit polyclonal anti-PDGFRB (sc-432, Santa Cruz Biotechnology), 10 µl of rabbit polyclonal anti-EGFR (sc-120, Santa Cruz Biotechnology), and 10 µl of mouse monoclonal anti-Her2/Neu (Ab-3, Calbiochem; MGR2 kindly provided by Dr. Tagliabue, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy).
Immunoblotting was performed using the anti-phosphotyrosine mouse monoclonal antibody (05–321, Upstate) diluted 1:3000 to reveal the absence or presence of activated/phosphorylated receptor. The filters were stripped and then incubated with the specific antibody in order to detect the degree of receptor expression: anti-PDGFRB (sc-432, Santa Cruz Biotechnology), anti-EGFR (sc-03, Santa Cruz Biotechnology), and anti-HER2/neu (Ab-3, Calbiochem). The filter of IP anti-PDGFRB was stripped again and incubated with anti-PI3-kinase p85α (sc-1637, Santa Cruz Biotechnology); the filters of IP anti-EGFR were incubated with anti-PDGFRB and/or anti-HER2/neu in order to detect the co-immunoprecipitated proteins.
For the EGFR IP/WB experiments, the protein bands detected on filters were densitometrically analyzed on X-ray film images using Quantity One Software (Bio-Rad). The densitometrically determined levels of phosphorylated EGFR were calculated as ratios of phosphorylated and expressed EGFR.
The expression and activation of the downstream targets were detected by means of direct Western blotting (WB) after loading 20 µg of total protein/lane using the antibodies anti-P-AKT (#4058S Ser473, Cell Signaling Technology), anti-AKT (#9272, Cell Signaling Technology), anti-P-MAPK (#4376S Thr202/Tyr204, Cell Signaling Technology), anti-MAPK (#9102 p44-42, Cell Signaling Technology), anti-P-mTOR (#2971S Ser2448, Cell Signaling Technology), anti-mTOR (#2972, Cell Signaling Technology), anti-P-S6 (#2215 Ser240/244 and #2211 Ser 235/236, Cell Signaling Technology), anti-P-4E-BP1 (#9455 Thr70, Cell Signaling Technology), anti-4E-BP1 (#9644 Cell Signaling Technology), anti-PTEN (#9559, Cell Signaling Technology), and anti-actin (A2066, Sigma). The negative control for PTEN WB was a cell line without PTEN protein expression (3962 M, kindly provided by Dr. Rodolfo, Fondazione IRCCS Istituto Nazionale dei Tumori).27
RNA Extraction and Real-Time PCR
Total RNA was extracted from fresh frozen tissue and reverse transcribed as previously described.28
RTK Ligand Detection. The presence or absence of EGFR ligand mRNA [EGF and transforming growth factor α (TGFα)] was verified by means of real-time PCR using specific EGF (Hs00153181_M1) and TGFα probes (Hs00608187_M1) (Applied Biosystems); Table 1 shows the experimental conditions. The mRNA was not quantified.
Table 1.
Experimental condition
| Real-time PCR | |||||
|---|---|---|---|---|---|
| Ligands | Probes | Conditions |
|||
| EGF | Hs00153181_M1 (Applied Biosystem) | 50°C 2′–95°C 10′–95°C 15′, 60°C 1′ × 40 cycles | |||
| TGFα | Hs00608187_M1 (Applied Biosystem) | ||||
| PTEN | Hs02621230_S1 (Applied Biosystem) | ||||
| S6 | Hs01058685_g1 (Applied Biosystem) | ||||
| Molecular analysis | |||||
| Gene | Exon | Sense | Antisense | Annealing temperature (°C) | |
| EGFR | 18 | ctgaggtgacccttgtctc | tacagcttgcaaggactctg | 60 × 40 cycles | |
| 19 | agatcactgggcagcatgtg | cagctgccagacatgagaa | |||
| 20 | cttctggccaccatgcgaa | ccaggcaaactctgctatcc | |||
| 21 | tcttcccatgatgatctgt | cctggtgtcaggaaaatgct | |||
| PI3K | 9 | gggaaaaatatgacaaagaaagc | ctgagatcagccaaattcagtt | 58 × 35 cycles | |
| 20 | ctcaatgatgcttggctctg | tggaatccagagtgagctttc | |||
| PTEN | 5 | tgcaacatttctaaagttacctact | gaggaaaggaaaaacatcaaaaa | 55 × 40 cycles | |
| 6 | tttttcaatttggcttctcttttt | tgttccaatacatggaaggatg | |||
| 7 | cagttaaaggcatttcctgtg | ttttggatatttctcccaatgaa | |||
| 8 | tgtcatttcatttctttttcttttc | aagtcaacaacccccacaaa | |||
| 9 | tcatggtgttttatccctcttg | tgagtcatatttgtgggtttca | |||
| K-Ras | 1 | tggtggagtatttgatagtgt | catgaaaatggtcagaga | 56 × 35 cycles | |
| 2 | ggtgcactgtaataatccagac | tgatttagtattatttatggc | 50 × 45 cycles | ||
| B-Raf | 11 | tccctctcaggcataaggtaa | cgaacactgaatatttcctttgat | 55 × 40 cycles | |
| 15 | tcataatgcttgctctgatagga | ggccaaaaatttaatcagtgga | 56 × 40 cycles | ||
| Immunohistochemical analysis | |||||
| Antibody | Clone | Company | Dilution | Antigen retrieval | Positive control |
| PDGFRB | sc-339 | Santa Cruz Biotecnology | 1:100 | 6′ at 95°C mM citrate buffer, pH 6 | Human breast carcinoma |
| EGFR | K1492 | Dako | Procedures performed in accordance with manufacturer's protocol | ||
| Her2 | Polyclonal | Dako | 1:2000 | 6′ at 95°C, pH 6 | Human breast carcinoma |
TGFα, transforming growth factor α.
PTEN and S6. Real-time PCR of PTEN and S6 was performed on respectively, 4 and 6, representative samples using specific PTEN (Hs02621230_S1) and S6 probes (Hs01058685_g1), and the experimental conditions shown in Table 1.
DNA Extraction and Sequencing
DNA was digested by proteinase-K and underwent standard phenol extraction. Mutation analyses were made of exons 18–21 of EGFR, exons 9 and 20 of PI3KCA, exons 5–9 of PTEN, exons 1 and 2 of KRAS, and exons 11 and 15 of BRAF. Table 1 shows the primers and experimental conditions used for PCR amplification. After purification using the ExoSAP-IT Kit (USB Amersham), the DNA of the PCR products was automatically sequenced (ABI prism 377; Applied Biosystems).
Analyses of FFPE Material
RNA Extraction and RT-PCR
Total RNA from microdissected samples was isolated by means of the TRIzol method (Invitrogen), and reverse transcribed using Superscript reverse transcriptase (Invitrogen) and both oligo dT and random hexamers.
Ligand Detection: The presence of PDGFB mRNA was verified by means of RT-PCR as previously described.28
Immunohistochemistry
The antibodies and experimental conditions are shown in Table 1. The score for each marker is listed below.
PDGFRB. Cytoplasmic immunoreactivity was evaluated and scored on the basis of the percentage of immunoreactive cells (not less than 70%). PDGFRB staining intensity was evaluated using vessel wall immunoreactivity as an in-built control.
EGFR. The score was applied according to the manufacturer's instructions, combining the scores relating to staining intensity and the percentage of immunopositive cells.29 The resulting low, intermediate, or high/strong immunodecoration are reported in Table 2. In the cases scored as negative, none of the cells were immunodecorated.
Table 2.
Summary results
|
Upstream marker and method |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
PDGFRB |
PDGFB |
EGFR |
EGF | TGFα |
Her2/neu |
|||||||||
| Cases | RTK array | IP/WB | IHC | RT-PCR | RTK array | IP/WB | IHC | FISH | Real-time PCR | Real-time PCR | RTK array | IP/WB | IHC | FISH |
| 1 | + | +P | + | + | + | − | − | − | + | + | + | − | − | |
| 2 | +P | + | + | +P | − | − | + | + | − | − | ||||
| 3 | +P | + | + | +P | +M | − | + | + | − | − | ||||
| 4 | + | +P | + | + | + | +P | +S | + | + | + | + | +P | − | + |
| 5 | + | +P | + | + | + | +P | − | NE | + | + | + | +P | − | NE |
| 6 | + | +P | + | + | + | +P | − | + | + | + | − | − | − | + |
| 7 | +P | + | + | +P | − | + | + | + | − | + | ||||
| 8 | + | +P | + | + | − | − | − | − | + | + | − | − | − | − |
| 9 | +P | + | + | − | − | − | + | + | − | − | ||||
| 10 | +P | + | + | − | − | − | + | + | − | − | ||||
| 11 | +P | + | + | − | NE | − | + | + | − | − | ||||
| 12 | + | +P | + | + | + | +P | − | NE | + | + | + | − | − | NE |
| 13 | + | +P | + | + | + | +P | +S | − | + | + | + | − | − | − |
| 14 | +P | + | +P | + | + | − | ||||||||
| 15 | +P | + | + | +P | − | − | + | + | ±P | − | − | |||
| 16 | − | + | + | +W | − | − | + | + | − | − | − | |||
| 17 | +P | + | + | +W | − | − | + | + | ± | − | − | |||
| 18 | +P | + | + | +P | +L | − | + | + | +P | − | − | |||
| 19 | +P | + | + | +P | +S | − | + | + | − | − | + | |||
| 20 | +P | + | + | +P | +M | − | + | + | + | − | − | |||
| 21 | +P | + | + | +P | +S | NE | + | + | NE | NE | ||||
| 22 | +P | + | + | +P | − | − | + | + | − | − | − | |||
|
Downstream marker and method |
||||||||||||||
| P85 | AKT | PTEN | ERK 1/2 | mTOR | S6 | PS6 Thr240/244 | PS6 Thr235/236 | S6 | 4EBP1 | |||||
| Cases | IP/WB | WB | WB | FISH | Real-time | WB | WB | WB | WB | WB | FISH | Real-time | WB | |
| 1 | + | +P | + | M(80) | + | +P | +P | + | − | ±P | D | + | +P | |
| 2 | + | +P | + | D | + | +P | +P | + | ±P | ±P | D | +P | ||
| 3 | + | +P | + | M(70) | + | +P | +P | + | ±P | ±P | D | + | +P | |
| 4 | + | +P | + | HP | +P | +P | + | +P | +P | D | + | +P | ||
| 5 | + | +P | + | NE | +P | +P | + | ±P | +P | NE | +P | |||
| 6 | + | +P | + | HP(10) | +P | +P | − | − | − | D | + | +P | ||
| 7 | + | +P | + | M(50) | +P | +P | ± | − | ±P | D | + | |||
| 8 | + | +P | + | D | + | +P | − | − | − | − | LP | + | +P | |
| 9 | + | − | − | D | +P | ±P | − | − | − | D | + | |||
| 10 | + | +P | + | M(50) | +P | +P | + | ±P | − | D | + | +P | ||
| 11 | + | +P | + | D | +P | +P | − | − | − | + | ||||
| 12 | + | +P | + | M(50) | +P | +P | + | ±P | +P | +P | ||||
| 13 | + | +P | + | D | +P | +P | ± | − | ±P | D | +P | |||
| 14 | + | +P | + | +P | +P | + | ±P | + | ||||||
| 15 | + | +P | + | D | +P | +P | + | ±P | M | +P | ||||
| 16 | − | +P | + | D | +P | +P | − | − | D | + | ||||
| 17 | + | +P | + | M(60) | +P | +P | + | ±P | NE | + | ||||
| 18 | + | +P | + | D | +P | +P | − | − | D | +P | ||||
| 19 | + | +P | + | D | +P | +P | − | − | D | ± | ||||
| 20 | + | +P | + | M(90) | +P | +P | + | ±P | D | +P | ||||
| 21 | + | +P | + | NE | +P | +P | − | − | NE | − | ||||
| 22 | + | +P | + | D | +P | +P | + | ±P | + | |||||
The empty spaces correspond to not performed analyses. Abbreviations: IP, immunoprecipitation; WB, Western blotting; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; NE, not evaluable; P, phosphorilated; W, weak; D, disomic; HP, high polisomy (percentage); M, monosomic (percentage). For EGFR IHC: S, strong; M, moderate; L, low.
HER2/neu. The same parameters considered for PDGFRB were applied for this receptor. As for EGFR, in the cases scored as negative, none of the cells were positive.
For all these markers, the inter-observer correlation for the assessment of the staining was concordant (100%).
Fluorescence in Situ Hybridization
Fluorescence in situ hybridization analyses were used to investigate the gene status of EGFR, HER2/neu, PTEN, and S6 as previously described.26,30,31 In particular, the BAC clones RP11-126H20 and RP11-624N8 were used as probes for S6, and Spectrum Green(SG)-labeled CEP9 (Vysis) was used as the control probe. The dual-color FISH mixture was prepared using Spectrum Orange (SO)-labeled S6+ SG-labeled CEP9. The corresponding gene status was evaluated as previously described.32 To simplify the FISH/IHC correlations, the high EGFR and HER2/neu polysomy/amplification group was classified as FISH positive (FISH + ), and the remaining groups (disomy, trisomy, or low polysomy) as FISH negative (FISH−), whereas the PTEN and S6 gene status patterns and loss/gain percentages were detailed in each case.
Results
The profiles of the 8 recurrent tumors were almost identical to those of the 14 primary tumors. All of the data are summarized in Table 2.
Upstream Targets
RTK Array Assays
The RTK array experiments used the 7 chordomas for which adequate total protein extract was available (Patients 1, 4–6, 8, 12, and 13). RTK activation was evaluated by comparing the intensity of the labeled spots relating to each tumoral protein extract with that of those relating to the positive and negative controls on the array membrane.
All 7 patients showed activated PDGFRB (although the level of activation was low in case Patient 13) and, to a lesser extent, FLT3 and CSF1R of the PDGFR family. EGFR was highly activated in 4 patients (Patients 1, 4, 12, and 13), activated in 2 (Patients 5 and 6), and negative in 1 (Patient 8). HER2/neu was detected in 5 cases (Patients 1, 4, 5, 12, and 13), and HER4 in 3 (Patients 4, 5, and 12). Of the “non-druggable” RTKs, Axl and Dtk were activated in all of the patients. The negative controls were always undetectable. Figure 2A shows 4 representative cases (Patients 4, 5, 12, and 13). Overall, the profiles of the 42 RTKs evaluated on the array membrane indicated significant activation of members of the PDGFR family (PDGFRB, FLT3, and CSF1R) and the EGFR families (EGFR, HER2/neu, and HER4).
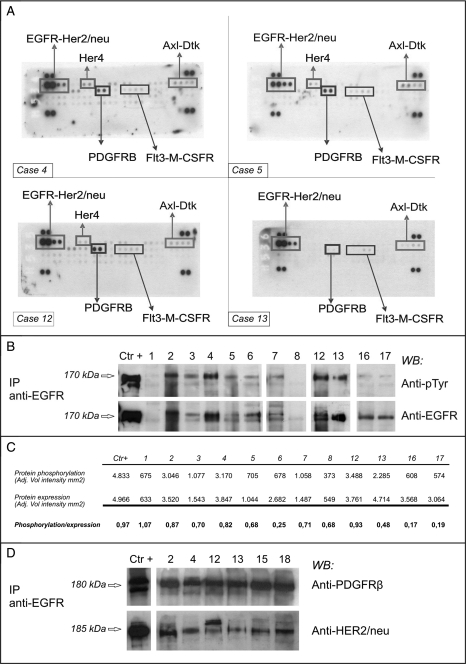
Fig. 2.
Expression and activation of RTKs. The sample numbers correspond to the cases listed in Table 2. (A) RTK arrays. Equal amounts of total protein extracts from 4 representative cases were incubated with the arrays. The rectangles indicate the presence of activated receptors. (B) EGFR IP/Western blot analysis of 12 representative cases. To confirm the presence of activated EGFR, proteins were immunoprecipitated with a specific antibody and blotted onto a membrane. The anti-pTyr panel identified the phosphorylated EGFR; the anti-EGFR panel indicates the expression of the corresponding receptor. The A431 cell line (American Type Culture Collection, Manassas, VA) was used as the positive control. (C) Densitometric analysis of levels of EGFR activation. The bands representing phosphorylated EGFR and total EGFR levels were quantified, and the ratios between phosphorylated and expressed EGFR were calculated, the ratio relating to the A431 cell line (positive control) was 0.97. The low level of phosphorylation detected in samples #16 and #17 may have been due to methodological problems (non-optimal cryopreservation). (D) Co-IP of EGFR/PDGFRB and EGFR/HER-2/neu. Total protein extracts from 6 EGFR-positive cases were immunoprecipitated with anti-EGFR antibody and run on a gel. The filter was incubated with anti-PDGFRB antibody, then stripped and re-incubated with anti-HER2/neu antibody. Extracts from the 2N5A cell line (immunoprecipitated with anti-PDGFRB) and SKBR3 cell line (immunoprecipitated with anti-HER2/neu) were respectively used as positive controls for PDGFRB and HER2/neu protein. The 2N5A cell line (derived from the NIH3T3 cell line and expressing the COL1-PDGFB fusion that characterized dermatofibrosarcoma protuberans) was kindly provided by Dr. Greco, Fondazione IRCCS Istituto Nazionale dei Tumori; the SKBR3 came from the American Type Culture Collection.
Confirmatory Biochemical Analysis of PDGFRB, EGFR, and HER2/Neu
All of the case material was analyzed by means of IP/WB in order to confirm the RTK array data. PDGFRB was activated in all but 1 case, in line with our previous data,15 whereas EGFR was activated in 17 and not activated in 5 (Patients 1 and 8–11); Fig. 2B shows 12 representative cases. The results obtained using the 2 methods perfectly overlapped, except in Patient 1. The EGFR WB results were quantified by densitometry in order to define the activation level of the receptor more precisely (Fig. 2C).
Total protein extracts from 6 cases expressing activated EGFR (Patients 2, 4, 12, 13, 15, and 18) were immunoprecipitated with anti-EGFR and then incubated with anti-PDGFRB: PDGFRB co-immunoprecipitated with EGFR in all cases (Fig. 2D).
Immunoprecipitation and Western blotting analysis of HER2/neu using antibody directed against the NH2 terminal showed that all of the cases were negative, which is consistent with the unrecognizable structural conformation of the receptor's extracellular domain. However, an antibody specific for the C-terminal applied to the 14 cases for which material was still available revealed HER2/neu expression/phosphorylation in 4 cases (Patients 4, 5, 15, and 18) and HER2/neu expression alone in 2 (Patients 17 and 20). The IP/WB and array data matched in 4 cases (Patients 4–6 and 8) and did not match in 2 (Patients 12 and 13); insufficient material prevented a comparison in 1 case (Patient 1).
The stripped membranes with EGFR immunoprecipitated proteins were re-incubated with a HER2/neu antibody, which revealed the presence of HER2/neu in 9/22 cases, thus suggesting that the 2 members of the family may be involved in heterodimers. The result was confirmed by further incubation of the stripped filter with the EGFR/PDGFRB co-IP experiment (Fig. 2D, upper panel) and with anti-HER2/neu antibody (Fig. 2D, lower panel).
Analysis of the Mechanisms Sustaining PDGFRB and EGFR Activation
We have previously demonstrated the absence of PDGFRB-activating mutations in chordomas;15 in this study, we analyzed the EGFR gene to exclude a mutation-based activation mechanism. EGFR was wild type in all of the samples.
Real-time PCR showed that all of the cases highly expressed both EGFR ligands (EGF and TGFα) and as expected,15 PDGFB mRNA was expressed in all of the analyzed cases (Table 2). We thus concluded that, most likely, the activation of these 2 receptors is sustained by an autocrine/paracrine loop.
Downstream Effectors
We biochemically and/or molecularly investigated the PI3K/AKT (the p85 and p110 subunits of PI3K, AKT, and PTEN) and MAPK pathways (RAS, BRAF, and Erk1/2), and then analyzed the mTOR kinase and its S6 and 4E-BP1 downstream effectors. The analysis of PTEN and S6 was complemented by the FISH analysis of FFPE material.
All of the data are summarized in Table 2.
PI3K/AKT Pathway
As a first confirmatory step of receptor activation, we verified the binding of the p85 subunit of PI3K to activated PDGFRB: co-IP was detected in all of the samples. The p110 subunit gene (PI3KCA) was investigated by looking for activating mutations, and the hot spots corresponding to exons 9 and 20 were sequenced; no mutations were detected in any of the cases.
Western blotting showed that AKT was activated to different extents in all of the analyzed samples (Fig. 3A upper panel), except Patient 9.
Fig. 3.
Expression of downstream signaling markers. (A) Western blots showing the presence of phosphorylated/activated and total AKT and ERK1/2. PTEN analyses: (B) FISH. The chromosome 10 probe is labeled with Spectrum Green and the PTEN locus-specific probe with Spectrum Orange. Case 1: monosomic PTEN in 80% of the cells. Case 2: disomic PTEN. (C) Western blotting. PTEN expression in 12 representative monosomic or disomic cases: monosomic cases were Patients 1, 3, 7, 12, and 17. The different electrophoretic mobility observed in Patient 16 may have been due to protein hyper-phosphorylation, which cannot be confirmed by the antibody used because it does not discriminate between phosphorylated and non-phosphorylated PTEN. The A431 and 3962 M cell lines were, respectively, used as positive and negative controls, and anti-actin antibody was used to normalize the results. (D) Real-time PCR in the disomic and monosomic cases. Lymphocyte mRNA was used for calibration purposes.
PTEN phosphatase reverses the effects of p110-PI3K. We sequenced the mutation hot spot (from exon 5 to exon 9) but did not detect any alterations.
As gene copy number is 1 of the mechanisms responsible for PTEN destabilization, we used FISH to analyze matched FFPE samples and found alterations in 9 (47%) of the 19 successfully analyzed cases: chromosome 10 monosomy in 7 cases (1 of which is shown in Fig. 3B) and a high degree of polysomy in 2 cases. However, WB evaluating the overall expression of PTEN protein indicated no differences in expression levels (Fig. 3C; the results were validated using a cell line null for PTEN expression). Given the discrepancy between the FISH analysis and protein expression, we further analyzed PTEN mRNA expression in 2 disomic (Patients 2 and 8) and 2 monosomic cases (Patients 1 and 3); the biochemical results were confirmed by real-time PCR as no differences were found between the monosomic and disomic cases (Fig. 4D).
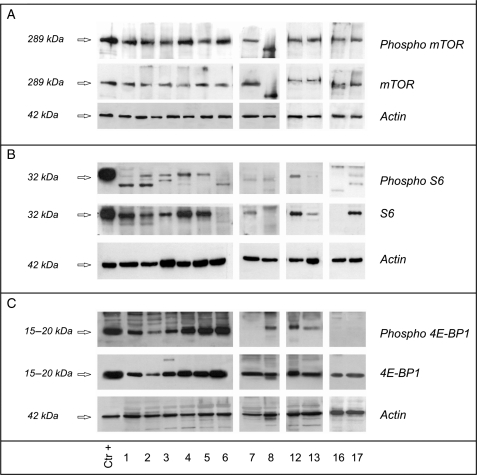
Fig. 4.
mTOR expression and its downstream effectors. mTOR (panel A), S6 (panel B), and 4E-BP1 (panel C) in the same 12 representative cases analyzed and reported in Figure 3, panel A. Twenty micrograms of total protein extract was loaded for each sample. The A431 cell line was used as the positive control and anti-actin antibody was used to normalize the results.
MAPK Pathway
Analysis of the KRAS and BRAF mutation hot spots revealed no activating mutations.
Direct WB showed that both p44 and p42 (Erk1/2) were much more highly expressed and phosphorylated than any other of the analyzed proteins (Fig. 3A lower panel).
mTOR, S6, and 4E-BP1
mTOR is activated by both pathways and, as both cascades are activated in chordomas, we expected to find an activated kinase in all 22 samples. WB revealed a phosphorylated protein in all but 2 cases (Patient 9 was only weakly positive; Patient 8 was negative) (Fig. 4A).
The activation status of S6 was investigated using Ser235–236 and Ser240–244 phospho-antibodies. Ser240–244 residues are preferentially phosphorylated in response to the mTOR/PI3K pathway, and S6 was found to be expressed and activated to a low/very low extent in 11 cases, not expressed in 8, and expressed but not phosphorylated in 3 (Fig. 4B). The same trend (a low level of activation) was observed in the 13 cases that were analyzed using the Ser235–236 antibody, which also detects the residues phosphorylated in response to the mTOR-independent RAS/MAPK pathway.
As the low level of protein expression/activation despite mTOR activation suggested that S6 plays an oncosuppressing role, we used FISH (on FFPE material) and real-time PCR to ascertain its gene status and mRNA expression. The FISH results showed that all but 1 of the analyzed cases had a disomic or low polysomic pattern (Patient 15 was monosomic, with positive protein expression). However, it is possible that our probe does not detect the very small 3979 bp deletion of the S6 gene,33 and that the loss in Patient 15 (which also carried monosomy of the p16 gene closely mapping on the same chromosome; data not shown) was attributable to a longer deletion encompassing both genes, although a second probe gave the same results (data not shown). Nevertheless, real-time PCR showed the same level of S6 transcript in the samples with (Patients 1, 3, 4, and 10) and without S6 protein expression (Patients 6 and 8).
To investigate 4E-BP1, we used the Thr70 phospho-antibody to highlight protein hyper-phosphorylation, which, by abrogating the association between 4E-BP1 and eIF4E, releases translational repression. 4E-BP1 was hyper-phosphorylated in 13 cases, not expressed in 1, and expressed but not phosphorylated in 8 (Fig. 4C), which means that eIF4E was released in 14 cases, thus favoring protein synthesis, cell proliferation, and tumor progression. In 4 cases (Patients 9, 11, 16, and 19), 4E-BP1 was expressed but not phosphorylated (and thus eIF4E was repressed), and the fact that S6 was null supports the hypothesis that malignant progression may be sustained by the absence of S6, which may act as an oncosuppressing gene as it does in chondrosarcomas.23 The level of S6 activation was low in the remaining 4 cases in which eIF4E was repressed (Patients 7, 14, 17, and 22). In Patient 8, in which 4E-BP1 was hyper-phosphorylated in the absence of mTOR activation and Ser235–236/240–244 S6 phosphorylation, it is possible to assume activation through the RAS/MAPK cascade mediated by a still unknown factor,19 although many oncogenic events may lead to 4E-BP1 phosphorylation.34
Taken together, our findings downstream of mTOR suggest that both PI3K/AKT and RAS/MAPK signaling are involved in regulating protein transduction and that the 4E-BP1 pathway drives protein synthesis, whereas the hypothesized oncosupprossing role of S6 remains undetermined and will require further experiments.
Methods of Selecting Patients for Possible Targeted Treatments
Our previous positive experience with PDGFRB immunophenotyping in chordoma,15 and the recent favorable results obtained in non-small cell lung cancer using gene and protein profiling,35,36 prompted us to find a screening method capable of identifying predictive markers for TK inhibitors that can be easily applied to FFPE samples (frozen material is not frequently available). To this end, we investigated the sensitivity of IHC and FISH analyses (alone or combined), taking the biochemical results as the gold standard.
IHC. In all the cases (100%), more than 80% of the cells showed PDGFRB cytoplasmatic immunoreactivity, with the intensity of the staining being stronger than that of the vessel wall used as the control.
EGFR was expressed in 7/20 cases (4 classified as strong, 2 as moderate, and 1 as low) showing a decoration mainly related to the cell membrane. In the remaining cases, scored as negative, none of the tumoral cells were positive for EGFR.
All cases were negative for HER2/neu as all tumor cells were negative.
FISH. As amplification is one of the mechanisms responsible for receptor activation in the case of EGFR family genes, we used FISH to analyze the EGFR and HER2/neu genes. Of the 18 evaluable cases, the EGFR gene was disomic in 11 and low polysomic in 4 (Patients 10 and 17–19) (FISH−) and high polysomic in 3 (Patients 4, 6, and 7) (FISH+).
The HER2/neu gene was disomic in 10 and low polysomic in 4 (Patients 10, 11, 17, and 18) (FISH−), and was high polysomic in 3 (Patients 4, 7, and 19) or amplified in some areas (Patient 6) (FISH+).
All 7 cases with deregulated EGFR also showed deregulated HER2/neu; they all also had the same deregulation pattern except for Patients 19 and 6.
Correlations Between IP/WB, IHC, and FISH
EGFR. We compared the IP/WB results with those of IHC and FISH in 17 evaluable patients. The IP/WB and IHC data correlated in 9 cases: in particular, all of the IP/WB-negative cases were confirmed by IHC, whereas only 5 of the 13 IP/WB-positive cases (38%) were also IHC positive, thus indicating that IHC is less sensitive than IP/WB but does not generate false positives.
The combination of IHC and FISH increased the number of “targetable” patients by recovering 2 FISH+ cases from the 8 IHC-negative and IP/WB-positive patients (7/13 cases; 54%).
HER2/neu. This receptor was identified by IP/WB in 6/14 evaluable cases (43%), all of which were IHC negative. The lack of any correlation between a gain in gene copy number and HER2/neu expression suggests that IP/WB positivity may be due to heterodimer formation.
Discussion
The aim of this study was to use biochemical analyses (RTK array cards that can provide an overview of phosphorylated RTK in small amounts of total protein extracts and IP/WB) of cryopreserved material to demonstrate and validate the presence of a reproducible panel of activated receptors from mainly the 2 RTK families of PDGFR and EGFR. We have previously shown that PDGFRB is activated by means of a ligand-dependent mechanism;15 we now confirm the PDGFRB data and show that the pathogenesis of chordoma also involves EGFR (Table II, IP/WB column, Fig. 2B and C).
The co-expression and co-immunoprecipation of PDGFRB and EGFR has already been described at the preclinical level, and it has been demonstrated that PDGFB stimulates the activation of PDGFRB and subsequently stimulates EGFR unidirectionally.37,38 Our results demonstrate the co-IP of EGFR and PDGFRB in surgical specimens and suggest heterodimer formation. If these findings are confirmed, they would explain the existence of imatinib-insensitive chordomas which, as they are also addicted for EGFR, can be expected to escape imatinib treatment. It is interesting to note in this regard that a response to cetuximab (an EGFR-specific monoclonal antibody) has been reported in 2 chordoma patients.39,40
In addition to the interaction between the 2 families, our co-IP data also indicate the possible existence of heterodimerization between individual members of the EGFR family at least in some cases, as supported by the finding that the receptor was only bound by a specific C-terminal antibody, and not by an N-terminal antibody. This suggests that the use of bi-specific inhibitors such as lapatinib, which blocks both EGFR and HER2/neu,41–43 may offer more clinical benefit than mono-specific treatments.
Finally, the phospho-RTK-array approach has shown that other members of the PDGFR family are activated, including CSF1R and FLT3, which have been reported to be inhibited by imatinib.44,45
After the upstream RTK analysis, we biochemically and/or molecularly and cytogenetically investigated some downstream effectors, and the results indicated that both the PI3K/AKT and MAPK pathways were activated in the absence of activating mutations. The co-IP experiments showed that the p85 subunit of PI3K directly binds the activated/dimerized PDGFRB receptor and, in line with this, AKT was phosphorylated (Fig. 3A). Molecular and FISH analyses of the PTEN oncosuppressor gene showed the absence of inactivating mutations and the presence of PTEN alterations (mainly chromosome 10 monosomy) in 7 of 19 cases, although the latter do not seem to affect protein function because direct WB did not reveal any difference in the expression of the phosphatase between the monosomic and disomic cases (Fig. 3B and C). The WB results were further confirmed by real-time PCR. We also detected highly expressed and phosphorylated ERK1 and ERK2 (Fig. 3A), which is consistent with activation of the EGFR family, in the presence of wild-type KRAS and BRAF genes (but these 2 markers were not biochemically analyzed). Finally, in line with previously published data indicating that both AKT and ERK1/2 can activate mTOR, the protein was phosphorylated in all but 1 case (Fig. 4A), which suggests that it may be a further “druggable” target.
Taken together, our results show that mTOR activation is due to the growth factor-mediated activation of PI3K and MAPK, with no contribution from PTEN. This means that our findings only partially overlap those of a recent WB study of the growth-factor deprived, chordoma-derived U-CHI cell line and IHC investigations of sporadic sacral chordomas samples, on the basis of which the authors suggested that partial or complete PTEN deficiency may be responsible for mTOR signaling activation.46
Although needing further confirmation, our findings concerning the 2 regulators of the translational machinery downstream of mTOR raise important speculative and practical issues. Unlike that in alveolar sarcomas,47 protein synthesis in chordomas seems to be harmonized by 4E-BP1 rather than S6, and this reinforces the idea that the translational apparatus may differently regulate protein production (and thus the cell cycle and tumor progression) in different histotypes.24 It is currently believed that the 4E-BP1/eIF4E pathway plays a key role in tumorigenesis and the 4E-BP1 is a “funnel effector” that is relevant to prognosis and treatment response.34 Our finding that the 2 effectors have selective and histotype-restricted functional roles raise doubts about the use of the phospho-S6 antibody as a biomarker for the activation of the PI3K/mTOR pathway in everyday practice, and indicates a need to extend the analysis to the phospho-4E-BP1 antibody.
Finally, the low level of S6 activation suggests that S6 may act as an oncosuppressor gene, even if the experimental proof here reported do not indicate any gene copy loss or any decrease in mRNA levels in the protein–negative cases.
As previous IHC investigations have shown that PDGFRB predicts receptor activation in 100% of cases,15 we analyzed matched FFPE samples for EGFR and HER2/neu by means of IHC and interphase FISH (taking the biochemical results as the gold standard) in an attempt to acquire insights into identifying patients who may benefit from targeted EGFR and HER2/neu treatments. The results indicated that after combining IHC and FISH (2 techniques that are commonly used in the case of FFPE material), EGFR sensitivity increases to 54% without any false positives and so, despite the constant HER2/neu null immunophenotype (given the co-expression of EGFR and HER2/neu), this may be sufficient for patient selection. As HER2/neu is involved in EGFR dimer formation, its null immunophenotype is expected and has previously been reported in chordomas. Cumulatively, our results are in agreement with previously reported IHC data48 for PDGFRB and Her2/neu, while a lower percentage of positivity was observed for EGFR. This discrepancy, however, could be due to the different experimental conditions applied (different EGFR antibody).
In conclusion, our data indicate new avenues for combining chordoma treatments49 by targeting upstream and downstream effectors. In addition to the PDGFR family, which is inhibited by imatinib, the presence of activated receptors of the EGFR family may provide a rationale for treating these patients with small molecules that are effective on both EGFR and HER2/neu. Moreover, the strong activation of downstream signaling supports the idea that a combination of upstream TK antagonists and mTOR and MAPK25 inhibitors could lead to the better control of tumor growth by avoiding the so-called negative feedback loop,17 lapatinib resistance,50 and the PI3K-dependent feedback loop.25 However, further studies are needed to clarify the multifaceted regulation of downstream mTOR effectors in different histotypes.
Conflict of interest statement. None declared.
Funding
This study was supported by a grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC) to Dr. Pilotti and Dr. Tamborini, and a grant from the Lyddi Shriver Foundation.
References
- 1.McMaster ML, Goldstein AM, Bromley CM, Ishibe N, Parry DM. Chordoma: incidence and survival patterns in the United States, 1973–1995. Cancer Causes Control. 2001;12:1–11. doi: 10.1023/a:1008947301735. [DOI] [PubMed] [Google Scholar]
- 2.Mirra JM, Nelson SD, Della Rocca C, Mertens F. Chordoma. In: Fletcher CDM, Unni KK, Mertens F, editors. Pathology and Genetics of Tumors of Soft Tissue and Bone. World Health Organization Classification of Tumors. Lyon: IARC Press; 2002. pp. 316–317. [Google Scholar]
- 3.Chugh R, Tawbi H, Lucas DR, Biermann JS, Schuetze SM, Baker LH. Chordoma: the nonsarcoma primary bone tumor. Oncologist. 2007;12(11):1344–1350. doi: 10.1634/theoncologist.12-11-1344. [DOI] [PubMed] [Google Scholar]
- 4.Miozzo M, Dalprà L, Riva P, et al. A tumor suppressor locus in familial and sporadic chordoma maps to 1p36. Int J Cancer. 2000;87(1):68–72. [PubMed] [Google Scholar]
- 5.Kelley MJ, Korczak JF, Sheridan E, Yang X, Goldstein AM, Parry DM. Familial chordoma, a tumor of notochordal remnants, is linked to chromosome 7q33. Am J Hum Genet. 2001;69(2):454–460. doi: 10.1086/321982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riva P, Crosti F, Orzan F, et al. Mapping of candidate region for chordoma development to 1p36.13 by LOH analysis. Int J Cancer. 2003;107(3):493–497. doi: 10.1002/ijc.11421. [DOI] [PubMed] [Google Scholar]
- 7.Börgel J, Olschewski H, Reuter T, Miterski B, Epplen JT. Does the tuberous sclerosis complex include clivus chordoma? A case report. Eur J Pediatr. 2001;160(2):138. doi: 10.1007/s004310000645. [DOI] [PubMed] [Google Scholar]
- 8.Dutton RV, Singleton EB. Tuberous sclerosis: a case report with aortic aneurysm and unusual rib changes. Pediatr Radiol. 1975;3(3):184–186. doi: 10.1007/BF01006909. [DOI] [PubMed] [Google Scholar]
- 9.Schroeder BA, Wells RG, Starshak RJ, Sty JR. Clivus chordoma in a child with tuberous sclerosis: CT and MR demonstration. J Comput Assist Tomogr. 1987;11(1):195–196. doi: 10.1097/00004728-198701000-00048. [DOI] [PubMed] [Google Scholar]
- 10.Storm PB, Magge SN, Kazahaya K, Sutton LN. Cervical chordoma in a patient with tuberous sclerosis presenting with shoulder pain. Pediatr Neurosurg. 2007;43(2):167–169. doi: 10.1159/000098396. [DOI] [PubMed] [Google Scholar]
- 11.Lee-Jones L, Aligianis I, Davies PA, et al. Sacrococcygeal chordomas in patients with tuberous sclerosis complex show somatic loss of TSC1 or TSC2. Genes Chromosomes Cancer. 2004;41(1):80–85. doi: 10.1002/gcc.20052. [DOI] [PubMed] [Google Scholar]
- 12.Casali PG, Messina A, Stacchiotti S, et al. Imatinib mesylate in chordoma. Cancer. 2004;101(9):2086–2097. doi: 10.1002/cncr.20618. [DOI] [PubMed] [Google Scholar]
- 13.Stacchiotti S, Bertulli R, Grosso F, et al. Antitumor activity of imatinib plus cisplatin in three patients with advanced chordoma. Proc. 12th Ann Connective Tissue. Oncology Society Meet. :58. Abs. # 725. [Google Scholar]
- 14.Stacchiotti S, Marrari A, Tamborini E, et al. Response to imatinib plus sirolimus in advanced chordoma. Ann Oncol. 2009;20(11):1886–1894. doi: 10.1093/annonc/mdp210. [DOI] [PubMed] [Google Scholar]
- 15.Tamborini E, Miselli F, Negri T, et al. Molecular and biochemical analyses of platelet-derived growth factor receptor (PDGFR) B, PDGFRA, and KIT receptors in chordomas. Clin Cancer Res. 2006;12(23):6920–6928. doi: 10.1158/1078-0432.CCR-06-1584. [DOI] [PubMed] [Google Scholar]
- 16.Weinberger PM, Yu Z, Kowalski D, et al. Differential expression of epidermal growth factor receptor, c-Met, and HER2/neu in chordoma compared with 17 other malignancies. Arch Otolaryngol Head Neck Surg. 2005;131(8):707–711. doi: 10.1001/archotol.131.8.707. [DOI] [PubMed] [Google Scholar]
- 17.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441(7092):424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 18.Roux PP, Shahbazian D, Vu H, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282(19):14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert TP, Tee AR, Proud CG. The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J Biol Chem. 2002;277:11591–11596. doi: 10.1074/jbc.M110367200. [DOI] [PubMed] [Google Scholar]
- 20.Azar R, Najib S, Lahlou H, Susini C, Pyronnet S. Phosphatidylinositol 3-kinase-dependent transcriptional silencing of the translational repressor 4E-BP1. Cell Mol Life Sci. 2008;65(19):3110–3117. doi: 10.1007/s00018-008-8418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolli-Derkinderen M, Machavoine F, Baraban JM, Grolleau A, Beretta L, Dy M. ERK and p38 inhibit the expression of 4E-BP1 repressor of translation through induction of Egr-1. J Biol Chem. 2003;278(21):18859–18867. doi: 10.1074/jbc.M211696200. [DOI] [PubMed] [Google Scholar]
- 22.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31(6):342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Rozeman LB, Szuhai K, Schrage YM, et al. Array-comparative genomic hybridization of central chondrosarcoma: identification of ribosomal protein S6 and cyclin-dependent kinase 4 as candidate target genes for genomic aberrations. Cancer. 2006;107(2):380–388. doi: 10.1002/cncr.22001. [DOI] [PubMed] [Google Scholar]
- 24.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA. 2008;105(45):17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carracedo A, Ma L, Teruya-Feldstein J, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamborini E, Casieri P, Miselli F, et al. Analysis of potential receptor tyrosine kinase targets in intimal and mural sarcomas. J Pathol. 2007;212(2):227–235. doi: 10.1002/path.2177. [DOI] [PubMed] [Google Scholar]
- 27.Daniotti M, Oggionni M, Ranzani T, et al. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23(35):5968–5977. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- 28.Lagonigro MS, Tamborini E, Negri T, et al. PDGFRalpha, PDGFRbeta and KIT expression/activation in conventional chondrosarcoma. J Pathol. 2006;208(5):615–623. doi: 10.1002/path.1945. [DOI] [PubMed] [Google Scholar]
- 29.Perrone F, Suardi S, Pastore E, et al. Molecular and cytogenetic subgroups of oropharyngeal squamous cell carcinoma. Clin Cancer Res. 2006;12(22):6643–6651. doi: 10.1158/1078-0432.CCR-06-1759. [DOI] [PubMed] [Google Scholar]
- 30.Dagrada GP, Mezzelani A, Alasio L, et al. HER-2/neu assessment in primary chemotherapy treated breast carcinoma: no evidence of gene profile changing. Breast Cancer Res Treat. 2003;80(2):207–214. doi: 10.1023/A:1024579206250. [DOI] [PubMed] [Google Scholar]
- 31.Perrone F, Lampis A, Orsenigo M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20(1):84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 33.Antoine M, Fried M. The organization of the intron-containing human S6 ribosomal protein (rpS6) gene and determination of its location at chromosome 9p21. Hum Mol Genet. 1992;1(8):565–570. doi: 10.1093/hmg/1.8.565. [DOI] [PubMed] [Google Scholar]
- 34.Armengol G, Rojo F, Castellví J, et al. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67(16):7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 35.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97(9):643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 36.Hirsch FR, Varella-Garcia M, Cappuzzo F, et al. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2007;18(4):752–760. doi: 10.1093/annonc/mdm003. [DOI] [PubMed] [Google Scholar]
- 37.Liu P, Anderson RGW. Spatial organization of EGF receptor transmodulation by PDGF. Biochem Biophysic Res Comm. 1999;261:695–700. doi: 10.1006/bbrc.1999.1082. [DOI] [PubMed] [Google Scholar]
- 38.He H, Levitzki A, Zhu HJ, Walker F, Burgess A, Maruta H. Platelet-derived growth factor requires epidermal growth factor receptor to activate p21-activated kinase family kinases. J Biol Chem. 2001;276:26741–26744. doi: 10.1074/jbc.C100229200. [DOI] [PubMed] [Google Scholar]
- 39.Hof H, Welzel T, Debus J. Effectiveness of cetuximab/gefitinib in the therapy of a sacral chordoma. Onkologie. 2006;29:572–574. doi: 10.1159/000096283. [DOI] [PubMed] [Google Scholar]
- 40.Lindén O, Stenberg L, Kjellén E. Regression of cervical spinal cord compression in a patient with chordoma following treatment with cetuximab and gefitinib. Acta Oncol. 2009;48:158–159. doi: 10.1080/02841860802266672. [DOI] [PubMed] [Google Scholar]
- 41.Xia W, Mullin RJ, Keith BR, et al. Anti-tumor activity of GW572016: a dual tyrosine kinase inhibitor blocks EGF activation of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene. 2002;21:6255–6263. doi: 10.1038/sj.onc.1205794. [DOI] [PubMed] [Google Scholar]
- 42.Burris HA., III Dual kinase inhibition in the treatment of breast cancer: initial experience with the EGFR/ErbB-2 inhibitor lapatinib. Oncologist. 2004;9(Suppl 3):10–15. doi: 10.1634/theoncologist.9-suppl_3-10. [DOI] [PubMed] [Google Scholar]
- 43.Johnston SR, Leary A. Lapatinib: a novel EGFR/HER2 tyrosine kinase inhibitor for cancer. Drugs Today (Barc). 2006;42:441–453. doi: 10.1358/dot.2006.42.7.985637. [DOI] [PubMed] [Google Scholar]
- 44.Dewar AL, Cambareri AC, Zannettino AC, et al. Macrophage colony-stimulating factor receptor c-fms is a novel target of imatinib. Blood. 2005;105:3127–3132. doi: 10.1182/blood-2004-10-3967. [DOI] [PubMed] [Google Scholar]
- 45.Sternberg DW, Licht JD. Therapeutic intervention in leukemias that express the activated fms-like tyrosine kinase 3 (FLT3): opportunities and challenges. Curr Opin Hematol. 2005;12:7–13. doi: 10.1097/01.moh.0000147891.06584.d7. [DOI] [PubMed] [Google Scholar]
- 46.Han S, Polizzano C, Nielsen GP, Hornicek FJ, Rosenberg AE, Ramesh V. Aberrant hyperactivation of Akt and mammalian target of rapamycin complex 1 signaling in sporadic chordomas. Clin Cancer Res. 2009;15(6):1940–1946. doi: 10.1158/1078-0432.CCR-08-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stacchiotti S, Tamborini E, Marrari A, et al. Response to sunitinib malate in advanced alveolar soft part sarcoma. Clin Cancer Res. 2009;15(3):1096–1104. doi: 10.1158/1078-0432.CCR-08-2050. [DOI] [PubMed] [Google Scholar]
- 48.Fasig HJ, Dupont WD, LaFleur BJ, Olson SJ, Cates JMM. Immunohistochemical analysis of receptor tyrosine kinase signal transduction activity in chordoma. Neuropathol Appl Neurobiolog. 2008;34:95–104. doi: 10.1111/j.1365-2990.2007.00873.x. [DOI] [PubMed] [Google Scholar]
- 49.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 50.Vazquez-Martin A, Oliveras-Ferraros C, Colomer R, Brunet J, Menendez JA. Low-scale phosphoproteome analyses identify the mTOR effector p70 S6 kinase 1 as a specific biomarker of the dual-HER1/HER2 tyrosine kinase inhibitor lapatinib (Tykerb) in human breast carcinoma cells. Ann Oncol. 2008;19:1097–1109. doi: 10.1093/annonc/mdm589. [DOI] [PubMed] [Google Scholar]