Abstract
IGF-1 receptor signaling contributes to the growth of many solid tumors, including glioblastoma. This study analyzed the sensitivity of 8 glioblastoma cultures to the IGF-1 receptor inhibitor NVP-AEW541. Growth reduction, caused by a combination of antiproliferative and proapoptotic effects, varied between 20% and 100%. Growth-inhibitory effects of IGF-1 receptor siRNA were also demonstrated in 2 of the cultures. Activating mutations in PIK3CA were found in 2 cultures, and 2 other cultures displayed ligand-independent Akt phosphorylation. Growth inhibition was significantly reduced in cultures with PIK3CA mutations or ligand-independent Akt phosphorylation. PTEN siRNA experiments supported the notion that the status of the PI3K/PTEN/Akt pathway is involved in determining NVP-AEW541 sensitivity. Combination treatments with either PI3 kinase or mTOR inhibitors together with NVP-AEW541 were performed. These experiments demonstrated the effects of NVP-AEW541 in cells not responding to mono-treatment with the IGF-1 receptor inhibitor, when used together with either of the 2 other inhibitors. Together, the studies support continued clinical development of IGF-1 receptor antagonists for glioblastomas and identify links between PI3K/PTEN/Akt status and sensitivity to mono-treatment with NVP-AEW541. Furthermore, the studies suggest that NVP-AEW541 is also active together with PI3 kinase and mTOR inhibitors in cultures with a dysregulated PI3K/PTEN/Akt pathway. These studies should assist in future clinical development of IGF-1 receptor antagonists for glioblastoma and other tumors.
Keywords: Akt, glioblastoma multiforme, IGF-1 receptor, NVP-AEW541, PIK3CA, PTEN
A series of experimental studies have demonstrated that IGF-1 receptor signaling contributes to the growth and survival of malignant cells from many types of solid tumors.1,2 IGF-1 receptor expression is commonly observed on the epithelial cells of solid tumors,3 and studies with different types of IGF-1 receptor antagonists have demonstrated proapoptotic as well as antiproliferative effects in human cancer cells following the IGF-1 receptor inhibition.4,5 A critical role of IGF-1 receptor signaling in the growth and survival of malignant cells is also supported by the observation that IGF-1 receptor-deficient fibroblasts are resistant to oncogene-mediated transformation.6
In contrast to many other cancer-related receptor tyrosine kinases, the IGF-1 receptor does not appear to be a common target for mutational activation through, for example, amplification or point mutations. However, a number of different types of IGF-1 receptor inhibitors, including antibodies and low-molecular–weight tyrosine kinase inhibitors, have entered clinical trials with promising results in tumor types where IGF-1 receptor mutations have not been described.7,8
Glioblastoma is the most common human brain tumor. Recent studies have demonstrated improvement in survival by treatment combining surgery, radiation, and temozolomide. However, median survival with this treatment is less than 15 months.9 Additional treatments are therefore highly warranted. Studies on the genetics of glioblastomas have indicated that inactivation of the p53 and Rb pathways appears in most, if not all, tumors.10–14 Additionally, perturbations of growth factor pathways are common. Amplification of EGF receptors occur in about 40% of glioblastomas where about 60% have genetic alterations, which can give rise to the EGFRvIII variant, which lacks part of the extracellular domain.15,16 Furthermore, mutations of PTEN or PIK3CA have been demonstrated to occur in gliomas at frequencies of about 30% and 10%–30%, respectively.17–19
A role of IGF-1 receptor signaling in glioblastoma growth has previously been suggested based on the inhibitory effects of IGF-1 receptor inhibitors in cell and animal glioblastoma models.20,21 Although recent large-scale sequencing studies have not identified mutations in the IGF-1 receptor as a common event in glioblastoma, there is other evidence supporting a role of IGF-1 receptor signaling in glioblastoma growth. A recent study demonstrated that stimulation of glioblastoma cells induces proliferation and migration.22 Furthermore, upregulation of IGF-1 was identified as a candidate mechanism for erlotinib resistance in glioblastoma cultures.23 Together, these studies suggest that IGF-1 receptor activation, through paracrine or autocrine IGF-1 production, contributes to the pathology of glioblastoma.
NVP-AEW541 is a selective IGF-1 receptor low-molecular–weight inhibitor, which has shown antitumor activity in experimental tumor models.24–27 In cellular assays, this compound displays an IC50 for the IGF-1 receptor inhibition of 0.086 µm, which is more than 20-fold lower than the IC50 for insulin receptors.27 The IC50 for PDGFR and EGFR receptors in cellular assays is >10 µM.27
Receptor tyrosine kinase inhibitors show large variations in efficacy when used in model systems or patients. Accumulating results from the experimental and clinical studies suggest that mutational alterations in downstream signaling pathways, rendering cells less dependent on receptor signaling, confer lack of sensitivity. For example, KRAS mutations are associated with resistance to EGFR inhibitors in colorectal and lung cancer.28–32 Similarly, PTEN loss or mutational PI3 kinase activation has been implied as resistance mechanisms for HER2- and EGFR-targeting antibodies in breast, lung, and colorectal cancer.33–36 Finally, BRAF mutations also reduce sensitivity to EGFR-targeting antibodies.37 So far, the effects of the status of downstream signaling pathways on sensitivity to IGF-1 receptor inhibitors have not been reported.
Concerning determinants of glioblastoma sensitivities to other inhibitors of receptor tyrosine kinases, some studies have been reported.38–40 Characterization of tumor specimens from patients receiving therapy with EGF receptor inhibitors, and associated experimental studies identified PTEN expression, Akt phosphorylation, and the expression of the EGFRvIII variant as important determinants of response to these agents.38,40 Furthermore, PDGFR expression and activation correlate with imatinib sensitivity.39
In this study, we have analyzed the NVP-AEW541 sensitivity of a panel of high-grade–glioma cultures and combined these data with results from the analyses of IGF-1 receptor status and of the components of the PI3K/PTEN/Akt signaling pathway.
Material and Methods
Cell Culture
The establishment of the high-grade–glioma cultures and methods for the determination of their basal growth rate has been described previously.39
Drug-Induced Growth Inhibition
The cell cultures were analyzed with regard to the effect of NVP-AEW541, LY294002, or rapamycin. Stock solutions of 10 mM NVP-AEW541 (the drug was provided by Novartis), 20 mM LY29002 (Calbiochem), or 0.1 mM rapamycin (Calbiochem) were maintained at −20°C dissolved in DMSO. Cells were seeded at a density of 3000–4000 cells/well in 96-well plates (Sarstedt). After 1 day of culture, media were exchanged to media with or without 1 µM NVP-AEW541, 20 µM LY294002, or 1 nM rapamycin. After 4 days of incubation, cells were fixed for 30 minutes in cold 4% para-formaldehyde in PBS and stained for 30 minutes with 0.01% crystal violet in 4% ethanol. Samples were washed 3 times with distilled water, and air-dried for at least 30 minutes. Stained cells were dissolved in 100 µL 1% SDS and absorbance was quantified in a Victor ELISA reader (PerkingElmer) at 600 nm. NVP-AEW541 response shown in Fig. 1 was calculated as described previously39 and is expressed as a percent reduction in increase in cell amount during the 4-day period of treatment. In the experiments with combination treatments, growth inhibition was quantified by comparing the cell number in control-treated cultures, which was set to 1, with the cell number in cultures treated with 1 mM NVP-AEW541, 20 mMLY29002, or 1 nM rapamycin, which were used alone or in combination.
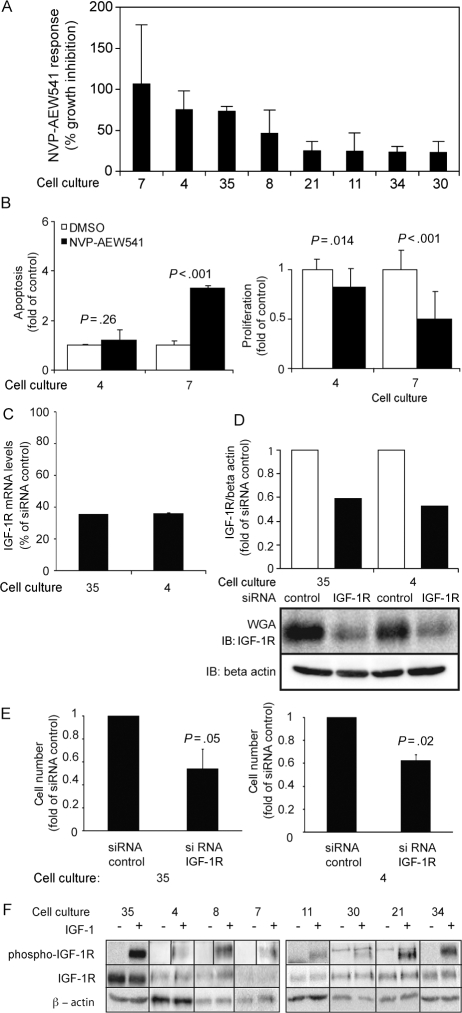
Fig. 1.
Analyses of the sensitivity to NVP-AEW541 and IGF-1 receptor siRNA and the characterization of IGF-1 receptor status. (A) Sensitivity of cultures to NVP-AEW541 treatment. Sensitivity was analyzed by comparing growth in tissue culture media containing 10% FCS, with and without the addition of 3 µM of NVP-AEW541; 100% growth inhibition corresponds to a situation with equal number of cells at the start and the end of the experiment. Results are means derived from 2 to 3 experiments for each cell culture. (B) Apoptotic and proliferative effects of NVP-AEW541 on 2 sensitive glioma cultures. (C and D) Efficacy of downregulation of IGF-1 receptor mRNA and protein levels upon IGF-1 receptor siRNA treatment. mRNA levels were determined after 3 days of siRNA treatment by qRT-PCR, and the receptor levels were determined in WGA fractions by IGF-1 receptor immunoblotting. Quantification of the receptor levels, normalized to β-actin levels, is shown as bar diagram above immunoblotting images. (E) IGF-1 receptor downregulation reduces the growth rate of cultures 35 and 4. Effects of IGF-1 receptor downregulation on growth were analyzed following 3 days of culture in 1% FCS. (B, C, and E) Results are means of 3 experiments for each cell culture and error bars indicate the standard deviation. The significance of changes was determined by Student's t-test. (F) IGF-1 receptor expression and phosphorylation in glioblastoma cultures. β-Actin amounts indicate input amount of the different cell lysates.
Analyses of IGF-1 Receptor Status
For the analyses of IGF-1 receptor protein expression and phosphorylation, 5 × 106 cells were plated in 10 cm dishes and grown in media containing 10% FCS. The next day, they were starved in 1% FCS overnight and stimulated with 10 ng/mL IGF-1 ligand (Peprotech) for 10 minutes at 37°C, followed directly by lysis on ice. To precipitate the IGF-1 receptor, cell lysates were either incubated with wheat germ agglutinin (WGA, Sigma-Aldrich) overnight or preincubated for 1 hour with an IGF-1Rβ antibody (Santa Cruz Biotechnologies) at 4°C during slow rotation, followed by overnight incubation with protein-G sepharose (GE Healthcare Bio-Science). The next day, the samples were washed with lysis buffer and denatured in sample buffer and subsequently subjected to SDS–polyacrylamide gel electrophoresis (SDS–PAGE) in 8% gels followed by transfer to Hybond-C-extra membranes (GE Healthcare Bio-Science). Membranes were blocked for 1 hour with 10% BSA and incubated overnight at 4°C with phospho-tyrosine antibody PY99 (sc-7020, Santa Cruz Biotechnologies) to detect phosphorylation of the IGF-1 receptor and with the same IGF-1Rβ antibody as above to detect total amounts of receptor.
NVP-AEW541-Induced Changes in Apoptosis and Proliferation
For the analysis of apoptosis, cells were seeded at a density of 20 000 cells/well in 24-well plates. For the proliferation assay, 10 000 cells/well in 96-well plates were seeded. After 1 day of culture, media were exchanged to media with 3 µM AEW541 or addition of the same amount of DMSO. Proliferation was measured after 1 day of incubation by incubating cells with 5-bromo-2′-deoxy-uridine (BrdU) for 5 hours followed by the analysis of incorporation with the BrdU Labeling and Detection Kit III (Roche). Apoptosis was measured after 2 days of incubation by the detection of cytoplasmic mono- and oligonucleosomes with the Cell Death Detection ELISA PLUS kit (Roche). Treatment with 25 µM etoposide was used as a positive control in both assays.
PCR, Sequencing, and Mutational Analysis
Previously described primers for PCR amplification and sequencing of PIK3CA, exon 9, were used (Sigma Genosys).18 Genomic DNA was extracted from selected cell cultures with the Genomic DNA Purification Kit (Fermentas). Fragments for sequencing were amplified by PCR with PfuI enzyme (Promega). The correct size of PCR products was confirmed by agarose gel electrophoresis. PCR products were cleared of single-strand products and free phosphate ends by incubation at 37°C for 45 minutes with 2 U exonuclease I (Fermentas) and 1 U calf intestine phosphatase (Fermentas). Reactions were terminated by heating to 80°C for 15 minutes. Following DNA precipitation, samples were sent for sequencing (Eurofins MWG Operon). Sequence data were analyzed and compared with the genomic sequence of PIK3CA in Sequencher 4.7 (Gene Codes Corporation).
Analyses of PTEN and Akt
For the analysis of PTEN expression, cells were plated in 12-well plates (Sarstedt) at a density of 250 000 cells/well. After 24 hours of culture in media containing 10% FCS, cell lysates were prepared and subjected to SDS–PAGE using 12% polyacrylamide gels followed by transfer to Hybond-C-extra membranes (Amersham Life Science). Immunoblotting was performed with commercially available antibodies (antihuman PTEN antibody [clone 6H2.1], Cascade BioScience). Immunoblotting signals were quantified using the AIDA software, version 3.10.039 (Raytest).
For the analysis of Akt expression and phosphorylation, a similar procedure was performed as for PTEN, but 1.6 million cells/well were plated in 6-well plates. After 24 hours of culture in 10% FCS, cells were subjected to overnight starvation in 1% FCS. The following day, they were stimulated for 10 minutes at 37°C with 10 ng/mL IGF-1 ligand. Immunodetection of Akt levels and phosphorylation was conducted in a similar way as for PTEN (Cell Signaling Technology).
siRNA-Mediated Downregulation of PTEN and IGF-1R
Cell culture 4 was used for analyzing the effects of PTEN downregulation. A total of 5000 cells/well were plated in 96-well plates in 1% or 10% FCS and 16 000 cells/well in 6-well plates. The following day, cells were transfected with either control or PTEN siRNA (Dharmacon RNA Technologies) using DharmaFect transfection reagent 3 (Dharmacon RNA Technologies) according to the manufacturer's instructions. NVP-AEW541 was added 48 hours after transfection, and at this time point, transfection was repeated. Experiments were stopped 48 hours after drug addition, and the cell number was determined as described above. For the analysis of siRNA effects on the expression and phosphorylation of PTEN and Akt, cells from the 6-well plates were harvested at the same time point as for the growth experiment. For the detection of PTEN levels and Akt levels, Akt phosphorylation immunoblotting was performed as described above.
For the analysis of siRNA effects on the IGF-1 receptor, 3000 cells/well were plated in 96-well plates and 16 000 cells in 6-well plates. Transfection was preformed as described above with either control or siRNA IGF-1R (siGenome Smart Pool, Dharmacon RNA Technologies). Experiments were stopped 72 hours after transfection, and cell number was determined as described above. For the analysis of siRNA effects on IGF-1 receptor expression, cells from the 6-well plates were harvested 72 hours after transfection and subjected to the analyses of IGF-1 receptor mRNA and protein.
RNA Isolation, CDNA Synthesis, and qRT-PCR
Isolation of RNA was performed with the RNeasy Mini Kit (Qiagen Inc.) according to the manufacturer's protocol. The RNA extracted was quantified with a Nano-Drop ND-1000 Spectrophotometer (Nano-Drop Technologies Inc.). cDNA was synthesized with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) using polydT primers according to the manufacturer's instructions. A 2.5-µL sample of the resulting cDNA was used for a Real-Time PCR assay, using the SYBRgreen Universal PCR Master Mix (Applied Biosystems). Primers were used at a concentration of 200 nM. The reaction was performed with the 7500 Real-Time PCR system (Applied Biosystems) under conditions recommended by the manufacturer. Expression levels were normalized to the expression of the house-keeping gene HPRT or to the amount of input cDNA, determined by analysis with the Nano-Drop ND-1000 spectrophotometer (Nano-Drop Technologies). Primer sequences are: IGF-1R forward: CCTCAACGCCAATAAGTTCGTC and IGF-1R reverse: TAAGTGGTGAAGACTCCATCCTTG.
Results and Discussion
Eight cultures from a previously described panel of high-grade gliomas were analyzed with regard to sensitivity to NVP-AEW541.39 As shown in Fig. 1A, sensitivity among the cultures showed large variation, ranging between 100% and 20% growth inhibition. Since the basal growth rate of these cultures show large differences, we analyzed whether a correlation existed between growth rate and sensitivity. A trend toward an association between the basal growth rate and the NVP-AEW541 sensitivity was observed, although it was not significant (data not shown). The mechanism causing the reduction in cell growth following treatment with NVP-AEW541 was further analyzed and demonstrated both antiproliferative and proapoptotic effects of the IGF-1 receptor inhibitor (Fig. 1B).
The importance of IGF-1 receptor signaling was also analyzed by IGF-1 receptor siRNA experiments performed on cultures 4 and 35. siRNA-mediated IGF-1 receptor downregulation was confirmed by qRT-PCR and immunoblotting (Fig. 1C and D). Quantification of immunoblots indicated downregulation with 40% and 47% in cultures 35 and 4, respectively (Fig. 1D). As shown in Fig. 1E, downregulation of IGF-1 receptors in cultures 35 and 4 induced a significant growth inhibition when cells were cultured at 1% FCS. Similar findings were made also at 10% FCS conditions (Supplementary Material, Fig. S1).
Microarray-derived gene expression data for these cultures grown under basal conditions have previously been collected.39 Hierarchical clustering of the 8 cultures used in this study was performed based on 67 features showing more than 4-fold differential expression in at least 2 samples and generated 2 clusters. No significant correlations were observed between drug sensitivity and the gene-expression–based clusters (data not shown). Array data were also used to investigate relationships between the levels of IGF-1 expression and the NVP-AEW541 sensitivity. Although IGF-1 expression was detected in all cultures, no associations were observed between IGF-1 levels and sensitivity to the IGF-1 receptor inhibitor.
IGF-1 receptor expression levels and phosphorylation of the receptors, in the absence or the presence of ligand, were also analyzed to investigate if any relationship existed between sensitivity to the inhibitor and IGF-1 receptor status. IGF-1 receptor expression was detected in all 8 cultures (Fig. 1F). In culture 4, the presence of activated IGF-1 receptors was also demonstrated by phospho-tyrosine immune-precipitation followed by IGF-1 receptor immunoblotting (Supplementary Material, Fig. S2). Expression levels varied between cultures, but no association was observed between the levels of receptor expression and drug sensitivity (data not shown). All cultures also displayed a 2- to 10-fold ligand-dependent increase in receptor phosphorylation (Fig. 1F). Quantifications of the basal receptor phosphorylation, and of the ligand-induced increase in phosphorylation, did not identify any consistent associations between these characteristics and drug sensitivity (data not shown).
To analyze whether any relationship existed between PIK3CA mutations and sensitivity to NVP-AEW541, all 8 high-grade–glioma cultures were analyzed with regard to PIK3CA mutations as well as basal and ligand-induced Akt phosphorylation.
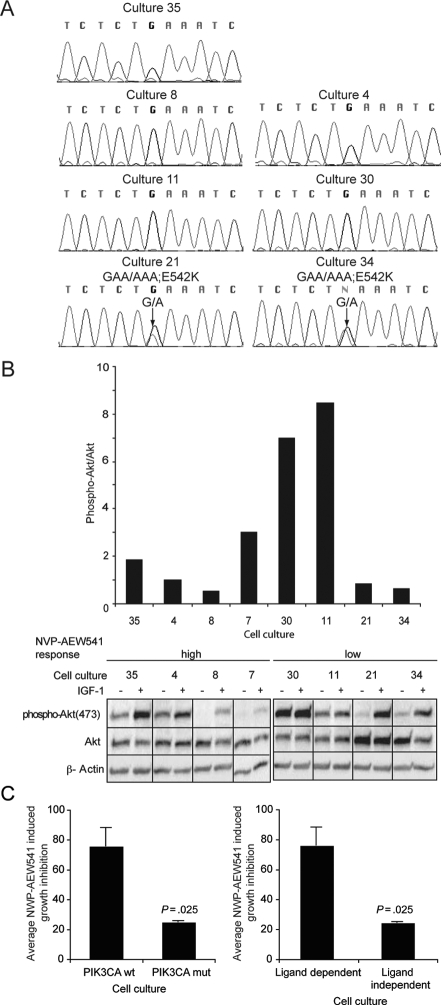
The mutation analyses of the catalytic p110 subunit of PIK3CA focused on exon 9, which has previously been shown to be the exon most frequently mutated in glioblastomas. As shown in Fig. 2A, this analysis revealed the presence of activating E542K mutations in cultures 21 and 34. Akt phosphorylation was analyzed in starved and IGF-1-stimulated cells. Quantification of basal Akt phosphorylation revealed that cultures 11 and 30 displayed high basal Akt phosphorylation levels (Fig. 2B). Furthermore, Akt phosphorylation remained largely unaffected by the addition of exogenous IGF-1 in these 2 cultures, in contrast to what was observed in the other 6 cultures (Fig. 2B).
Fig. 2.
Cultures with PIK3CA mutations, or constitutively high Akt phosphorylation, show significantly reduced sensitivity to NVP-AEW541. (A) PIK3CA mutations in glioblastoma cultures. Chromatograms show sequences derived from exon sequencing of genomic DNA. The chromatograms show the results from the sequencing of 5 cultures with wild-type PIK3CA and 2 cultures with activating E542K mutation. Positions of mutations in cultures 21 and 34 are indicated by an arrow, and the amino acid alterations are indicated above the chromatograms. (B) Basal and IGF-1-induced changes in Akt phosphorylation. The experiment was performed 4 times for each culture and yielded similar results. (C) Growth inhibition in different subsets of glioblastoma cultures. Results from Fig. 1A were used to compare the average growth inhibition in cultures without detected alteration in PI3K/PTEN/Akt, with activating mutations in PIK3CA and with ligand-independent Akt phosphorylation (Student's t-test).
To analyze the potential relationship between NVP-AEW541 sensitivity and PI3K mutations (cultures 21 and 34) or ligand-independent Akt phosphorylation (cultures 11 and 30), we compared the average growth inhibition in cultures with any of these types of aberrations with the other 4 cultures. As shown in Fig. 2C, these analyses revealed a significantly reduced growth inhibition in the cultures with PIK3CA mutations or ligand-independent Akt phosphorylation. Together, these findings suggest that these properties are associated with a reduced sensitivity to NVP-AEW541.
To extend these associations, we explored experimentally the consequences of a well-defined experimental perturbation of this pathway. For this purpose, we selected siRNA-mediated PTEN downregulation to mimic the loss of PTEN, which is commonly observed in GBM. For this experiment, culture 4 was used. This culture has the characteristics of NVP-AEW541 responsiveness, the absence of PIK3CA mutation, and IGF-1 sensitive Akt phosphorylation (Figs 1A, data not shown, and 2B).
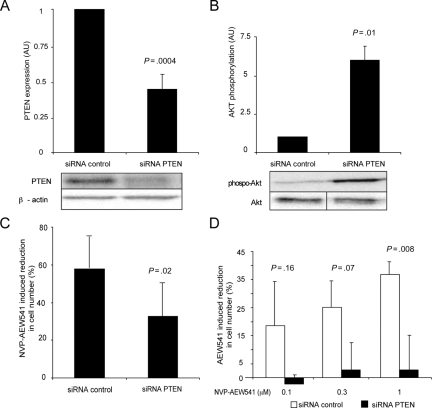
Transfection of PTEN siRNA resulted in consistent downregulation of PTEN protein (Fig. 3A), with a concomitant increase in Akt phosphorylation (Fig. 3B). siRNA-mediated downregulation also significantly reduced the growth-inhibitory response to NVP-AEW541 treatment (Fig. 3C). The relative inhibitory effect, induced by 3 µM NVP-AEW541, was reduced from 57% in the cells transfected with control siRNA to 32% in cells transfected with PTEN siRNA (Fig. 3C). It was noted that siRNA-mediated downregulation of PTEN also was associated with a reduction in the basal growth rate (Supplementary material, Fig. S3A). The negative effect on the NVP-AEW541 sensitivity and the basal growth rate by PTEN downregulation was also observed when assays were performed at low-serum conditions (Fig. 3D and Supplementary material, Fig. S3A).
Fig. 3.
Downregulation of PTEN reduces sensitivity to NVP-AEW541. (A and B) PTEN siRNA leads to a significant reduction in PTEN expression and an increase in Akt phosphorylation. Akt phosphorylation was analyzed, as in Fig. 2, 48 hours after siRNA transfection. PTEN expression was analyzed in the same cell lysates by immunoblotting. (C and D) Downregulation of PTEN reduces sensitivity to NVP-AEW541. Cells were kept in 10% FCS (C) or 1% FCS (D) and treated with 3 µM (C) or indicated concentrations (D) of NVP-AEW541, and effects on growth were determined after 4 days of drug treatment. Results in A–C are derived from 3 independent experiments, and the results in D from 2 experiments. Experiments in A–D were performed in cell culture 4. Error bars indicate the standard error of the mean. P values were determined by Student's t-test.
To extend these findings, similar experiments were performed in the NVP-AEW541-sensitive GBM cell line U343MG. Also in this cell line, it was noted that PTEN downregulation led to an increase in pAkt/Akt, with a concomitant reduction in the basal growth rate and the NVP-AEW541 sensitivity (Supplementary material, Fig. S3B and C).
These experiments thus support the notion that alterations in the PI3K/PTEN/Akt pathway are associated with a reduced sensitivity to mono-treatment with NVP-AEW541. However, it is also noted that the effects of PTEN downregulation on the basal growth rate of both culture 4 and U343MG cells introduce a confounding factor in these analyses.
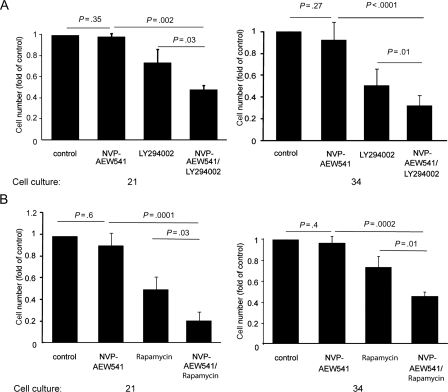
Previous studies have suggested that the combinations of receptor tyrosine kinase–targeting agents, with inhibitors of downstream signaling molecules, such as PI3K or mTOR, can exert synergistic effects or overcome resistance to receptor tyrosine kinase inhibitors.41,42 To test if such a mechanism could also be demonstrated in the case of NVP-AEW541 effects on GBM cells, we performed combination treatments with a PI3K inhibitor, or an mTOR inhibitor, and the IGF-1 receptor inhibitor. For this purpose, we used cultures 21 and 34, which are characterized by activating PIK3CA mutations (Fig. 2A) and lack of sensitivity to NVP-AEW541 (Fig. 1A). In agreement with previous experiments, mono-treatment with NVP-AEW541 did not affect the growth of these cells. However, a significant effect of the IGF-1 receptor inhibitor was observed in both the cultures when it was used together with the PI3K inhibitor LY294002 (Fig. 4A) or with the mTOR inhibitor rapamycin (Fig. 4B).
Fig. 4.
NVP-AEW541 is active in combination with PI3K or mTOR inhibitors in cultures with an activating PIK3CA mutation. Cell cultures 21 and 34 were treated with or without 1 µM NVP-AEW541, 20 µM PI3K inhibitor LY294002, or 1 nM mTOR inhibitor rapamycin. Growth inhibition was quantified by comparing final cell number in control-treated cultures, which was set to 1, with cell number in drug-treated cultures. The extent of growth inhibition was determined after 4 days of culture. Results are derived from 3 experiments. Error bars indicate the standard deviation. P values were determined by Student's t-test.
This finding suggests that the IGF-1 receptor inhibitors, when used in combination with other signal transduction inhibitors, might also have an effect in cells with a dysregulated PI3K/PTEN/Akt pathway.
In summary, our studies thus identify associations between PI3K/PTEN/Akt status and sensitivity to mono-treatment with NVP-AEW541. The studies also demonstrate that NVP-AEW541 is also active together with PI3 kinase and mTOR inhibitors in cultures with a dysregulated PI3K/PTEN/Akt pathway.
On the basis of the proposed PDGFR dependency of glioblastoma growth, 7 of the cultures of this study have previously been analyzed with regard to sensitivity to the PDGFR/c-kit/Abl-inhibitor imatinib.39 Among the 3 NVP-AEW541-sensitive cultures, which also had been tested with regard to imatinib sensitivity, 2 cultures showed moderate or high sensitivity to imatinib (7 and 8). Culture 35, which expressed very low levels of PDGFRs, did not respond to imatinib. Interestingly, 3 of the 4 low-responsive cultures were also insensitive to imatinib.
In the present study, Akt phosphorylation was augmented by the IGF-1 stimulation of the cultures expressing an activated PI3K variant (Fig. 2A and B). Previous biochemical analyses have indicated that the E542K and E545K mutants of the p110 part of PI3K (PIK3CA) in complex with p85 are not further activated by the addition of SH2 phospho-tyrosine ligands.43,44 Structural studies have suggested that these mutations lead to the disruption of an inhibitory interaction between the N-SH2 domain of p85 and the helical domain of PIK3CA.43,45 It is therefore likely that the ligand-dependent Akt phosphorylation in these cells is occurring through the activation of the wild-type variant of PIK3CA.
The present study strongly suggests that PI3K and PTEN status impacts on sensitivity to IGF-1 receptor antagonists. This is suggested by the observed associations between PI3K mutation status, as well as the ligand dependency of pAkt/Akt and NVP-AEW541 sensitivity. Furthermore, downregulation of PTEN led to a reduction in sensitivity, providing independent support for a role of PTEN as a sensitivity determinant. The associated finding that PTEN downregulation also reduced the basal growth rate remains to be better characterized. Possibly this phenomenon reflects a transient cell stress phenomenon, which would eventually be overcome following a longer time of culture.
In general, knowledge about the determinants of sensitivity and resistance to IGF-1 receptor inhibitors thus remains very limited. Interestingly, a recent study could demonstrate an association between the lack of sensitivity and the high levels of IGFBP-3 and -6.46 This study also showed that high expression of the IGF-1 receptor and IGF-1 and -2 was associated with high sensitivity. These findings, as well as those of the present study, should be further validated in clinical settings.
In summary, this study thus supports the evaluation of the effects of IGF-1 receptor inhibitors, used alone or in combination, in glioblastomas. It is furthermore expected that this characterization of determinants of IGF-1 receptor antagonist sensitivity will aid in patient selection and thereby facilitate the test of this type of drugs in glioblastomas. The analyses performed in this study should also be extended to other types of solid tumors to optimize the clinical development of the novel IGF-1 receptor inhibitors.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
A.Ö. and M.N. acknowledge support from the Swedish Cancer Society and the Cancer Society in Stockholm.
Acknowledgments
Members of the laboratory of A.Ö. and M.N. are acknowledged for critical discussions throughout the project.
Conflict of interest statement. A.Ö. and M.N. have received research from Novartis. A.Ö. and D.H. have received honorarium from Novartis. F.H. and C.G-E. are employed by Novartis.
References
- 1.Clemmons DR. Modifying IGF1 activity: an approach to treat endocrine disorders, atherosclerosis and cancer. Nat Rev Drug Discov. 2007;6:821–833. doi: 10.1038/nrd2359. doi:10.1038/nrd2359. [DOI] [PubMed] [Google Scholar]
- 2.Miller BS, Yee D. Type I insulin-like growth factor receptor as a therapeutic target in cancer. Cancer Res. 2005;65:10123–10127. doi: 10.1158/0008-5472.CAN-05-2752. doi:10.1158/0008-5472.CAN-05-2752. [DOI] [PubMed] [Google Scholar]
- 3.Hellawell GO, Turner GD, Davies DR, Poulsom R, Brewster SF, Macaulay VM. Expression of the type 1 insulin-like growth factor receptor is up-regulated in primary prostate cancer and commonly persists in metastatic disease. Cancer Res. 2002;62:2942–2950. [PubMed] [Google Scholar]
- 4.Arteaga CL, Kitten LJ, Coronado EB, Jacobs S, Kull FC, Jr, Allred DC, et al. Blockade of the type I somatomedin receptor inhibits growth of human breast cancer cells in athymic mice. J Clin Invest. 1989;84:1418–1423. doi: 10.1172/JCI114315. doi:10.1172/JCI114315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Sun Y. Insulin-like growth factor receptor-1 as an anti-cancer target: blocking transformation and inducing apoptosis. Curr Cancer Drug Targets. 2002;2:191–207. doi: 10.2174/1568009023333863. doi:10.2174/1568009023333863. [DOI] [PubMed] [Google Scholar]
- 6.Sell C, Dumenil G, Deveaud C, Miura M, Coppola D, DeAngelis T, et al. Effect of a null mutation of the insulin-like growth factor I receptor gene on growth and transformation of mouse embryo fibroblasts. Mol Cell Biol. 1994;14:3604–3612. doi: 10.1128/mcb.14.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hewish M, Chau I, Cunningham D. Insulin-like growth factor 1 receptor targeted therapeutics: novel compounds and novel treatment strategies for cancer medicine. Recent Pat Anticancer Drug Discov. 2009;4:54–72. doi: 10.2174/157489209787002515. doi:10.2174/157489209787002515. [DOI] [PubMed] [Google Scholar]
- 8.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. doi:10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 9.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. doi:10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 10.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. doi:10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer_Genome_Atlas_Research_Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. doi:10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reifenberger G, Collins VP. Pathology and molecular genetics of astrocytic gliomas. J Mol Med. 2004;82:656–670. doi: 10.1007/s00109-004-0564-x. doi:10.1007/s00109-004-0564-x. [DOI] [PubMed] [Google Scholar]
- 14.Fults D, Brockmeyer D, Tullous MW, Pedone CA, Cawthon RM. p53 mutation and loss of heterozygosity on chromosomes 17 and 10 during human astrocytoma progression. Cancer Res. 1992;52:674–679. [PubMed] [Google Scholar]
- 15.Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci USA. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. doi:10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki H, Fukui Y, Ueyama Y, Tamaoki N, Kawamoto T, Taniguchi S, et al. Amplification of the structurally and functionally altered epidermal growth factor receptor gene (c-erbB) in human brain tumors. Mol Cell Biol. 1988;8:1816–1820. doi: 10.1128/mcb.8.4.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. doi:10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 18.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. doi:10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann C, Bartels G, Gehlhaar C, Holtkamp N, von Deimling A. PIK3CA mutations in glioblastoma multiforme. Acta Neuropathol (Berl) 2005;109:639–642. doi: 10.1007/s00401-005-1000-1. doi:10.1007/s00401-005-1000-1. [DOI] [PubMed] [Google Scholar]
- 20.Trojan J, Cloix JF, Ardourel MY, Chatel M, Anthony DD. Insulin-like growth factor type I biology and targeting in malignant gliomas. Neuroscience. 2007;145:795–811. doi: 10.1016/j.neuroscience.2007.01.021. doi:10.1016/j.neuroscience.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 21.Resnicoff M, Sell C, Rubini M, Coppola D, Ambrose D, Baserga R, et al. Rat glioblastoma cells expressing an antisense RNA to the insulin-like growth factor-1 (IGF-1) receptor are nontumorigenic and induce regression of wild-type tumors. Cancer Res. 1994;54:2218–2222. [PubMed] [Google Scholar]
- 22.Schlenska-Lange A, Knupfer H, Lange TJ, Kiess W, Knupfer M. Cell proliferation and migration in glioblastoma multiforme cell lines are influenced by insulin-like growth factor I in vitro. Anticancer Res. 2008;28:1055–1060. [PubMed] [Google Scholar]
- 23.Low S, Vougioukas VI, Hielscher T, Schmidt U, Unterberg A, Halatsch ME. Pathogenetic pathways leading to glioblastoma multiforme: association between gene expressions and resistance to erlotinib. Anticancer Res. 2008;28:3729–3732. [PubMed] [Google Scholar]
- 24.Moser C, Schachtschneider P, Lang SA, Gaumann A, Mori A, Zimmermann J, et al. Inhibition of insulin-like growth factor-I receptor (IGF-IR) using NVP-AEW541, a small molecule kinase inhibitor, reduces orthotopic pancreatic cancer growth and angiogenesis. Eur J Cancer. 2008;44:1577–1586. doi: 10.1016/j.ejca.2008.04.003. doi:10.1016/j.ejca.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Manara MC, Landuzzi L, Nanni P, Nicoletti G, Zambelli D, Lollini PL, et al. Preclinical in vivo study of new insulin-like growth factor-I receptor–specific inhibitor in Ewing's sarcoma. Clin Cancer Res. 2007;13:1322–1330. doi: 10.1158/1078-0432.CCR-06-1518. doi:10.1158/1078-0432.CCR-06-1518. [DOI] [PubMed] [Google Scholar]
- 26.Tanno B, Mancini C, Vitali R, Mancuso M, McDowell HP, Dominici C, et al. Down-regulation of insulin-like growth factor I receptor activity by NVP-AEW541 has an antitumor effect on neuroblastoma cells in vitro and in vivo. Clin Cancer Res. 2006;12:6772–6780. doi: 10.1158/1078-0432.CCR-06-1479. doi:10.1158/1078-0432.CCR-06-1479. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Echeverria C, Pearson MA, Marti A, Meyer T, Mestan J, Zimmermann J, et al. In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell. 2004;5:231–239. doi: 10.1016/s1535-6108(04)00051-0. doi:10.1016/S1535-6108(04)00051-0. [DOI] [PubMed] [Google Scholar]
- 28.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. doi:10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. doi:10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 30.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. doi:10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 31.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. doi:10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 32.Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. doi:10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 33.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. doi:10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 34.Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. doi:10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita T, Doihara H, Kawasaki K, Takabatake D, Takahashi H, Washio K, et al. PTEN activity could be a predictive marker of trastuzumab efficacy in the treatment of ErbB2-overexpressing breast cancer. Br J Cancer. 2006;94:247–252. doi: 10.1038/sj.bjc.6602926. doi:10.1038/sj.bjc.6602926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. doi:10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 37.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. doi:10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 38.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. doi:10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 39.Hagerstrand D, Hesselager G, Achterberg S, Wickenberg Bolin U, Kowanetz M, Kastemar M, et al. Characterization of an imatinib-sensitive subset of high-grade human glioma cultures. Oncogene. 2006;25:4913–4922. doi: 10.1038/sj.onc.1209497. doi:10.1038/sj.onc.1209497. [DOI] [PubMed] [Google Scholar]
- 40.Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 41.Eichhorn PJ, Gili M, Scaltriti M, Serra V, Guzman M, Nijkamp W, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. doi:10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.She QB, Solit D, Basso A, Moasser MM. Resistance to gefitinib in PTEN-null HER-overexpressing tumor cells can be overcome through restoration of PTEN function or pharmacologic modulation of constitutive phosphatidylinositol 3′-kinase/Akt pathway signaling. Clin Cancer Res. 2003;9:4340–4346. [PubMed] [Google Scholar]
- 43.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. doi:10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 44.Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, et al. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem J. 2008;409:519–524. doi: 10.1042/BJ20070681. doi:10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 45.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. doi:10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 46.Huang F, Greer A, Hurlburt W, Han X, Hafezi R, Wittenberg GM, et al. The mechanisms of differential sensitivity to an insulin-like growth factor-1 receptor inhibitor (BMS-536924) and rationale for combining with EGFR/HER2 inhibitors. Cancer Res. 2009;69:161–170. doi: 10.1158/0008-5472.CAN-08-0835. doi:10.1158/0008-5472.CAN-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]