Abstract
Glioblastoma multiforme (GBM) is one of the deadliest tumors afflicting humans, and the mechanisms of its onset and progression remain largely undefined. Our attempts to elucidate its molecular pathogenesis through DNA copy-number analysis by genome-wide digital karyotyping and single nucleotide polymorphism arrays identified a dramatic focal amplification on chromosome 1q32 in 4 of 57 GBM tumors. Quantitative real-time PCR measurements revealed that HDMX is the most commonly amplified and overexpressed gene in the 1q32 locus. Further genetic screening of 284 low- and high-grade gliomas revealed that HDMX amplifications occur solely in pediatric and adult GBMs and that they are mutually exclusive of TP53 mutations and MDM2 amplifications. Here, we demonstrate that HDMX regulates p53 to promote GBM growth and attenuates tumor response to chemotherapy. In GBM cells, HDMX overexpression inhibits p53-mediated transcriptional activation of p21, releases cells from G0 to G1 phase, and enhances cellular proliferation. HDMX overexpression does not affect the expression of PUMA and BAX proapoptotic genes. While in GBM cells treated with the chemotherapeutic agent 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU), HDMX appears to stabilize p53 and promote phosphorylation of the DNA double-stranded break repair protein H2AX, up-regulate the DNA repair gene VPX, stimulate DNA repair, and confer resistance to BCNU. In summary, HDMX exhibits bona fide oncogenic properties and offers a promising molecular target for GBM therapeutic intervention.
Keywords: chemoresistance, GBM, HDMX, oncogene, p53
Glioblastoma multiforme (GBM) is the most common and aggressive primary brain tumor in adults and is associated with a median survival of 12 months.1,2 Current therapy includes surgery, radiation, and adjuvant chemotherapy. An important component of multiple agent adjuvant chemotherapy regimens in brain and other tumors has been the DNA alkylating agent 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU), which exerts its cytotoxicity mainly by alkylating DNA at the O6 position of guanine and by forming DNA interstrand crosslinks. Patients with newly diagnosed malignant gliomas who were treated with BCNU wafers have noted survival benefits.3,4 The emergence of resistance to BCNU, however, has hampered its use in GBM patients.5,6 Variations in multidrug resistance genes and DNA repair proteins such as O6-methylguanine-DNA methyltransferase, glutathione S-transferase, and intracellular glutathione content could all bestow BCNU chemoresistance.7–10
The protein encoded by TP53 controls multiple cellular functions, including cell proliferation, DNA repair, senescence, and apoptosis.11 Tumor resistance to chemotherapeutic agents has also been reported to arise through failed p53 signaling.12 Genes involved in the p53 signaling pathway are frequently altered in GBMs: TP53 mutations are found in 28% of primary GBMs;2 the murine double minute 2 protein (MDM2), a negative regulator of p53,13,14 is amplified and overexpressed in 8%–10% of GBMs;15 and p14ARF, which inhibits p53 degradation by sequestering MDM2 to the nucleolus and rendering it inactive, undergoes deletion in 70% of GBMs.1,3,15–18 Additionally, p53 may enhance chemosensitivity by promoting apoptosis through either the intrinsic or the extrinsic pathways. It may also undermine chemosensitivity by promoting cell-cycle arrest, DNA repair, differentiation, and the transcription of antiapoptotic genes.18 The role of p53 in the GBM response to BCNU, however, remains controversial. Inactivation of p53 was found to sensitize tumor cells to BCNU,7–9 whereas another study showed that exogenous expression of wild-type p53 resulted in an increased sensitivity to BCNU.19 In another study, TP53 status was found to be an unreliable prognostic marker of tumor sensitivity to BCNU.20
HDMX was originally identified as a homologue of MDM221,22 and HDMX is a potent antagonist of p53 transcriptional activity.23 HDMX is amplified or overexpressed in 10%–20% of lung, colon, stomach, and breast cancers and in 65% of retinoblastomas.13 Amplification of the 1q32 chromosomal region encompassing HDMX has been observed in GBMs,24–29 implicating HDMX as a candidate oncogene in the amplicon. Though HDMX is a negative regulator of p53,13,30–32 its functional role and the mechanisms whereby it modulates p53 activity during GBM pathogenesis and its effects on the response of GBMs to chemotherapeutic agents have yet to be fully delineated.
In this study, we investigated the genomic amplification and overexpression of the HDMX gene in GBMs and their correlation with the TP53 status and MDM2 amplification. We also investigated the oncogenicity of HDMX in GBMs and how the overexpression of HDMX affected the response of GBM cells to BCNU in vitro and in vivo. We found that HDMX amplifications occurred mutually exclusively of TP53 mutation and MDM2 amplification. The overexpression of HDMX drove GBM growth and enhanced tumor cell survival against BCNU. We found that the induction of HDMX stimulated the repair of BCNU-inflicted DNA damage. These novel findings cast HDMX as an attractive therapeutic target with potential clinical benefits.
Materials and Methods
Cell Lines and Tissue Samples
Two colorectal carcinoma cell lines, HCT-116/TP53+/+ (wild-type TP53) and HCT-116/TP53−/− (TP53-deficient), were provided by Dr Bert Vogelstein at Johns Hopkins University. Human GBM cell lines D54 and D245, along with frozen primary tumor samples and matched blood pairs, were obtained from the Duke University Brain Tumor Center Tissue Bank. Acquisition of tissue specimens was approved by the Duke University Health System Institutional Review Board and was performed in accordance with HIPAA regulations. Genomic DNA extracts were generated from 21 grade I astrocytomas, 21 grade II and III astrocytomas, 37 grade II and 39 grade III oligodendrogliomas, 23 pediatric GBMs, and 143 adult GBMs.
Digital Karyotyping and Single Nucleotide Polymorphism Arrays
Digital karyotyping library construction and data analysis of 25 GBM samples were performed as described.33,34 The protocols and software for extraction and analysis of genomic tags are available at http://www.digitalkaryotyping.org. Single nucleotide polymorphism (SNP) genotyping on genomic DNA from 32 GBM samples, including 7 pediatric GBMs, was performed using the Illumina HumanHap550 Genotyping BeadChip array (Illumina). Raw data from the SNP chips were collected and subjected to copy-number analysis using Nexus Copy Number Professional software (BioDiscovery Inc.). LogR ratio data sets were processed with Nexus algorithms to identify altered copy-number loci. These identified loci were compared across the entire sample set to determine regions of overlap.
Quantitative Real-Time PCR and Fluorescence in Situ Hybridization
The genomic DNA content and mRNA expression levels of genes of interest within tumor and normal cells were quantified by quantitative real-time polymerase chain reaction (Q-PCR).33 Genomic DNA from normal blood cells served as controls, and genomic DNA content was normalized to that of Line-1. For the mRNA expression measurement, cDNA from normal human adult cortex was used as the control and cDNA content was normalized to that of GAPDH. Additionally, dual-color fluorescence in situ hybridization (FISH) analysis was performed on tissue sections from primary GBM samples and tissue microarrays. Bacterial artificial chromosome-derived probes targeting HDMX (1q32.1, RP11-970B14, Invitrogen) and a chromosome 1 control (1q21.2, RP11-458I7, Invitrogen) were labeled with biotin- and digoxigenin-dUTP, respectively, for hybridization. We evaluated the FISH signals with a Nikon TE2000-E fluorescent microscope.
Generation of Stable HDMX-Inducible GBM Cell Lines
The lentivirus vector expressing HDMX, pLenti4/TO/V5-DEST-HDMX, was generated by using the ViraPower T-Rex Lentiviral Expression System according to the manufacturer's instructions (Invitrogen). D54 cells, which contain neither TP53 mutation nor HDMX or MDM2 amplification, were selected as the parental cells. Stable D54-HDMX cells containing the lentivirus vector with tetracycline-inducible HDMX expression were established after selections with appropriate antibiotics. Tetracycline (Sigma) was dissolved in PBS and used (1 μg/mL) for HDMX induction.
Luciferase Assays
The luciferase reporter vectors (pLG2, pLG2-p21P, pLG2-p21PΔp53, pLG2-p21PΔp800, and pLG2-p21PΔ1.1) were previously described35 and provided by Dr Xiaofan Wang at Duke University. We performed the luciferase assays using D54-HDMX, HCT-116/TP53+/+, or HCT-116/TP53−/− cells transiently transfected with Renilla constructs (as an internal control) or plasmids pLG2, pLG2-p21p, pLG2-p21PΔp53, pLG2-p21PΔp800, pLG2-p21PΔ1.1, or pLenti6.2-HDMX using the Dual Luciferase Assay system following the manufacturer's instructions (Promega). All luciferase activity readings were normalized relative to the activity of the Renilla luciferase control. All experiments were performed in triplicate.
Small Interfering RNA Transfection Assays
For small interfering RNA (siRNA) knockdown of HDMX, we used siRNA #931 (sense: r[GCUGCUGAUACUGAACAAA]dTdT; antisense: r[UUUGUUCAGUAUCAGCAGC]dGdA) and siRNA #1325 (sense: r[CCUGCAACUCAGUGGAAUU]dTdT; antisense: r[AAUUCCACUGAGUUGCAGG]dGdA) (Ambion). Cells were collected at 48 hours after siRNA transfection for immunoblotting analysis.
Cell Proliferation and Clonogenic Assays
For cell proliferation studies, cells were seeded at a density of 1 × 105 per well on 6-well plates and treated with tetracycline. After 8 days, cell viability was assessed by the tetrazolium-based semiautomated colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay. For clonogenic survival assays, cells were plated in 100-mm culture dishes and treated with 0 or 1 µg/mL tetracycline for 24 hours and with different concentrations of BCNU. After 2 weeks, cell colonies were stained with 0.05% crystal violet (Sigma). For drug treatments, BCNU (Sigma) was dissolved in dimethyl sulfoxide and freshly prepared each time before use.
Cell-Cycle Analysis
D54-HDMX cells were induced with 0 or 1 µg/mL tetracycline for 24 hours. For fluorescence-activated cell sorting analysis, treated cells were collected at 24 hours posttreatment, fixed in 70% ice-cold ethanol for 24 hours, and stained with 20 µg/mL propidium iodide, 0.1% (v/v) Triton X-100, and 100 µg/mL DNase-free RNase A.
GBM Xenografts
For xenograft experiments, 1 × 107 D54-HDMX or D54 control cells were injected subcutaneously in the right flank of athymic mice. When the implanted tumors reached a median tumor volume of 200–300 mm3, 5 mice per arm were selected for the following treatments. The mice were fed either a standard or a doxycycline-containing diet. For the drug resistance study, BCNU was administered intraperitoneally at a dose of 13 mg/kg 3 days after the doxycycline treatment. Tumor growth was assessed as previously described.36 The growth delay was expressed as T–C, the difference between the median times required for tumors in treated mice (T group) and control mice (C group) to reach a volume 5 times that at the initiation of therapy and more than 1000 mm3. The Wilcoxon's rank-sum test was applied for statistical analysis.
Immunoblotting Assays
For immunoblotting experiments, conducted as previously described,37 the antibodies against the following proteins were used: HDMX/MDM4 (Bethyl Laboratories), GADPH, FAS, MDM2, p53, and Bax (Santa Cruz Biotechnology), p21 (Cell Signaling Technology), PUMA (ProSci Incorporated), and anti-γ-H2AX antibody (serine 139) (Abcam). The immunofluorescence microscopy was performed as previously described.38
Results
HDMX is Amplified and Overexpressed in GBMs
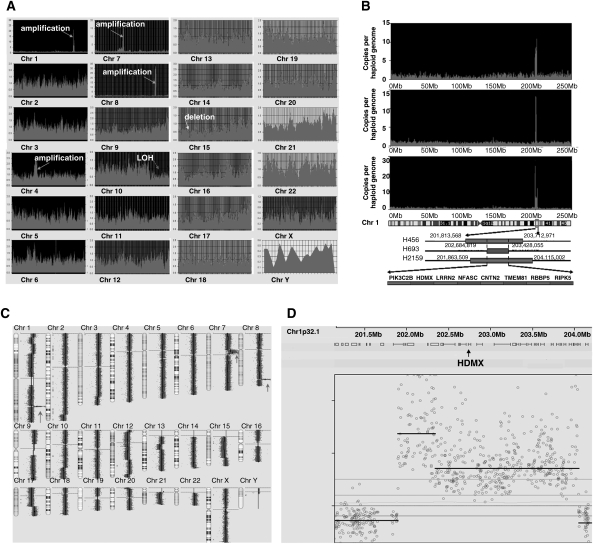
To identify cancer-specific genetic changes in GBMs, we applied 2 genomic approaches, digital karyotyping and the Illumina HumanHap550 Genotyping BeadChip array, to interrogate the GBM genome for gene copy-number variations in 57 human patient GBM samples. We identified a dramatic focal amplification of the chromosome 1q32 region encompassing HDMX in 7% of the samples (Fig. 1A–D). Q-PCR analysis of 284 human brain tumor samples revealed that HDMX amplifications occurred exclusively in pediatric and adult GBMs (Supplementary Material, Tables S1 and S2). Dual-color FISH analysis of primary GBM tissue sections using an HDMX-specific probe highlighted a substantial amplification of the HDMX locus (Supplementary Material, Fig. S1). Moreover, Q-PCR analysis of genomic DNA and cDNA extracts from available GBM samples revealed that HDMX is the most frequently amplified and overexpressed gene in the 1q32 amplicon (Supplementary Material, Table S3). All the GBM samples with HDMX amplification also displayed high-level HDMX expression (Supplementary Material, Fig. S2). These findings suggest HDMX as the GBM-specific target gene of amplification in the 1q32 locus.
Fig. 1.
HDMX is amplified in primary GBMs. (A) A tag density map from digital karyotyping of GBM sample D456 revealed subchromosomal changes. For all chromosomes (labeled 1–22, x, and y), Y-axis values indicate genome copies per haploid genome, while X-axis values represent positions along the chromosome, in Mb. Prominent large-scale genomic amplifications, including chromosome 1q32, 7p11, and 8q24, are clearly observed. (B) Genomic amplifications encompassing HDMX in chromosome 1q32 in 3 GBMs. (C) Genome-wide copy-number measurements of genomic DNA from one GBM xenograft by using Illumina HumanHap550 Genotyping BeadChip array. Three prominent, large-scale genomic amplifications are observed at 1q32, 7p11, and 8q24. The red arrows point to the 3 distinguishing regions of amplification. (D) A drill-down map of the SNPs displays high copy-number gain of multiple SNPs in a region of chromosome 1q32 containing HDMX.
HDMX Amplifications and Inactivation of P53 Signaling in Gliomas
We further investigated the correlation between HDMX amplifications and TP53 and MDM2 status. We sequenced the TP53 gene in WHO grade I–IV gliomas and showed that TP53 was mutated in 11 (52%) of the 21 WHO grade II and III astrocytomas, 7 (30%) of the 23 pediatric GBMs, and 45 (31%) of the 143 adult GBMs (Supplementary Material, Tables S1 and S2). TP53 mutations, however, were rarely present in WHO grade I astrocytomas or WHO grade II and III oligodendrogliomas. Q-PCR analysis of these tumor samples revealed the amplification of MDM2 and HDMX in the higher grade astrocytomas, including GBMs (Supplementary Material, Table S1 and S2), but not in WHO grade I astrocytomas or WHO grade II and III oligodendrogliomas. In 89 samples with genetic aberrations associated with p53 pathway inactivation, only samples displaying wild-type TP53 and normal MDM2 copy number exhibited HDMX amplifications (Supplementary Material, Table S1). This indication that these HDMX amplifications were mutually exclusive of TP53 mutations and MDM2 amplifications supports the hypothesis that HDMX, MDM2, and p53 function in the same oncogenic pathway in GBM development.
HDMX Promotes GBM Cell Proliferation
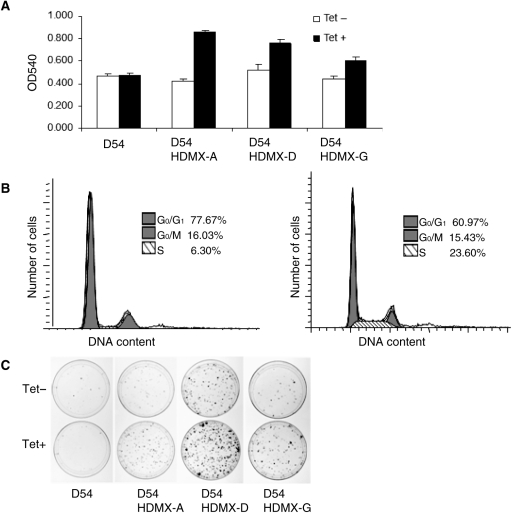
In order to study the pathological role of HDMX in glioma tumorigenesis, we established multiple HDMX-inducible GBM cell lines, named D54-HDMX lines (Supplementary Material, Fig. S3). We assessed the proliferative activities of D54-HDMX cells and detected a substantial increase in the viable cell populations following HDMX induction by tetracycline as revealed by the MTT assays (Fig. 2A). HDMX induction was also associated with a more than 3-fold increase in the proportion of GBM cells undergoing S phase replication (Fig. 2B) and a robust formation of colonies (Fig. 2C).
Fig. 2.
HDMX promotes GBM cell growth. (A) HDMX promotes GBM cell proliferation. D54-HDMX clones (also see Supplementary Material, Fig. S2) were induced with 0 or 1 µg/mL tetracycline and their growth measured by MTT assays. (B) HDMX facilitates the release of GBM cells from the G0/G1 phase to S phase. Solid areas, G1 phase or G2/M phase; diagonal lines, S phase. (C) HDMX promotes colony formation. D54-HDMX clones were induced with tetracycline and D54 parental cells served as control.
HDMX Regulates p53 Transcriptional Activity
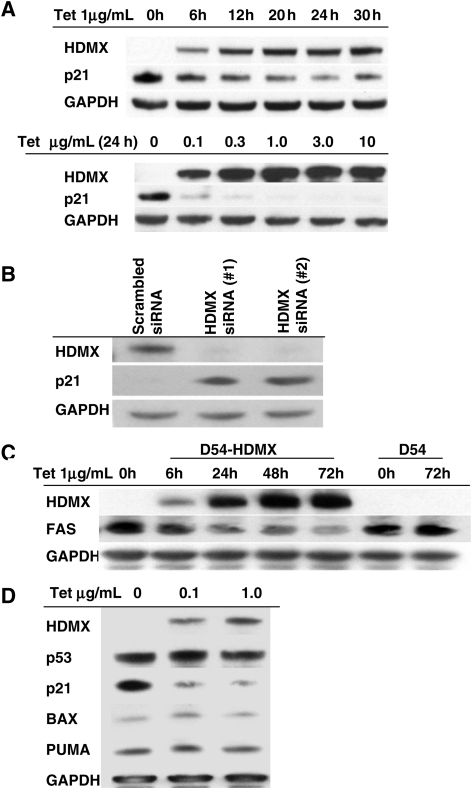
As HDMX is a homologue of MDM2, which functions by binding to p53 and thus regulates the transcriptional activity of p53, and as our above findings verified that HMDX, MDM2, and p53 may function in the same oncogenic pathway during GBM development, we sought to determine whether HDMX promotes cell proliferation through regulating downstream target genes of p53. We first examined the effect of HDMX induction on the expression of several canonical p53 target genes, including p21, FAS, Bax, and PUMA. We observed that the induction of HDMX depressed p21 expression in a dose- and time-dependent manner in D54-HDMX cells (Fig. 3A). Furthermore, in D245 cells, which express high levels of HDMX, siRNA knockdown of HDMX caused an elevated expression of p21 (Fig. 3B). Induction of HDMX also reduced the expression of FAS, a proapoptotic gene critical to the extrinsic pathway,39 in D54-HDMX cells (Fig. 3C). However, we did not observe any alteration in the expression of Bax or PUMA following HDMX induction (Fig. 3D).
Fig. 3.
HDMX selectively depresses p21 and Fas expression. (A) HDMX reduces p21 expression. D54-HDMX cells were treated with tetracycline at the indicated concentrations and time points. HDMX and p21 expression were analyzed by immunoblotting. (B) siRNA knockdown of HDMX significantly lowers HDMX expression while elevating p21 expression. GBM D245 cells transfected with HDMX-specific siRNAs were analyzed by immunoblotting for HDMX and p21. (C) HDMX downregulates FAS expression. D54-HDMX cells were treated with 1.0 µg/mL tetracycline for the indicated time points and were subsequently examined by immunoblotting. (D) HDMX does not affect Bax or PUMA expression. The expression of HDMX, p21, p53, Bax, and PUMA was analyzed in D54-HDMX cells treated with the indicated concentrations of tetracycline.
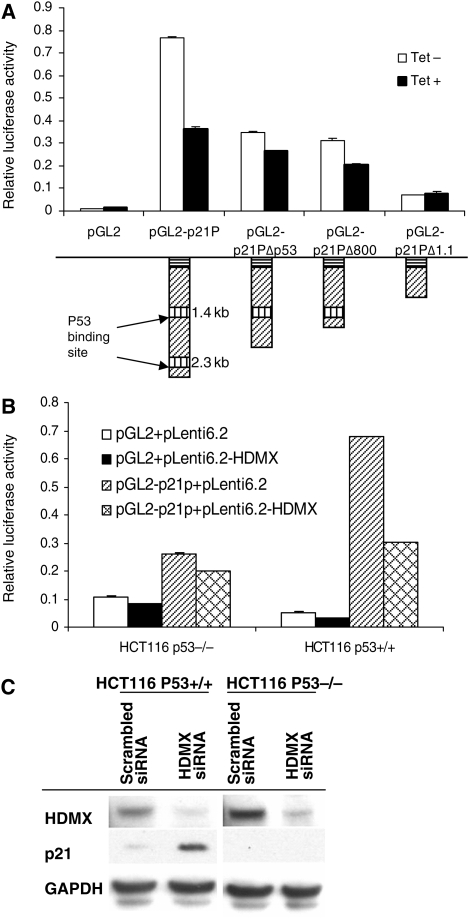
We next used luciferase reporter assays to examine whether the downregulation of p21 expression by HDMX depended upon p53. HDMX induction in D54-HDMX cells repressed greater than 50% of the luciferase activities in cells transfected with pGL2-p21P, which contains both proximal and distal p53 response elements, and roughly 25% of the luciferase activities in cells transfected with pGL2-p21PΔp53 or pGL2-p21PΔ800, both containing only one p53 binding site. In contrast, in cells transfected with pGL2-p21PΔ1.1, which lacks both p53 binding sites, HDMX induction exerted no effect on the luciferase activities (Fig. 4A). Furthermore, HDMX overexpression dramatically reduced the luciferase activities in TP53-proficient HCT-116/p53+/+ cells, while exerting no effect on p21 in p53-deficient HCT-116/p53−/− cells (Fig. 4B). Moreover, siRNA knockdown of HDMX upregulated p21 expression in HCT-116/p53+/+ cells, but failed to affect p21 expression in HCT-116/p53−/−cells (Fig. 4C). Taken together, the above findings revealed that HDMX attenuates p53 transcriptional activity and leads to a subsequent downregulation of p21 expression.
Fig. 4.
HDMX depresses p21 expression by binding to TP53 consensus sites within the p21 promoter. (A) Both proximal and distal TP53 response elements in the p21 promoter are required to fully mediate the p53-dependent effects of HDMX. D54-HDMX cells were transfected with the indicated vectors and subsequently treated with 1.0 μg/mL tetracycline. Relative luciferase activity was measured after 24 h. (B) Transient overexpression of HDMX significantly reduces p21 promoter activity. HCT-116/p53+/+ and HCT-116/p53−/− cells were transfected with the indicated plasmids. Luciferase activity was measured 24 h after transfection. (C) siRNA knockdown of HDMX elevates p21 expression in TP53 wild-type cells, but has no effect on p21 expression in TP53-deficient cells. HCT-116/p53+/+ and HCT-116/p53−/− cells were transfected with the indicated siRNAs and immunoblotted with anti-HDMX and anti-p21 antibodies.
HDMX Confers Resistance to BCNU Treatment
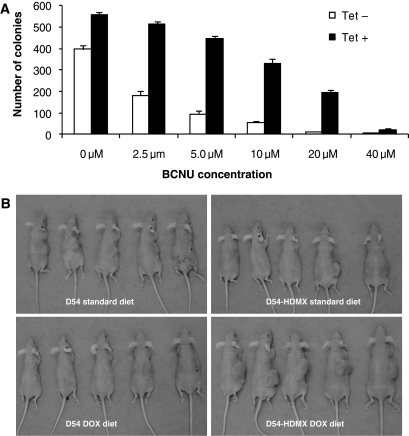
Our above findings demonstrated that HDMX promotes GBM cell proliferation and regulates p53 activities. Dysregulation of p53 has been reported to play a role in regulating tumor cell response to chemotherapeutic drugs. We therefore investigated whether HDMX overexpression modulated the response of GBM cells to BCNU, the standard cytotoxic drug for gliomas. We found that D54-HDMX cells exhibited greater clonogenic survival upon HDMX induction across all concentrations of BCNU treatment (Fig. 5A). Additionally, athymic mice bearing D54-HDMX xenografts treated with BCNU and fed with a doxycycline diet showed greater tumor growth than mice treated with BCNU and fed a regular diet (T–C of more than 5 days, P < 0.001). No mice bearing D54 xenografts developed substantial tumors, defined as exceeding 1000 mm3 in size, 40 days after BCNU treatment. Among mice with D54-HDMX-derived tumors fed with the doxycycline diet, 5 out of 5 mice grew substantial tumors after 40 days, whereas only 1 out of 5 mice fed with the standard diet developed tumors of that size within the same time frame (Fig. 5B). Moreover, we confirmed the BCNU-resistance phenotype conferred by HDMX in another human GBM cell line U87MG (Supplementary Material, Fig. S4). In contrast, HDMX overexpression did not provide temozolomide resistance (Supplementary Material, Fig. S5), indicating there is a specific mechanism underlying the HDMX-BCNU resistance.
Fig. 5.
HDMX confers resistance to BCNU in GBM cells. (A) HDMX prolongs GBM cell survival following BCNU treatment. D54-HDMX cells were treated with 0 or 1 µg/mL tetracycline for 24 h and with the indicated concentrations of BCNU for another 24 h. The number of colonies was counted after 2 weeks. (B) HDMX inhibits the tumoricidal effects of BCNU on GBM in vivo. Athymic mice bearing D54-HDMX and D54 xenografts were fed either a standard or doxycycline-containing diet and subsequently treated with 13 mg/kg of BCNU intraperitoneally.
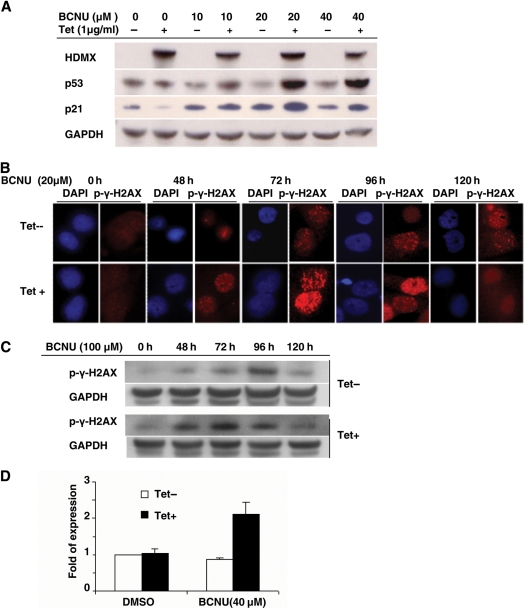
HDMX Potentiates DNA Repair in BCNU-Treated GBM Cells
The above findings suggest that expression of HDMX confers resistance to BCNU both in vitro and in vivo. To understand the mechanism of HDMX-mediated BCNU resistance, we studied the effect of HDMX overexpression on p53 signaling and DNA repair genes in BCNU-treated D54-HDMX cells. We first investigated the effect of HDMX overexpression on the expression of p53 in BCNU-treated GBM cells. In contrast to the result showing that HDMX expression repressed p21 expression without affecting the level of p53 in the unstressed cells (Fig. 3D), with BCNU treatment, we found that the HDMX induction resulted in a dose-dependent accumulation of p53 in response to BCNU and a concurrent upregulation of p21 (Fig. 6A). We further examined the effect of HDMX on DNA repair in BCNU-treated cells by immunofluorescence microscopy using an antibody against the phosphorylated form of H2AX, γ-H2AX, a marker of DNA double-stranded break (DSB) repair. The results showed that γ-H2AX formed distinct subnuclear foci in response to BCNU, which was potentiated by HDMX induction (Fig. 6B), suggesting the presence of a more vigorous repair of DNA damage induced by BCNU in the GBM cells with HDMX overexpression. Immunoblotting studies further revealed that the HDMX induction was associated with increased phosphorylation of the DNA repair protein H2AX (Fig. 6C). Additionally, we found that the xeroderma pigmentosum variant protein XPV, a major DNA repair gene regulated by p53, was upregulated in the HDMX-induced GBM cells treated with BCNU (Fig. 6D). These findings together suggest that HDMX may promote chemoresistance to BCNU by upregulating DNA repair activities in the GBM cells.
Fig. 6.
HDMX stabilizes p53 expression and stimulates DNA repair in BCNU-treated GBM cells. (A) HDMX enhances p53 and p21 expression following BCNU treatment. D54-HDMX cells were treated with 0 or 1 µg/mL tetracycline for 24 h and with the indicated doses of BCNU for 72 h. The expression of HDMX, p53, and p21 was detected by immunoblotting. (B and C) HDMX potentiates DSB repair. D54-HDMX cells were treated as described in (A), and DNA damage response to BCNU was examined by immunofluorescent microscopy (B) or immunoblotting (C) using an antibody against γ-H2AX. (D) XPV is overexpressed in HDMX-induced cells treated with BCNU. GBM D54-HDMX cells were treated with 0 or 1 µg/mL tetracycline for 24 h and then with the indicated doses of BCNU for 72 h. XPV expression was measured by Q-PCR.
Discussion
In this report, we demonstrated that HDMX, an upstream regulator of p53 signaling, acts as a bona fide oncogene in GBM and contributes to chemoresistance. We found that HDMX is frequently amplified and overexpressed in GBM samples. HDMX stimulates GBM cell proliferation in vitro and in vivo through p53-dependent inhibition of p21. Furthermore, we showed that HDMX overexpression confers GBMs with resistance to the DNA alkylator BCNU through promoting DNA repair capacities in response to BCNU treatment.
Our demonstration that HDMX amplification, MDM2 amplification, and TP53 mutation are mutually exclusive events in GBM development supports the hypothesis that HDMX amplification is a critical genetic event in the dysregulation of the p53 signaling pathway, one of the most frequently altered oncogenic pathways in GBMs. Apart from HDMX amplification in GBMs, increased HDMX copy number is present in 65% of retinoblastomas40 and in 19% of breast carcinomas.41 In all these cases, amplification of HDMX has been correlated with wild-type p53 status and lack of MDM2 amplification. HDMX amplification and overexpression therefore provide an alternative mechanism for cancer cells to deregulate the p53 pathway. Cells expressing HDMX and activated Ras (RasV12) were shown to be oncogenic in nude mice.41 Herein, we show that HDMX constitutes a true oncogene in GBM that promotes tumorigenesis by inhibiting p53 induction of cell-cycle arrest in tumors with wild-type p53. The central role of HDMX in the negative regulation of p53 suggests that HDMX may present an alternative target in the p53 signaling pathway and thus indicate a new therapy for cancer patients.
We found that the induction of HDMX enhanced cell proliferation and increased the number of cells actively undergoing cell-cycle progression. HDMX executes its growth-promoting capabilities by inhibiting p53-mediated p21 transactivation. Given the importance of p21 in regulating cell-cycle arrest, these results are consistent with previous mouse models showing that the mechanism of lethality in HDMX knockout mice was mainly attributed to cell-cycle arrest, and that the phenotype could be rescued via TP53 deletion.42,43 The p53 protein also tightly controls apoptosis, which is influenced by a series of quantitative and qualitative events that ultimately induce the activation of p53 signaling. In our study, we found that the induction of HDMX downregulated the expression of the p53 downstream proapoptotic gene FAS, but not that of PUMA or Bax, which suggests that HDMX may also modulate specific apoptotic gene expression through its interaction with p53. Interestingly, we observed that the phenomenon of HDMX-mediated BCNU drug resistance appears to be mechanistically distinct from the cell-cycle progression of unstressed cells. With BCNU treatment, we show here that HDMX overexpression stabilized p53 in the cells. In response to cellular stresses such as DNA damage or oncogene activation, p53 becomes stabilized and modulates the transcription of target genes.44 These effectors drive a variety of cellular responses to stress, including DNA repair, cell-cycle arrest, senescence, and apoptosis. Under different cellular environments or in different cell types, p53 may undergo a variety of posttranslational modifications to precisely coordinate its biological activity in response to stress. This diversity of modifications may provide one explanation for the varying downstream effects of HDMX overexpression observed in both unstressed cells and BCNU-treated cells. Future research needs to define the cellular response and the mechanism of HDMX function within the highly specific cellular environment of GBM in normal growth conditions as well as under exposure to particular chemotherapy treatments.
Several studies have shown that BCNU constitutes a successful treatment option for certain GBM patients.3,4 Batista et al. found that the BCNU treatment in glioma cells causes DSBs, and that p53 wild-type cells were capable of repairing DSBs while p53 mutant cells accumulated DNA damage.7–10 We have discovered that HDMX overexpression in BCNU-treated GBM cells stabilizes p53 levels. Furthermore, we have found that HDMX stimulates the repair of DNA damage induced by BCNU by potentiating the phosphorylation of H2AX and upregulating the DNA repair protein, XPV. These results suggest that resistance to BCNU in GBM cells with induced HDMX may arise through stabilizing p53 and stimulating DNA repair. These observations provide evidence for a potential novel role for HDMX as a reinforcer of p53 function under stress conditions and a potentiator of DNA repair under genotoxic stresses.
These observations present a robust rationale for the oncogenicity of HDMX, and for its clinical potential as an adjuvant therapeutic target that could enhance the tumoricidal effect of genotoxic drugs when antagonized. Our findings strongly suggest that an inhibitor of the p53-HDMX interaction would be valuable not only for facilitating tumor regression, but also for sensitizing tumors to more traditional chemotherapies such as BCNU. Further advancing our knowledge of the molecular mechanisms by which HDMX enhances GBM tumorigenesis and confers drug resistance will indeed prove critical as we strive to develop highly safe and effective therapies for GBM patients.
Supplementary Material
Supplementary Material is available at Neuro-Oncology online.
Funding
This work was supported by a National Brain Tumor Society Grant, a Pediatric Brain Tumor Foundation Institute Grant, and by the National Institutes of Health: [NINDS Grant 5P50 NS20023, NCI SPORE Grant 5P50 CA108786, NCI Merit Award R37 CA 011898].
Acknowledgements
We thank Melissa J. Ehinger, Diane L. Satterfield, and Jennifer D. Funkhouser for assistance with collecting clinical samples, and Joe Zeidner for assistance in generating the GBM xenografts.
Conflict of interest statement. None declared.
References
- 1.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. doi:10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. doi:10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart MG, Grant R, Garside R, Rogers G, Somerville M, Stein K. Chemotherapeutic wafers for High Grade Glioma. Cochrane Database Syst Rev. 2008;4:CD007294. doi: 10.1002/14651858.CD007294. [DOI] [PubMed] [Google Scholar]
- 4.Perry J, Chambers A, Spithoff K, Laperriere N. Gliadel wafers in the treatment of malignant glioma: a systematic review. Curr Oncol. 2007;14:189–194. doi: 10.3747/co.2007.147. doi:10.3747/co.2007.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang DI, Yin JH, Ju TC, Chen LS, Hsu CY. Nitric oxide and BCNU chemoresistance in C6 glioma cells: role of S-nitrosoglutathione. Free Radic Biol Med. 2004;36:1317–1328. doi: 10.1016/j.freeradbiomed.2004.02.010. doi:10.1016/j.freeradbiomed.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Wolff JE, Trilling T, Molenkamp G, Egeler RM, Jurgens H. Chemosensitivity of glioma cells in vitro: a meta analysis. J Cancer Res Clin Oncol. 1999;125:481–486. doi: 10.1007/s004320050305. doi:10.1007/s004320050305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu GW, Mymryk JS, Cairncross JG. Inactivation of p53 sensitizes astrocytic glioma cells to BCNU and temozolomide, but not cisplatin. J Neurooncol. 2005;74:141–149. doi: 10.1007/s11060-004-6601-3. doi:10.1007/s11060-004-6601-3. [DOI] [PubMed] [Google Scholar]
- 8.Xu GW, Mymryk JS, Cairncross JG. Pharmaceutical-mediated inactivation of p53 sensitizes U87MG glioma cells to BCNU and temozolomide. Int J Cancer. 2005;116:187–192. doi: 10.1002/ijc.21071. doi:10.1002/ijc.21071. [DOI] [PubMed] [Google Scholar]
- 9.Xu GW, Nutt CL, Zlatescu MC, Keeney M, Chin-Yee I, Cairncross JG. Inactivation of p53 sensitizes U87MG glioma cells to 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res. 2001;61:4155–4159. [PubMed] [Google Scholar]
- 10.Batista LF, Roos WP, Christmann M, Menck CF, Kaina B. Differential sensitivity of malignant glioma cells to methylating and chloroethylating anticancer drugs: p53 determines the switch by regulating xpc, ddb2, and DNA double-strand breaks. Cancer Res. 2007;67:11886–11895. doi: 10.1158/0008-5472.CAN-07-2964. doi:10.1158/0008-5472.CAN-07-2964. [DOI] [PubMed] [Google Scholar]
- 11.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. doi:10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 12.Gasco M, Crook T. p53 family members and chemoresistance in cancer: what we know and what we need to know. Drug Resist Updat. 2003;6:323–328. doi: 10.1016/j.drup.2003.11.001. doi:10.1016/j.drup.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Toledo F, Wahl GM. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int J Biochem Cell Biol. 2007;39:1476–1482. doi: 10.1016/j.biocel.2007.03.022. doi:10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- 15.Reifenberger G, Liu L, Ichimura K, Schmidt EE, Collins VP. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 1993;53:2736–2739. [PubMed] [Google Scholar]
- 16.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. doi:10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.TCGA collaborative work. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weller M. Predicting response to cancer chemotherapy: the role of p53. Cell Tissue Res. 1998;292:435–445. doi: 10.1007/s004410051072. doi:10.1007/s004410051072. [DOI] [PubMed] [Google Scholar]
- 19.Biroccio A, Bufalo DD, Ricca A, et al. Increase of BCNU sensitivity by wt-p53 gene therapy in glioblastoma lines depends on the administration schedule. Gene Ther. 1999;6:1064–1072. doi: 10.1038/sj.gt.3300935. doi:10.1038/sj.gt.3300935. [DOI] [PubMed] [Google Scholar]
- 20.Weller M, Rieger J, Grimmel C, et al. Predicting chemoresistance in human malignant glioma cells: the role of molecular genetic analyses. Int J Cancer. 1998;79:640–644. doi: 10.1002/(sici)1097-0215(19981218)79:6<640::aid-ijc15>3.0.co;2-z. doi:10.1002/(SICI)1097-0215(19981218)79:6<640::AID-IJC15>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 21.Shvarts A, Bazuine M, Dekker P, et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics. 1997;43:34–42. doi: 10.1006/geno.1997.4775. doi:10.1006/geno.1997.4775. [DOI] [PubMed] [Google Scholar]
- 22.Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YV, Wade M, Wong E, Li YC, Rodewald LW, Wahl GM. Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc Natl Acad Sci USA. 2007;104:12365–12370. doi: 10.1073/pnas.0701497104. doi:10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kros JM, van Run PR, Alers JC, et al. Genetic aberrations in oligodendroglial tumours: an analysis using comparative genomic hybridization (CGH) J Pathol. 1999;188:282–288. doi: 10.1002/(SICI)1096-9896(199907)188:3<282::AID-PATH355>3.0.CO;2-S. doi:10.1002/(SICI)1096-9896(199907)188:3<282::AID-PATH355>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 25.Muleris M, Almeida A, Dutrillaux AM, et al. Oncogene amplification in human gliomas: a molecular cytogenetic analysis. Oncogene. 1994;9:2717–2722. [PubMed] [Google Scholar]
- 26.Rickman DS, Bobek MP, Misek DE, et al. Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 2001;61:6885–6891. [PubMed] [Google Scholar]
- 27.Riemenschneider MJ, Buschges R, Wolter M, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59:6091–6096. [PubMed] [Google Scholar]
- 28.Riemenschneider MJ, Knobbe CB, Reifenberger G. Refined mapping of 1q32 amplicons in malignant gliomas confirms MDM4 as the main amplification target. Int J Cancer. 2003;104:752–757. doi: 10.1002/ijc.11023. doi:10.1002/ijc.11023. [DOI] [PubMed] [Google Scholar]
- 29.Schrock E, Thiel G, Lozanova T, et al. Comparative genomic hybridization of human malignant gliomas reveals multiple amplification sites and nonrandom chromosomal gains and losses. Am J Pathol. 1994;144:1203–1218. [PMC free article] [PubMed] [Google Scholar]
- 30.Gilkes DM, Chen J. Distinct roles of MDMX in the regulation of p53 response to ribosomal stress. Cell Cycle. 2007;6:151–155. doi: 10.4161/cc.6.2.3719. [DOI] [PubMed] [Google Scholar]
- 31.Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. doi:10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 32.Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. doi:10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 33.Di CH, Liao SH, Adamson DC, Parrett TJ, Broderick DK, Shi Q, et al. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans-retinoic acid. Cancer Res. 2005;65:919–924. [PubMed] [Google Scholar]
- 34.Wang TL, Maierhofer C, Speicher MR, et al. Digital karyotyping. Proc Natl Acad Sci USA. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. doi:10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datto MB, Yu Y, Wang XF. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem. 1995;270(48):28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- 36.Badruddoja MA, Keir ST, King I, et al. Activity of VNP40101M (Cloretazine) in the treatment of CNS tumor xenografts in athymic mice. Neuro Oncol. 2007;9:240–244. doi: 10.1215/15228517-2007-011. doi:10.1215/15228517-2007-011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui B, Johnson SP, Bullock N, Ali-Osman F, Bigner DD, Friedman HS. Bifunctional DNA alkylator 1,3-bis(2-chloroethyl)-1-nitrosourea activates the ATR-Chk1 pathway independently of the mismatch repair pathway. Mol Pharmacol. 2009;75:1356–1363. doi: 10.1124/mol.108.053124. doi:10.1124/mol.108.053124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldberg M, Stucki M, Falck J, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. doi:10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 39.Wajant H. The Fas signaling pathway: more than a paradigm. Science. 2002;296:1635–1636. doi: 10.1126/science.1071553. doi:10.1126/science.1071553. [DOI] [PubMed] [Google Scholar]
- 40.Laurie NA, Donovan SL, Shih CS, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. doi:10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 41.Danovi D, Meulmeester E, Pasini D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. doi:10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chavez-Reyes A, Parant JM, Amelse LL, de Oca Luna RM, Korsmeyer SJ, Lozano G. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 2003;63:8664–8669. [PubMed] [Google Scholar]
- 43.Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. doi:10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 44.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. doi:10.1038/nrc864. [DOI] [PubMed] [Google Scholar]