Abstract
Ski acts as a transcriptional co-repressor by multiple direct and indirect interactions with several distinct repression complexes. Ski represses retinoic acid (RA) signaling by interacting with, and stabilizing, key components of the co-repressor complex, namely, HDAC3. However, little is known as to how the Ski protein can stabilize HDAC3. In the present study, we identified the Siah2 protein as a potential E3 ubiquitin ligase that mediated proteasomal degradation of HDAC3. Reciprocal co-immunoprecipitation assays further revealed that Ski interacts with Siah2. Furthermore, co-expression of the Ski protein stabilized the level of Siah2 protein. Since Siah2 regulates its own level of expression by self-degradation, the stabilization of Siah2 by Ski is an indication that Ski association leads to inhibition of Siah2 E3 ubiquitin ligase activity. Only wild type Ski and Ski truncation mutants that were in the same complex with Siah2 could stabilize HDAC3 levels. Taken together, the results suggest that association with Ski leads to inhibition of Siah2 E3 ubiquitin ligase activity and in this way, the Ski protein inhibits Siah2-mediated proteasomal degradation of HDAC3.
Keywords: Ski, Siah2, HDAC3, co-repressor complex, proteasomal degradation
Introduction
Ski is the founding member of a family of proteins that includes itself and the Ski-related protein Sno. The Ski protein can form complexes with N-CoR, SMRT, HDAC3 and mSin3, and is involved in transcriptional regulation of several nuclear receptors [1; 2; 3; 4]. Ski can interact with retinoic acid receptor α (RARα) and block its transactivation activity [1; 2]. RA-induced co-repressor complex degradation is important for the subsequent recruitment of the co-activator complex and transcription initiation of target genes [5]. Ski has been shown to repress RA signaling pathway by stabilizing HDAC3 and RARα [6], however the mechanisms involved in this process have not been characterized.
Retinoic acid receptors (RARs) are degraded by the 26S proteasome system in response to RA [7; 8; 9; 10; 11; 12; 13]. Until now, the E3 ligases that are responsible for the RARs recognition and degradation are unknown. Recently, it has been shown that the HECT domain and Ankyrin repeat containing E3 ubiquitin-protein ligase (HACE1) can interact with the A/B region of RARβ3 [14]. Even though HACE1 was initially identified as an E3 ubiquitin ligase [15], surprisingly, it inhibited RA-induced RARβ3 degradation. The manner of interaction between HACE1 and RARβ3 might block the E3 ubiquitin ligase activity of HACE1 and prevent its function as an E3 ligase for RARβ3. Therefore, the exact E3 ligase responsible for degradation of RARs is still unknown.
HDAC3 also undergoes proteasomal degradation upon RA treatment [5; 6]. TBL1, an F box/WD-40-containing protein, is partially responsible for N-CoR and HDAC3 degradation by recruiting the ubiquitin-proteasome complex, and this degradation is required for thyroid hormone receptor (TR), estrogen receptor (ER), and peroxisome proliferator-activated receptors (PPAR)-mediated transcriptional activation [5]. RARα-mediated transcriptional activation might utilize different E3 ubiquitin ligase(s) to mediate proteasomal degradation of the co-repressor complex. A possible candidate is Siah2 protein. Siah (Seven in Absentia Homolog) proteins are the homologs of SINA (Seven in Absentia) in drosophila, and are RING finger domain containing proteins that function as E3 ubiquitin ligases. Siah2 interacts with N-CoR and is responsible for proteasomal degradation of N-CoR [16]. Since N-CoR is an important component of the nuclear receptor co-repressor complex, we hypothesize that Siah2 might be also involved in proteasomal degradation of other components in the co-repressor complex, namely HDAC3.
In this study, we investigated the effects of Siah2 expression on HDAC3 and RARα degradation and determined how co-expression of Ski influenced this degradation. The data indicated that Siah2 was involved in the degradation of HDAC3 via the proteasome and Ski stabilized HDAC3 levels by inhibiting Siah2 function.
Materials and Methods
Cell culture, Reagents, and Transfection
COS-1 cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco/Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, penicillin G (100 and streptomycin (100 units/ml), μg/ml). Oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA). Expression plasmids were introduced into cells using TransIT-LT1 (Mirus Bio LLC, Madison, WI, USA) as described by the manufacturer.
Plasmid constructs
Flag-HDAC3, Myc-HDAC3, T7-Ski, T7-Ski (1-491), Flag-Ski (1-491), Flag-Ski, and Flag-Ski (491-728) were described previously [3; 6; 17; 18; 19]. T7-Ski truncation mutants Ski (1-261), Ski (1-371), and Ski (Δ101-240) were generated by inserting PCR-generated human Ski cDNA fragments into BglII and SalI sites of pCMV-T7 vector. The primers used for generation of the 1-261 fragment were: 5′-TTTTAGATCTATGGAGGCGGCGGCAGG-3′ and 5′-TTTTCTCGAGCACCACGAACTTGTGC-3′. The primers used for generation of the 1-371 fragment were: 5′-TTTTAGATCTATGGAGGCGGCGGCAGG-3′ and 5′-TTTTCTCGAGGTGAACACAGCCCAGG-3′. The primers used for generation of the 1-100 fragment were 5′-TTTTAGATCTATGGAGGCGGCGGCAGG-3′ and 5′-CGGGATCCGCGCTCGGTGGAGCGG-3′. The primers used for generation of the 241-728 fragment were 5′-CGGGATCCAGCGCCGCCTGCATCCAGTGCC-3′ and 5′-TTTTCTCGAGCTACGGCTCCAGCTCCGC-3′. Fragments 1-100 and 241-728 were ligated together and inserted into the BglII and SalI sites of pCMV-T7 vector.
Whole cell lysates preparation
Cells were washed with PBS twice and scraped into 1ml Nonidet P-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, and 10% glycerol) containing protease inhibitor phenylmethylsulfonylfluoride (PMSF), phosphatase inhibitors NaF (10mM) and sodium vanadate (1mM), and one complete protease inhibitor cocktail tablet. Then the collected cells were sonicated briefly on Branson sonifier 450 (VWR scientific, West Chester, PA, USA) at 2.5 output and precleared at 13,000rpm for 10 min at 4°C. The precleared extracts were taken as whole cell lysates and stored at −20°C for future use.
Co-immunoprecipitation assays and Western blot analysis
All co-immunoprecipitation assays and western blot analysis were performed as described [3]. Antibodies used for immunoprecipitation and western blot analysis were: anti-Myc (9E10, Santa Cruz Biotechnology, CA, USA), anti-Flag mouse monoclonal or anti-Flag rabbit polyclonal antibody (Sigma, St Louis, MO, USA), anti-T7 mouse monoclonal antibody (Novagen, Madison, WI, USA), anti-Ski rabbit polyclonal antibody (H-329, Santa Cruz Biotechnology), anti-Ski mouse monoclonal antibody (G8, Cascade Bioscience, Winchester, MA, USA), anti-β-Actin antibody (Sigma), and anti-α-Tubulin antibody (Sigma). Proteins were detected with the appropriate secondary antibodies (GE Healthcare, Piscataway, NJ, USA) by chemiluminescence (Perkin Elmer, Shelton, CT, USA).
Proteasome inhibition and protein stability assay
The proteasome inhibitor MG132 (Sigma) was used at a concentration of 25 μM for 5 hrs after 24 hrs transfection. In order to determine the degradation of HDAC3 and RARα, COS-1 cells were transiently transfected with plasmids as indicated. 24 hrs after transfection, cycloheximide (CHX, 50μg/ml) (Sigma) and/or retinoic acid (RA, 1μM) (Sigma) were added into the culture medium. Cells were lysed at the indicated times after addition of CHX and/or RA. Lysates were analyzed by western blot analysis.
Results and Discussion
Siah2 interacts with HDAC3 and targets HDAC3 for proteasomal degradation
In order to investigate how Ski can stabilize HDAC3 and RARagr;, we characterized the mechanism involved in the proteasomal degradation of these two proteins. E3 ubiquitin ligases play critical roles in the degradation of substrate proteins through the ubiquitin-proteasome pathway due to their abilities to specifically recognize substrates and recruit target proteins for ubiquitination [20]. A possible E3 ubiquitin ligase responsible for HDAC3 proteasomal degradation is Siah2, since Siah2 interacts with N-CoR, an important component in the co-repressor complex, and targets N-CoR for proteasomal degradation [16]. Furthermore, analysis of the turnover of the co-repressor complex indicated that HDAC3 protein levels were affected by Siah2 overexpression [5]. To further investigate whether Siah2 could also function as an E3 ubiquitin ligase for HDAC3 degradation, we checked the levels of HDAC3 in the presence of Siah2. We found that expression of Siah2 could enhance HDAC3 degradation (Figure 1A, lane 2), and this enhanced degradation could be inhibited by addition of the proteasome inhibitor MG132 (Figure 1A, lane 3). Since Siah2 is self-ubiquitinated and then rapidly degraded, it is barely detectable due to this rapid turnover [21; 22]; however, in the presence of MG132, it is readily detected (Figures 1A, lane 3 and 1B, lane 2). These data indicate that expression of Siah2 leads to enhanced proteasomal degradation of HDAC3.
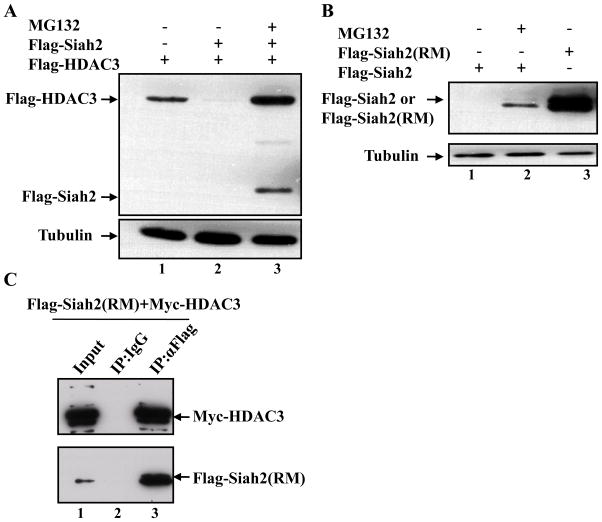
Figure 1. Siah2 is in the same complex with HDAC3 and mediates HDAC3 degradation through the ubiquitin-proteasome pathway.
(A) COS-1 cells were transfected with Flag-HDAC3 alone or together with Flag-Siah2. After 24 hrs transfection, cells were treated with MG132 for 5 hrs as indicated in the figure, and the levels of HDAC3 were detected by western blot analysis using an anti-Flag antibody. Tubulin was used as a loading control. (B) COS-1 cells were transfected with Flag-Siah2 or Flag-Siah2 (RM), and treated with MG132 after 24 hrs transfection as indicated in the figure. Flag-Siah2 and Flag-Siah2 (RM) were detected by western blot analysis using an anti-Flag antibody. Siah2 (RM): A ring finger mutant, H99A/C102A. (C) COS-1 cells were transfected with Flag-Siah2 (RM) and Myc-HDAC3. Whole cell lysates were prepared 24 hrs post-transfection for the co-immunoprecipitation assay. Flag-Siah2 (RM) was immunoprecipitated with a rabbit anti-Flag antibody, and a rabbit normal IgG was used as a control. The immunocomplexes and 10% input were analyzed by western blot analysis using a mouse anti-Myc antibody to detect Myc-HDAC3 or a mouse anti-Flag antibody to detect Flag-Siah2 (RM).
If Siah2 is an E3 ligase for HDAC3, it should bind to HDAC3 as part of the mechanism of ubiquitination. Interactions of Siah2 with its substrates are difficult to detect due to the rapid self-directed degradation of Siah2. However, a RING finger domain mutant (RM) of Siah2 with double point mutations (H99A/C102A) has been shown to be stable due to defective E2 conjugating enzyme recruitment capability [23] (Figure 1B, lane 3). To further characterize the association between Siah2 and HDAC3, we took advantage of this RING finger mutant Siah2 (RM), and co-expressed Siah2 (RM) with HDAC3 in COS-1 cells. By performing a co-immunoprecipitation assay, we found that Siah2 and HDAC3 were in the same complex (Figure 1C, lane 3). Since Siah2 could mediate the proteasomal degradation of HDAC3 and interacted with HDAC3, we reasoned that Siah2 might be an E3 ubiquitin ligase responsible for the degradation of HDAC3. We further determined whether Siah2 is also responsible for RARα degradation. Even though we found that Siah2 expression could accelerate RARα proteasome-dependent degradation under the same experimental conditions as shown above to show HDAC3 interaction (Figure 1C), we could not detect any interaction between RARα and Siah2 (RM) by the co-immunoprecipitation assay (data not shown). These data suggest that either the effects of the Siah2 protein on RARα degradation might be indirect or an additional unidentified protein is required to mediate the interaction between Siah2 and RARα. During transcriptional activation by RA, the co-repressor complex containing HDAC3 undergoes degradation to allow the recruitment of co-activators and transcription. Following transcription, the RARα protein is degraded by unknown means to complete the process and allow other rounds of transcription [5; 24]. Therefore, it is possible that the enhanced degradation of HDAC3 by Siah2 indirectly influences the subsequent degradation of RARα.
The Ski protein is a binding partner of Siah2
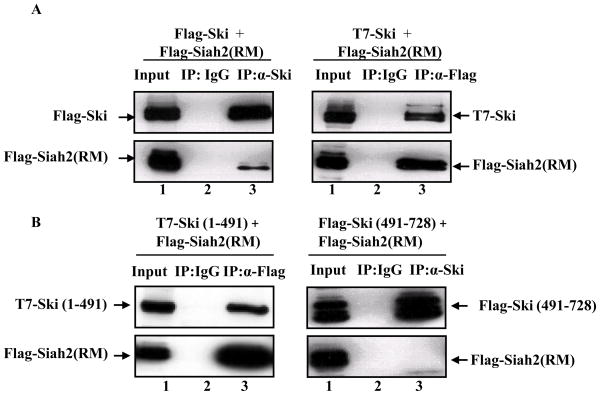
Given that Ski could stabilize HDAC3, and Siah2 could induce HDAC3 degradation via the ubiquitin-proteasome pathway, we next investigated the relationship between Siah2 and Ski. First, we performed reciprocal co-immunoprecipitation assays to determine whether they could interact with each other. As shown in Figure 2A, we could demonstrate the interaction of Siah2 (RM) with Ski by immunoprecipitating either Ski using anti-Ski antibody (left panel, lane 3) or Siah2 (RM) using anti-Flag antibody (right panel, lane 3). We could also demonstrate the interaction between wild type Siah2 and Ski by treating cells with MG132 to inhibit Siah2 auto-degradation (Data not shown). Since the interaction between Siah2 and Ski might help us to understand the mechanism of HDAC3 stabilization by Ski, we further characterized the region on Ski responsible for Siah2 interaction. We made the N-terminal and C-terminal Ski truncation mutants, T7-Ski (1-491), which encoded the first 491 amino acids of Ski, and Flag-Ski (491-728), which encoded the C-terminus of Ski from amino acids 491 to 728. By performing co-immunoprecipitation assays, we found that the N-terminus of Ski (1-491) was co-immunoprecipitated with Siah2 (RM) and responsible for Siah2 (RM) interaction (Figure 2B, left panel, lane 3), while the C-terminal Ski protein was not in the same complex with Siah2 (RM) (Figure 2B, right panel, lane 3).
Figure 2. The Ski protein is an interacting partner of Siah2.
(A) Reciprocal co-immunoprecipitation assays were performed to detect the interaction between Ski and Siah2 (RM). COS-1 cells were transfected with either Flag-Ski and Flag-Siah2 (RM) (left panel), or T7-Ski and Flag-Siah2 (RM) (right panel). Then, we used either an anti-Ski antibody to pull down Ski or an anti-Flag antibody to pull down Siah2 (RM), and checked whether the other protein was present in the immunocomplexes by western blot analysis using appropriate antibodies. (B) N-terminal Ski truncation mutant, T7-Ski (1-491), or C-terminal Ski truncation mutant, Flag-Ski (491-728), were co-transfected with Flag-Siah2 (RM) into COS-1 cells. Flag-Siah2 (RM) was immunoprecipitated from extracts of COS-1 cells using a rabbit anti-Flag antibody. Co-immunoprecipitation was determined by western blot analysis using an anti-T7 antibody to detect T7- Ski (1-491) (left panel). C-terminal Ski fragment was immunoprecipitated from extracts of co-transfected COS-1 cells using an anti-Ski antibody. The co-immunoprecipitation of Siah2 with C-terminal Ski fragment was determined by using an anti-Flag antibody (right panel).
The stabilizing effects of Ski on RARα and HDAC3 correlate with the ability of Ski to interact with Siah2
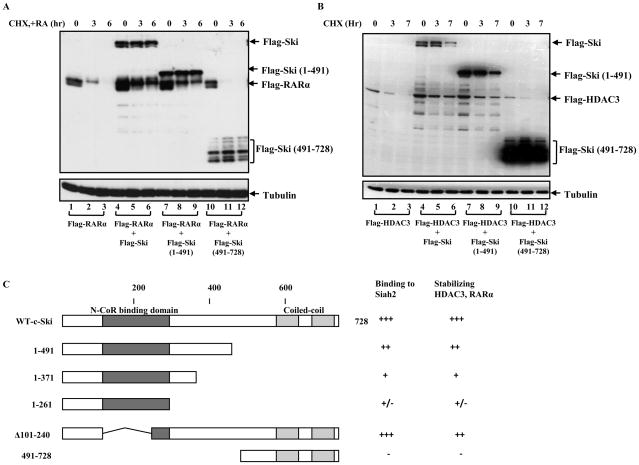
To investigate whether the two Ski truncation mutants have similar abilities to prevent the degradation of HDAC3 and RARα, we treated transfected cells with cycloheximide, CHX, to inhibit protein synthesis and checked the levels of HDAC3 and RARα under different conditions. Consistent with our previous reports [6], HDAC3 and RARα both decreased with time after CHX and/or RA addition (Figure 3A, lanes 1–3 and 3B, lanes 1–3), and this degradation was inhibited by co-expressed Ski (Figure 3A, lanes 4–6, and 3B, lanes 4–6). Interestingly, we found that N-terminal Ski (1-491) protein could also stabilize HDAC3 and RARα (Figure 3A, lanes 7–9 and 3B, lanes 7–9), whereas the C-terminal Ski (491-728) protein, which did not associate with Siah2, could not prevent HDAC3 and RARα degradation (Figure 3A, lanes 10–12, and 3B, lanes 10–12). To characterize this phenomenon further, we investigated if there was any correlation between the Ski-Siah2 interaction and Ski’s ability to stabilize HDAC3 and RARα by creating more Ski truncation mutants and analyzing them as above. We found that the stabilization effects of Ski truncation mutants on HDAC3 and RARα directly correlated with their ability to bind to Siah2 as summarized in Figure 3C. Consistent with the results shown above, the N-terminal region of Ski (most likely from amino acid 261 to 491) appeared to be responsible for Ski’s ability to bind to Siah2 and stabilize HDAC3 and RARα. Importantly, the extent of stabilization effects appeared to be correlated with Ski’s binding ability for Siah2. These results strongly suggest that the stabilization effect on HDAC3 and RARα by Ski is dependent on the Ski-Siah2 interaction.
Figure 3. The N-terminal Ski is responsible for the stabilization effects on HDAC3 and RARα.
(A) COS-1 cells were transfected with Flag-RARα alone or co-transfected with Ski (wild type, N-terminal and C-terminal truncation mutants) and Flag-RARα as indicated. After 24 hrs transfection, cells were treated with CHX and RA for different times as shown in the figure and Flag-RARα levels were determined and compared by western blot analysis using an anti-Flag antibody. (B) The HDAC3 levels were detected using similar methods described in (A) under conditions indicated in the figure. (C) Schematic summary of the relationship between the Ski-Siah2 interaction and the ability of Ski to stabilize HDAC3 and RARα.
Ski expression can increase the expression level of Siah2
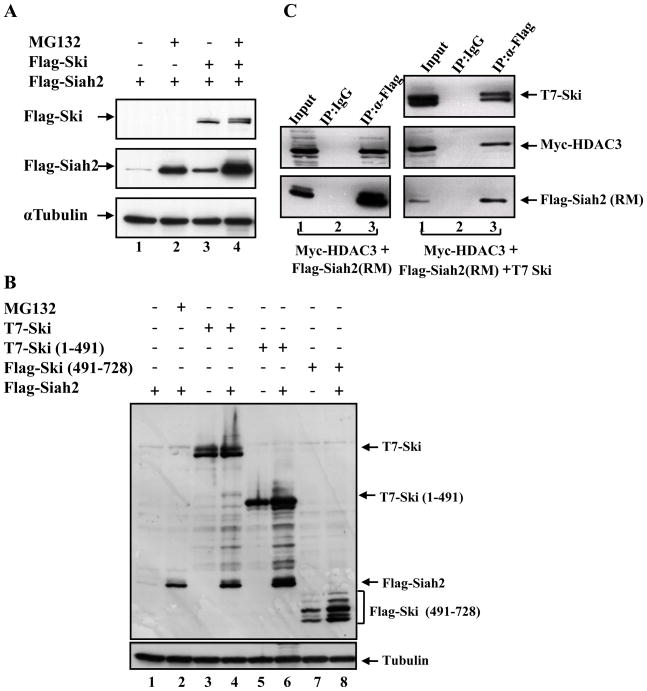
Since the stabilization effects of Ski on RARα and HDAC3 correlated with its Siah2 binding ability, we wondered whether Ski could affect Siah2 function by interacting with Siah2. To determine whether Ski could affect Siah2 ubiquitin ligase activity, the effect of Ski on Siah2 levels was investigated as a measure of the self-degradation ability of Siah2. As shown in Figure 4A, the Siah2 protein level was barely detectable due to its potent self-ubiquitination and rapid proteasomal degradation (Figure 4A, lane 1). MG132 treatment could block Siah2 degradation and allow its detection (Figure 4A, lane 2). Surprisingly, co-expressed Ski also increased the Siah2 protein level (Figure 4A, lane 3) indicating that Ski had an inhibitory effect on Siah2 self-ubiquitination and degradation. Since the self-ubiquitination and degradation of Siah2 is a sign of its ubiquitin ligase activity [25; 26], these observations indicate an inhibitory effect of Ski on the activity of Siah2. Interestingly, we found that the expression level of Ski was essentially unaffected by Siah2 even though Ski can interact with Siah2 (Figure 4B, lanes 3 and 4). These data suggest that Ski is a regulator rather than a substrate of Siah2. We further checked the effects of Ski N-terminal and C-terminal truncation mutants on Siah2 protein level and found that the N-terminal Ski (1-491) protein, which could bind Siah2, also stabilized Siah2 (Figure 4B, lanes 6). The C-terminal Ski (491-728) protein could not bind to Siah2 and had no effect on the self-degradation of Siah2 (Figure 4B, lanes 8). Together these data suggest that Ski increases the levels of HDAC3 (and/or RARα indirectly) by interacting with Siah2 and inhibiting Siah2 ubiquitin ligase activity.
Figure 4. The Ski protein can stabilize Siah2.
(A) COS-1 cells were transfected with Flag-Siah2, or Flag-Siah2 and Flag-Ski together. Where indicated, we treated the transfected cells with 25 μM MG132 for 5 hrs after 24 hrs transfection. Total cell lysates were analyzed by western blot analysis with an anti-Flag antibody. (B) The wild type Ski, N-terminal and C-terminal Ski truncation mutants were transfected alone or together with Siah2 as indicated. Total cell lysates were analyzed by western blot analysis with an anti-Flag antibody to determine the Flag-Siah2 levels under various conditions. (C) COS-1 cells were transfected with Flag-Siah2 (RM) and Myc-HDAC3 without or with T7-Ski. Whole cell lysates were prepared 24 hrs post-transfection for the co-immunoprecipitation assay. Flag-Siah2 (RM) was immunoprecipitated with a rabbit anti-Flag antibody, and a rabbit normal IgG was used as a control. The immunocomplexes and 10% input were analyzed by western blot analysis using a mouse anti-Myc antibody to detect Myc-HDAC3, a mouse anti-Flag antibody to detect Flag-Siah2 (RM), and an anti-T7 antibody to detect T7-Ski.
Recent studies have shown that Siah proteins can be inhibited by several proteins, such as Dab1, Sunphilin-1A, and a fragment derived from a drosophila protein (Phyllopod) [21; 27; 28]. The mechanism of how these different proteins inhibit Siah function is not clear. However, analysis of the inhibitory effects of a Phyllopod fragment on Siah activity found that this small fragment interfered with Siah-substrate interactions by occupying the substrate-binding groove on the Siah protein, which led to stabilization of Siah substrates [21]. In order to investigate how Ski might inhibit Siah2 activity, we checked whether the presence of Ski might interfere with the interaction between HDAC3 and Siah2 by performing co-immunoprecipitation assays. Our data showed the interaction between HDAC3 and Siah2 (RM) (Figure 4C, left panel, lane 3), and that Ski expression had no significant effects on the interaction between Siah2 (RM) and HDAC3 (Figure 4C, right panel, lane 3). Thus, it does not appear that Ski inhibits Siah2’s effects on HDAC3 levels by interfering with Siah2-HDAC3 interaction. The mechanism by which Ski inhibits Siah2 activity needs to be further examined.
Our previous studies demonstrated that Ski could repress RARα-mediated target gene transactivation by stabilizing key components of the co-repressor complex, HDAC3 [6]. In the present study, we provide evidence to show that Siah2 acts as an E3 ubiquitin ligase to mediate HDAC3 degradation. The Ski protein was also identified as another Siah2 interactor, however, the data suggested that Ski was a regulator rather than a substrate of Siah2 and functioned as an inhibitor of Siah2. Potentially, Ski could stabilize components in the co-repressor complex by interfering with Siah2 E3 ubiquitin ligase activity, thus providing a plausible mechanism for the repressive effects of Ski on RA signaling. Regulation of Siah2 E3 ubiquitin ligase activity by Ski provides new insights into the role of Ski in transcriptional regulation and the linkage between the ubiquitin-proteasome system and transcriptional regulation. The potential role of Ski as a Siah2 inhibitor also raises the possibility that Ski might function not only in transcriptional regulation but also in the ubiquitin-proteasome system-related cellular events, which may help explain why the Ski protein plays roles in a variety of biological systems.
Acknowledgments
We thank the members of the Hayman laboratory for helpful discussions and criticisms on the manuscript. This work was supported by U.S. Public Health Service Grant CA42573 from the National Cancer Institute to M.J.H. We also thank Dr. D. Bar-Sagi from New York University for kindly providing Flag-Siah2 and Flag-Siah2 (RM) constructs.
Abbreviations
- HDAC3
histone deacetylase-3
- RA
retinoic acid
- TGFβ
transforming growth factor-β
- N-CoR
nuclear receptor co-repressor
- SMRT
silencing mediator for retinoid and thyroid receptors
- TBL1
transducin beta-like protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dahl R, Kieslinger M, Beug H, Hayman MJ. Transformation of hematopoietic cells by the Ski oncoprotein involves repression of retinoic acid receptor signaling. Proc Natl Acad Sci U S A. 1998;95:11187–92. doi: 10.1073/pnas.95.19.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ritter M, Kattmann D, Teichler S, Hartmann O, Samuelsson MK, Burchert A, Bach JP, Kim TD, Berwanger B, Thiede C, Jager R, Ehninger G, Schafer H, Ueki N, Hayman MJ, Eilers M, Neubauer A. Inhibition of retinoic acid receptor signaling by Ski in acute myeloid leukemia. Leukemia. 2006;20:437–43. doi: 10.1038/sj.leu.2404093. [DOI] [PubMed] [Google Scholar]
- 3.Ueki N, Hayman MJ. Signal-dependent N-CoR requirement for repression by the Ski oncoprotein. J Biol Chem. 2003;278:24858–64. doi: 10.1074/jbc.M303447200. [DOI] [PubMed] [Google Scholar]
- 4.Nomura T, Khan MM, Kaul SC, Dong HD, Wadhwa R, Colmenares C, Kohno I, Ishii S. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;13:412–23. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–26. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 6.Zhao HL, Ueki N, Marcelain K, Hayman MJ. The Ski protein can inhibit ligand induced RARalpha and HDAC3 degradation in the retinoic acid signaling pathway. Biochem Biophys Res Commun. 2009;383:119–24. doi: 10.1016/j.bbrc.2009.03.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudjelal M, Wang Z, Voorhees JJ, Fisher GJ. Ubiquitin/proteasome pathway regulates levels of retinoic acid receptor gamma and retinoid X receptor alpha in human keratinocytes. Cancer Res. 2000;60:2247–52. [PubMed] [Google Scholar]
- 8.Kopf E, Plassat JL, Vivat V, de The H, Chambon P, Rochette-Egly C. Dimerization with retinoid X receptors and phosphorylation modulate the retinoic acid-induced degradation of retinoic acid receptors alpha and gamma through the ubiquitin-proteasome pathway. J Biol Chem. 2000;275:33280–8. doi: 10.1074/jbc.M002840200. [DOI] [PubMed] [Google Scholar]
- 9.Osburn DL, Shao G, Seidel HM, Schulman IG. Ligand-dependent degradation of retinoid X receptors does not require transcriptional activity or coactivator interactions. Mol Cell Biol. 2001;21:4909–18. doi: 10.1128/MCB.21.15.4909-4918.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka T, Rodriguez de la Concepcion ML, De Luca LM. Involvement of all-transretinoic acid in the breakdown of retinoic acid receptors alpha and gamma through proteasomes in MCF-7 human breast cancer cells. Biochem Pharmacol. 2001;61:1347–55. doi: 10.1016/s0006-2952(01)00600-1. [DOI] [PubMed] [Google Scholar]
- 11.Gianni M, Bauer A, Garattini E, Chambon P, Rochette-Egly C. Phosphorylation by p38MAPK and recruitment of SUG-1 are required for RA-induced RAR gamma degradation and transactivation. EMBO J. 2002;21:3760–9. doi: 10.1093/emboj/cdf374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gianni M, Tarrade A, Nigro EA, Garattini E, Rochette-Egly C. The AF-1 and AF-2 domains of RAR gamma 2 and RXR alpha cooperate for triggering the transactivation and the degradation of RAR gamma 2/RXR alpha heterodimers. J Biol Chem. 2003;278:34458–66. doi: 10.1074/jbc.M304952200. [DOI] [PubMed] [Google Scholar]
- 13.Bour G, Lalevee S, Rochette-Egly C. Protein kinases and the proteasome join in the combinatorial control of transcription by nuclear retinoic acid receptors. Trends Cell Biol. 2007;17:302–9. doi: 10.1016/j.tcb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Zhang Z, Vucetic Z, Soprano KJ, Soprano DR. HACE1: A novel repressor of RAR transcriptional activity. J Cell Biochem. 2009;107:482–93. doi: 10.1002/jcb.22146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, Leach S, Marra MA, Brooks-Wilson AR, Penninger J, Sorensen PH. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hacel, in sporadic Wilms’ tumor versus normal kidney. Hum Mol Genet. 2004;13:2061–74. doi: 10.1093/hmg/ddh215. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Guenther MG, Carthew RW, Lazar MA. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 1998;12:1775–80. doi: 10.1101/gad.12.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritter M, Kattmann D, Teichler S, Hartmann O, Samuelsson MK, Burchert A, Bach JP, Kim TD, Berwanger B, Thiede C, Jager R, Ehninger G, Schafer H, Ueki N, Hayman MJ, Eilers M, Neubauer A. Inhibition of retinoic acid receptor signaling by Ski in acute myeloid leukemia. Leukemia. 2006 doi: 10.1038/sj.leu.2404093. [DOI] [PubMed] [Google Scholar]
- 18.Marcelain K, Hayman MJ. The Ski oncoprotein is upregulated and localized at the centrosomes and mitotic spindle during mitosis. Oncogene. 2005;24:4321–9. doi: 10.1038/sj.onc.1208631. [DOI] [PubMed] [Google Scholar]
- 19.Ueki N, Zhang L, Hayman MJ. Ski can negatively regulates macrophage differentiation through its interaction with PU.1. Oncogene. 2008;27:300–7. doi: 10.1038/sj.onc.1210654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 21.Moller A, House CM, Wong CS, Scanlon DB, Liu MC, Ronai Z, Bowtell DD. Inhibition of Siah ubiquitin ligase function. Oncogene. 2009;28:289–96. doi: 10.1038/onc.2008.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Depaux A, Regnier-Ricard F, Germani A, Varin-Blank N. Dimerization of hSiah proteins regulates their stability. Biochem Biophys Res Commun. 2006;348:857–63. doi: 10.1016/j.bbrc.2006.07.092. [DOI] [PubMed] [Google Scholar]
- 23.Habelhah H, Frew IJ, Laine A, Janes PW, Relaix F, Sassoon D, Bowtell DD, Ronai Z. Stress-induced decrease in TRAF2 stability is mediated by Siah2. EMBO J. 2002;21:5756–65. doi: 10.1093/emboj/cdf576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis AP, Haq RU, Nawaz Z. Importance of the regulation of nuclear receptor degradation. Front Biosci. 2001;6:D954–9. doi: 10.2741/dennis. [DOI] [PubMed] [Google Scholar]
- 25.Hu G, Fearon ER. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999;19:724–32. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szargel R, Rott R, Eyal A, Haskin J, Shani V, Balan L, Wolosker H, Engelender S. Synphilin-1A inhibits seven in absentia homolog (SIAH) and modulates alpha-synuclein monoubiquitylation and inclusion formation. J Biol Chem. 2009;284:11706–16. doi: 10.1074/jbc.M805990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park TJ, Hamanaka H, Ohshima T, Watanabe N, Mikoshiba K, Nukina N. Inhibition of ubiquitin ligase Siah-1A by disabled-1. Biochem Biophys Res Commun. 2003;302:671–8. doi: 10.1016/s0006-291x(03)00247-x. [DOI] [PubMed] [Google Scholar]