When we hear the terms mutagenesis and DNA damage, most scientists naturally think about nuclear genomic DNA. However, eukaryotic cells also contain an additional genome within the mitochondria. The mitochondrial genome is susceptible to damage to a greater extent than the nuclear genome, as a result of its proximity to the production of reactive chemical species and the absence of histones, yet has a more limited capacity for DNA repair. And while mutagenesis of mitochondrial genes may not be as directly related to carcinogenesis as some nuclear genes, mutations in mitochondrial genes are clearly linked to human disease and tumorigenesis. This Special Issue addresses these aspects of mitochondrial genome and proteome function, with an emphasis on the role of this organelle in disease and as a potential target for therapy. Here, as an introduction to the Special Issue, we present a brief primer on mitochondrial function, including an evolutionary perspective.
The concept of endosymbiosis and the origin of cell organelles (e.g. plastids and mitochondria) goes back to the late 19th century, when Schimper mused that green plants may have originated from the unification of a colorless organism with an organism colored green by chlorophyll (Schimper, 1885). Soon thereafter, Altman uncovered structures he termed “bioblasts” (mitochondria), which could be stained by fuchsin and were present in nearly all cell types (Altmann, 1890). In 1927, Wallin proposed that the mitochondria of eukaryotic cells are in fact descendants of ancient, once free-living bacteria (Wallin, 1927). This proposal was based largely on the work of Mereschkowsky, who expanded upon the thoughts of Schimper that chloroplasts closely resemble cyanobacteria and that chromatophores developed from free-living blue-green algae which adapted to life within a host organism (Mereschkowsky, 1905). Today, the endosymbiotic events that occurred ca. 2,200-1,500 million years ago in the oceans to give rise to the first eukaryotic cells are described within the framework of the “Serial Endosymbiosis Theory” (SET) (Taylor, 1979; Margulis, 1993). In brief, the theory holds that the cellular evolution of eukaryotes arose through the endosymbiotic union of an engulfed ancient α-proteobacterium with a precursor nucleus-containing (eukaryotic) host cell that resembled extant amitochondriate protists. This “capture” occurred only once, and following a subsequent intracellular domestication process, potentially driven by the emergence of oxygen in the environment, the formerly free-living bacterium was reduced to an organelle, the mitochondrion, for the production and export of energy-rich adenosine triphosphate (ATP). The SET is supported by a sound body of empirical data and has become the most widely accepted explanation for the origin of mitochondria and chloroplasts, although detractors still exist (reviewed in (Lang et al., 1999) and (Kutschera and Niklas, 2005)).
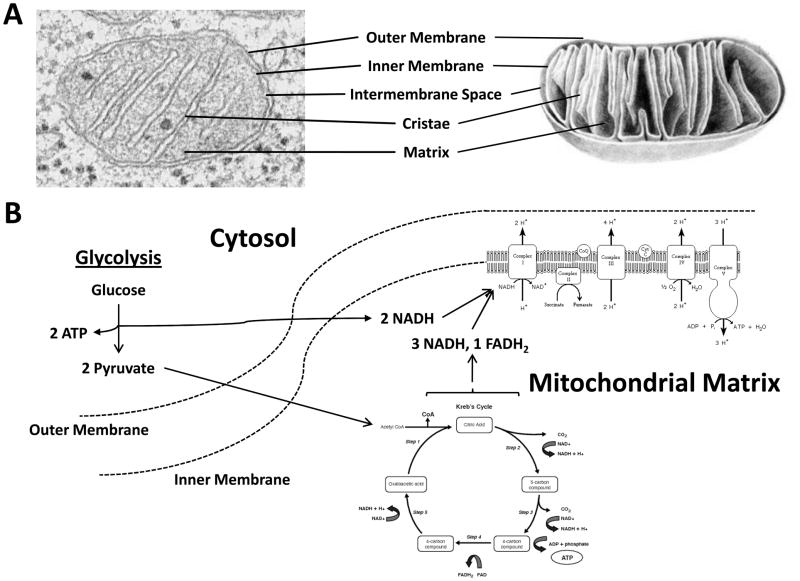
Mitochondria are composed of specialized compartments or regions that include the outer membrane, the intermembrane space, the inner membrane, and the cristae and matrix (Figure 1A). The main function of mitochondria is to provide energy for a range of activities, including movement, the regulation of signaling, cellular differentiation and cell death, and the control of cell cycle and cell growth. The outer membrane is a relatively simple phospholipid bilayer, containing protein structures called porins, which render the membrane permeable to molecules of about 10 kDa or less (i.e. ions, ATP, and some of the smallest proteins). The inner membrane, on the other hand, is freely permeable only to oxygen, carbon dioxide, and water, and thus contains integral proteins for the active transport of specific metabolites across the membrane in a highly regulated manner. The inner membrane also harbors the components of the electron transport chain and the ATP synthetase complex, which are involved in cellular respiration and energy production, and is organized to include internal compartments (or folds) called cristae to increase the surface area to accommodate the numerous structures above. The intermembrane space, as implied, is the region between the inner and outer membranes. The main function of the intermembrane space is oxidative phosphorylation, the final stage of aerobic cellular respiration in which ATP is formed as the result of transfer of electrons from NADH or FADH2 to O2 by a series of electron carriers located in the inner membrane (Figure 1B). Finally, the matrix contains the mitochondrial genome (see below), the ribosomes and other components necessary for translation, and the enzymes responsible for the citric acid (a.k.a. Krebs) cycle reactions. In aerobic organisms, the citric acid cycle is part of a metabolic pathway involved in the chemical conversion of carbohydrates, fats and proteins into carbon dioxide and water to generate usable and convertible forms of energy, such as GTP/ATP or NADH.
Figure 1. Mitochondrial structural organization.
Left, electron microscopy image of a representative mitochondrion. Right, schematic of a single mitochondrion. The different structural components of a mitochondrion are denoted. Aerobic cellular respiration. Glycolysis refers to a series of biochemical reactions by which a single molecule of glucose is oxidized to two molecules of pyruvate. In addition to conversion of glucose to pyruvic acid, two molecules of oxidized NAD are reduced to NADH, two molecules of ADP are phosphorylated to ATP, and two molecules of water and two protons are produced. The process of glycolysis occurs within the cytosol of a eukaryotic cell, under either aerobic or anaerobic conditions. When oxygen is present, pyruvate, which serves as a key intersection in several metabolic pathways, moves out of the cytosol and crosses into the mitochondrial matrix. Here, pyruvate is converted into acetyl-coenzyme A, which is the main input for a series of reactions known as the citric acid cycle (also known as the tricarboxylic acid cycle (TCA cycle) or the Krebs cycle). From a single molecule of acetyl-CoA, in the presence of sufficient oxygen, the eight reactions of the citric acid cycle produce three NADH and one flavin adenine dinucleotide (FAD/FADH2), along with one molecule of ATP. NADH and FADH2 molecules act as electron carriers and are used to generate the bulk of ATP during oxidative phosphorylation. Oxidation of NADH and phosphorylation of ADP to form ATP are processes supported by the electron transport chain assembly and ATP synthase, which are integral protein complexes of the inner mitochondrial membrane.
Although most eukaryotic DNA is packaged in chromosomes within the nucleus, the mitochondrion has its own independent genome, which shows substantial similarity to bacterial genomes. Each mitochondrion is estimated to contain 2–10 copies of double-stranded circular mitochondrial DNA (mtDNA), with each mammalian mtDNA molecule consisting of 15,000–17,000 base pairs. The two strands of mtDNA are differentiated by their nucleotide content, such that the guanine rich strand is referred to as the heavy strand and the cytosine rich strand is referred to as the light strand. The heavy strand encodes 28 genes, and the light strand encodes 9 genes, all of which are essential for normal mitochondrial function. Thirteen of the 37 mtDNA genes provide the instructions for making the protein subunits of the respiratory chain complexes involved in oxidative phosphorylation. Of the remaining genes, 22 provide instructions for making transfer RNAs (tRNA) and 2 encode the small and large subunits of ribosomal RNA (rRNA). Although not covered explicitly in detail in this Special Issue, the predominant theory is that the mitochondrial genome is replicated in a strand-displacement mechanism, whereby leading-strand synthesis starts at a fixed point and advances approximately two-thirds of the way around the molecule before second-strand (or lagging-strand) DNA synthesis is initiated (reviewed in (Falkenberg et al., 2007; Holt, 2009)).
Since all the mitochondria in the developing human embryo come from the egg (there are no mitochondria in the sperm head, which carries the paternal half of the nuclear genome), mtDNA is maternally inherited. Despite the diminutive size of the mitochondrial genome, a number of rare genetic diseases are caused by mutations in mtDNA, and the tissues primarily affected are those that rely heavily on cellular respiration, i.e. the nervous system, muscles, kidneys and liver. Although mitochondrial diseases are individually very rare, since hundreds of them exist, they collectively have a large impact, affecting at least 1 in 5,000 people. The classic mitochondrial diseases include Leber’s hereditary optic neuropathy, in which there is loss of visual acuity often combined with cardiac arrhythmia, and Kearns-Sayre syndrome, which involves paralysis of the eye muscles, dementia and seizures. In addition to a wide array of diseases originating in the mitochondria itself (see more extensive list in (Tuppen et al., 2009)), malfunctioning mitochondria may also contribute to complex disorders like Parkinson disease, Alzheimer disease, epilepsy and diabetes, among others, as well as to aging.
The contributors to this Special Issue will each address one or more aspects of mitochondrial genome and proteome dynamics, with a particular focus on the role of this organelle in disease manifestation and as a potential target in therapy. Specifically, Dr. Robert S. Balaban summarizes what is known about the mitochondrial proteome, which consists of both nuclear and mitochondrial DNA encoded gene products. Dr. Gerald S. Shadel describes the current picture of how mtDNA gene expression is regulated. As the major source of endogenous reactive oxygen and nitrogen species, which are produced by mitochondria as natural products during cellular respiration, Dr. Dean P. Jones reviews the data underscoring the importance of such chemical species in mitochondrial dysfunction. Dr. Ben Van Houten subsequently outlines how flawed mitochondrial activity can give rise to cancer and disease, and Dr. Vilmante Borutaite discusses the active participation of mitochondria in the apoptotic cell death response. Dr. Bruce Demple highlights how the mitochondrial genome protects itself against the deleterious consequences of DNA damage via a series of DNA repair mechanisms. Drs. Jason Bielas and Douglas C. Wallace review the association of mtDNA mutations with tumorigenesis, disease, and aging. The potential in being able to target mitochondria using a gene-therapy paradigm is discussed by Dr. Glenn L. Wilson, and Drs. Peter Wipf and David M. Hockenberry review strategies that target mitochondria in the treatment of disease and cancer. Collectively, we hope that these contributions will inform and interest the readership of Environmental and Molecular Mutagenesis to think about mitochondrial dynamics, and ultimately stimulate new research in this area.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging and National Institute on Alcohol Abuse and Alcoholism.
Reference List
- Altmann R. Die Elementarorganismen und ihre Beziehungen zu den Zellen. Leipzig: Verlag yon Veit &Comp; 1890. [Google Scholar]
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Holt IJ. Mitochondrial DNA replication and repair: all a flap. Trends Biochem Sci. 2009;34:358–365. doi: 10.1016/j.tibs.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Niklas KJ. Endosymbiosis, cell evolution, and speciation. Theory Biosci. 2005;124:1–24. doi: 10.1016/j.thbio.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- Margulis L. Microbial Communities in the Archean and Proterozoic Eons. New York: W. H. Freeman & Co; 1993. Symbiosis in Cell Evolution. [Google Scholar]
- Mereschkowsky C. Uber Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol Centralbl. 1905;25:593–604. [Google Scholar]
- Schimper AFW. Untersuchungen fiber die Chlorophyllk6rper und die ihnen homologen Gebilde. Jb wiss Botanik. 1885;16:1–247. [Google Scholar]
- Taylor FJ. Symbionticism revisited: a discussion of the evolutionary impact of intracellular symbioses. Proc R Soc Lond B Biol Sci. 1979;204:267–286. doi: 10.1098/rspb.1979.0027. [DOI] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Wallin IE. Symbionticism and the Origin of Species. London: Bailliere, Tindall & Cox; 1927. [Google Scholar]