Abstract
Cannabis is the most widely used illicit drug in the United States. There is ample evidence that cannabis use has a heritable component, yet the genes underlying cannabis use disorders are yet to be completely identified. This study's aims were to map susceptibility loci for cannabis use and dependence and two narrower cannabis-related phenotypes of “craving” and “withdrawal” using a family study design. Participants were 2524 adults participating in the University of California San Francisco (UCSF) Family Alcoholism Study. DSM-IV diagnoses of cannabis dependence, as well as indices of cannabis craving and withdrawal, were obtained using a modified version of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA). Genotypes were determined for a panel of 791 microsatellite polymorphisms. Multipoint variance component LOD scores were obtained using SOLAR. Genome-wide significance for linkage (LOD > 3.0) was not found for the DSM-IV cannabis dependence diagnosis, however, linkage analyses of cannabis “craving” and the cannabis withdrawal symptom of “nervous, tense, restless or irritable” revealed five sites with LOD scores over 3.0 on chromosomes 1, 3, 6, 7, 9. These results identify new regions of the genome associated with cannabis use phenotypes as well as corroborate the importance of several chromosome regions highlighted in previous linkage analyses for other substance dependence phenotypes.
Keywords: dependence, genome scan, heritability, marijuana
Introduction
National epidemiological samples have demonstrated that cannabis is the most commonly used illicit drug [Anthony et al., 1994; Stinson et al., 2005, 2006], and that its use has substantially increased over the last decade [Compton et al., 2004]. Chronic use of cannabis is associated with both physical and mental health problems. Persistent use poses health problems similar to those of tobacco [Taylor et al., 2000; Mittleman et al., 2001; Fisher et al., 2005; Hashibe et al., 2005; Tashkin, 2005]. Cannabis use has also been implicated in a syndrome characterized by apathy, loss of goal-directed behavior, and cognitive impairment termed the “amotivational syndrome” [Sharma, 1975; Pope et al., 2001; Solowij et al., 2002; Schuckit, 2006]. Cannabis use is associated with psychotic illness and depression [Degenhardt et al., 2003; Hall et al., 2004], as well as impaired educational and work performance [Kandel and Chen, 2000; Lynskey and Hall, 2000; Swift et al., 2001; Schuckit, 2006]. Use of cannabis, particularly by adolescents and young adults, may also facilitate progression to other illicit drug use (the “gateway” drug hypothesis) [Fergusson and Horwood, 2000; Lynskey et al., 2003].
Twin and family studies have consistently found that cannabis use and use disorders appear to in part have a genetic basis [for review see Agrawal and Lynskey, 2006]. Studies that have evaluated the role of genetic and environmental risk factors on cannabis abuse or cannabis dependence in twin samples have found estimates of heritability that range from 0.45 to 0.78 [see Kendler and Prescott, 1998; Tsuang et al., 1998; van den Bree et al., 1998; Maes et al., 1999; Kendler et al., 2000, McGue et al., 2000; Miles et al., 2001; Rhee et al., 2003]. Taken together, these studies suggest that there are substantial heritable influences on cannabis use disorders and that studies aimed at identifying genes that contribute to cannabis involvement may be warranted.
Disorders of cannabis use may be influenced by a number of genes that are difficult to detect because they may each have a small effect on the broad clinical phenotype. However, the genes influencing cannabis use disorders might be detected if they have a major effect on more narrowly defined phenotypes that more closely index the biological processes associated with addiction. Several biologically based theories have been suggested to explain the compulsion to use addictive drugs that might be profitably used to develop phenotypes useful for the study of cannabis use disorders. A general theory of addiction posits that the neurobiological mechanisms underlying the homeostatic regulation of appetitive drives and instincts becomes dysregulated during the process of drug exposure [Koob, 2000]. Some measures of the strength of this process include an increase or strong desire to take the drug often called ‘drug craving’ [see Anton, 1999], and ultimately the development of tolerance to and withdrawal from use of the drug [Schuckit, 1995]. Some studies have shown that requiring the presence of craving and withdrawal for the diagnosis of alcohol dependence leads to a better diagnostic distinction between abuse and dependence [De Bruijn et al., 2004]. In the case of cannabis dependence, recent data from large national epidemiologic samples have confirmed that cannabis withdrawal is prevalent, correlated with more symptoms of dependence, and associated with significant distress and impairment [Agrawal et al., 2008c; Hasin et al., 2008].
A few studies have conducted linkage analyses using cannabis dependence, cannabis dependence symptoms, and cannabis dependence subtypes as phenotypes [see Hopfer et al., 2007; Agrawal et al., 2008a,b; Ehlers et al., 2009], and some significant loci have been identified. Yet no studies to date have conducted linkage analyses using cannabis withdrawal and/or craving phenotypes. The aim of the present set of analyses was to identify loci associated with cannabis dependence, withdrawal and craving in the UCSF Family Alcoholism Study. The UCSF Family Alcoholism Study is a project that was designed to identify genetic loci that influence susceptibility to alcohol dependence and related phenotypes. It used a small family design, focusing primarily on sibling pairs and parent-child trios. The study enrolled 2454 individuals from 970 families from December 1995 through January 2003. Test-retest and inter-rater reliability for clinical data have been shown to be very good [Vieten et al., 2004]. Design, methods, recruitment strategies and sample demographics of the UCSF Family Alcoholism Study have been presented previously [see Seaton et al., 2004; Vieten et al., 2004], along with intrafamilial correlations for primary diagnostic phenotypes.
Methods
Participants
The UCSF family study recruited probands with lifetime DSM-IV alcohol dependence and the relatives of those probands, nation-wide. In addition, a community sample was ascertained in order to estimate population means and standard deviations for heritability and linkage analyses. Participants were recruited through the use of semi-targeted direct mail, a web site, press releases and advertisements and from alumni of treatment centers across the nation. Probands were invited to participate if they met screening criteria for alcohol dependence at some point in their lifetime and had at least one sibling or both parents available to participate in the study. With the permission of the proband, relatives were invited by mail to participate.
Probands reporting serious drug dependence (defined as use of stimulants, cocaine, or opiates daily for more than three months or weekly for more than six months), and those who reported any history of intravenous substance use were excluded. Probands were excluded if, upon screening, they reported a current or past diagnosis of schizophrenia, bipolar disorder, or other psychiatric illness involving psychotic symptoms (those with depressive and anxiety disorders were accepted); a life-threatening illness; or an inability to speak and read English.
Two thousand one hundred and fifty-four individuals were enrolled in the UCSF Family Alcoholism Study. The sample had a mean age of 48.8 ± 13.2 years, a mean educational level of 14.4 ± 2.9 years, and a mean annual income of $57,356 ± $54,656 (median, $45,000). Racial distribution was 92% Caucasian, 3% each African American and Hispanic, and 1% each Native American and other. No attempt was made to exclude or over sample minorities. Probands were 58% female. Relatives of probands were 38% alcohol dependent.
An unselected general population sample of 147 individuals was also recruited to assess phenotype base rates. Letters were sent to residents of the same geographical areas as the family samples, requesting participation in a study on “health behaviors and characteristics” to avoid a sample biased toward participation in a study on alcoholism. No inclusion/exclusion criteria were applied aside from the ability to respond to the telephone interview and complete the questionnaires. The details of recruitment of all participants have been previously published [see Seaton et al., 2004; Veiten et al., 2004].
Clinical measures and diagnoses
A remote data collection procedure was developed allowing for blood samples and other questionnaires to be returned by mail, and structured diagnostic interviews to be conducted by telephone, making nationwide data collection possible. Potential participants first had the study explained and gave written informed consent. A modified version of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) [Bucholz et al., 1994] was used to collect demographic, medical, psychiatric, alcohol, nicotine, cannabis, and other drug use history. The SSAGA is a fully structured, poly-diagnostic psychiatric interview that has undergone both reliability and validity testing. Modifications of the SSAGA for the present study included changes in diagnostic criteria and items to approximate DSM-IV criteria. Only the sections of the SSAGA relevant to substance abuse, demographics and medical history were utilized. We have previously demonstrated that a number of cannabis usage phenotypes are significantly heritable including: having ever used cannabis, used more than 21 times in a single year, DSM-IV cannabis dependence, individual criteria of DSM-IV cannabis dependence [Ehlers et al., unpublished work]. Two phenotypes with the highest heritability were cannabis “craving” and withdrawal. Therefore, the phenotypes chosen for the present linkage analyses were: (1) a DSM-IV cannabis dependence diagnosis; (2) cannabis “craving” defined as endorsing: “In situations where you couldn't use marijuana, did you ever have such a strong desire for it that you couldn't think of anything else”; and (3) a measure of cannabis withdrawal defined as: “Did stopping or cutting down after a period of regular use of marijuana ever cause you to feel nervous, tense, restless or irritable”. The number of participants with each phenotype, with and without co-morbid alcohol dependence, is presented in table 1. Twenty percent of those enrolled did not complete all study requirements. Details of the interview process and inter-rater reliability of the SSAGA in this study have been reported previously [Vieten et al., 2004].
Table 1.
Number of subjects with and without Alcohol Dependence that were genotyped for the linkage analysis of MJ phenotypes
| No AlcDep Dx | Yes AlcDep Dx | Total | |
|---|---|---|---|
| DSM-IV MJ Dependence | 763 | 850 | 1613 |
| MJ Withdrawal | 761 | 850 | 1611 |
| MJ Craving | 754 | 847 | 1601 |
DNA collection and genotyping
The DNA extraction procedure and genotyping protocol have been previously described [Wilhelmsen et al., 2003]. Briefly, DNA was isolated from whole blood using a commercial kit (Gentra, Minneapolis, MN), and genotypes for a panel of microsatellite polymorphisms were generated using fluorescently labeled polymerase chain reaction (PCR) primers (HD5, version 2.0; Applied Biosystems, Foster City, CA). The HD5 panel set consisted of 811 markers with an average marker-to-marker distance of 4.6 cM (maximum, 14 cM) and an average heterozygosity of greater than 77%. A small subset of markers was omitted from the panel because of null alleles, irregular allele spacing or other problems with reproducibility. None of the omitted markers were adjacent to other omitted markers. The sizes of the PCR products for the markers were determined from electropherograms produced with an ABI 3700 (Applied Biosystems, Foster City, CA), a 96-channel capillary electrophoresis device.
The sizes of marker amplimers were determined (blinded to pedigree structure and subject characteristics) from the electropherogram using the Genotyper software package (ABI). All electropherograms were visually inspected and exported from Genotyper in base pair sizes relative to the standard measured to one hundredth of a base pair. Fragment sizes were binned to alleles by using an automated algorithm developed by one of the authors (K.C.W.), which assumes that the distribution of allele sizes will have a sine-squared distribution with a fitted periodicity near two base pairs. The program determines the best periodicity and phase for the modeled distribution relative to the observed distribution. Fragments that are distributed between minimums of the modeled distribution are assumed to be the same allele. Allele frequencies observed in the founders were used for all analysis. The sex-averaged marker map order obtained from the manufacturer was used and verified with the family data from the current sample.
The genotypes for all of the autosomal markers were analyzed for each family by using Pedigree Relationship Statistical Test (PREST) [see McPeek and Sun, 2000] to detect sample and pedigree structure errors. DNA was re-isolated from a stored frozen blood specimen, and the genotyping was repeated for any individual for whom PREST detected a probable error. If regenotyping failed to resolve the error, the problematic genotype was subsequently treated as missing. The program Pedcheck was used to detect non-Mendelian inheritance [O'Connell et al., 1998]. Markers with a high frequency of Mendelian segregation errors were excluded from analysis, and for isolated Mendelian errors, the genotypes for the entire family were excluded for the specific marker that yielded the error. To further reduce errors, the probability that each genotype was correct was assessed by using the error-checking algorithm implemented in Merlin [Abecasis et al., 2002], in which genotypes that had a probability of less than 0.025 of being correct were removed from further consideration.
Data analyses
Both genotype and phenotype data were available for 1647 individuals, and phenotype but not genotype data was available for an additional 720 individuals. Of these 720 individual 87 were relatives that could not b e contacted or did not provide DNA samples and 462 were probands that did not provide a sufficient number of genetically informative relatives for inclusion and the remaining 171 subjects were the relatives of these probands. Seven hundred and thirteen families were considered genetically informative for the heritability analyses. Families that contained sibling, half-sibling, avuncular or cousin pairs were included as being potentially genetically informative. These families ranged in size from 3 to 20 subjects (average 4.63 ± 2.13). The data includes: 563 parent-child, 1085 sibling, 40 half-sibling, 17 grandparent-grandchild, 238 avuncular, and 32 cousin relative pairs. An additional 177 families contained only a single individual with phenotype data. These individuals were included within some analyses to the extent that they contribute information about trait means and variance and the impact of covariates.
Analyses were previously conducted to estimate the heritability of the three phenotypes of interest: DSM-IV cannabis dependence, cannabis craving, and irritable/anxious during cannabis withdrawal using SOLAR [http://solar.sfbrgenetics.org/] [Almasy and Blangero, 1998] [Ehlers et al., unpublished work]. Participant's age at the time of evaluation and sex were evaluated as potential covariates and retained if they accounted for at least 5% of the total variance. The total additive genetic heritability (h2) and its standard error were estimated, and the probability that h2 was greater than zero was determined using a Student's t-test for each scale, and all three phenotypes were found to be heritable and as such suitable for linkage analyses.
The variance components method implemented in SOLAR [http://solar.sfbrgenetics.org/] was then used to calculate multipoint LOD scores across the genome at 1 cM intervals for the three cannabis phenotypes. All traits were analyzed under a continuous trait model rather than using the latent threshold model as the former approach provided a more conservative estimate of linkage in the present study. A LOD threshold of 3.0 was used to identify loci that provided evidence for linkage, and a LOD threshold of 2.0 was used to identify loci yielding suggestive evidence of linkage. For each reported linkage peak, simulation analyses were then conducted using SOLAR to derive nominal p-values. Specifically, a genetic locus was simulated under the null hypothesis of no linkage across 20,000 trials, and the distribution of LOD scores across those trials was used to derive a nominal p-value for the observed LOD score.
Because the current sample was selected for alcohol dependence, it should be noted that prevalence rates for the cannabis dependence diagnoses and respective symptoms in the unselected control sample were estimated and included in the tested models to correct for ascertainment bias when calculating h2 and linkage. In addition, the high marker density of the HD5 microsatellite panel allows for the extraction of a large proportion of IBD information among relative pairs as participants' genotypes can be almost completely phased, thus reducing the reliance on population allele frequencies to estimate allele-sharing probabilities. As a result, and given the very low number of ethnic minority participants in the UCSF sample, we chose to include all participants in the analyses without correcting for ethnic background.
Results
Demographics
The demographic characteristics of this population have been presented previously [see Vieten et al., 2004]. In brief, the UCSF family sample had a mean age of 48.4 ± 13.4 years, a mean educational level of 14.4 ± 2.9 years, and an annual income of $57,356 ± $54,656. Racial distribution was: 92% Caucasian, 3% each African American and Hispanic, and 1% each Native American and other. Fifty-four percent were married. Probands were 58% female, with 97% of probands meeting lifetime criteria for alcohol dependence, whereas, 38% of the relatives of probands were found to be alcohol dependent. The random sample had similar demographic characteristics to the Family Study sample with 15% having a lifetime history of DSM-IV alcohol dependence. The prevalence rates for DSM-IV lifetime cannabis dependence were .16 for probands and .07 for the random sample. Prevalence rates for cannabis withdrawal and cannabis craving were .09 in the probands and .03 in the random sample.
Linkage analyses
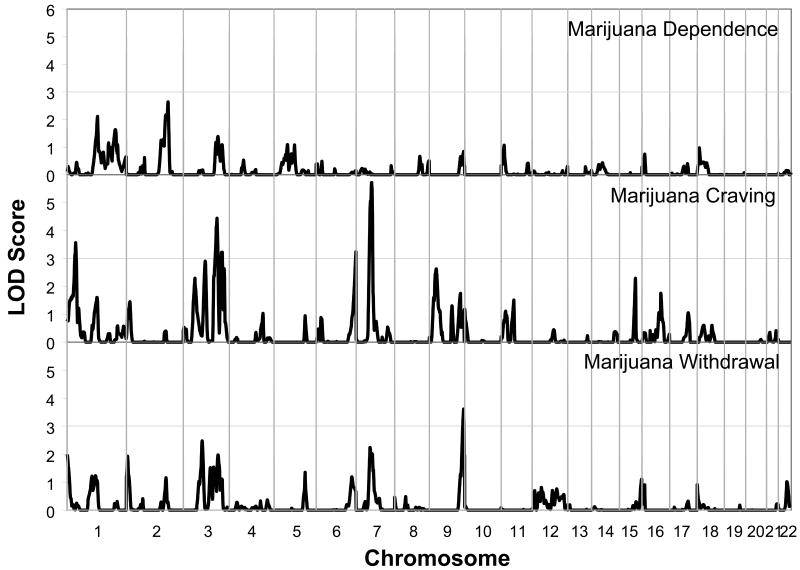
Analyses of multipoint variance component LOD scores for the dichotomous DSM-IV cannabis dependence phenotype did not yield any peaks that achieved genome-wide significance. However, two sites were identified with LOD scores over 2.0, a site on chromosome 1 between 142 and 159 cM with a peak at 154 cM (LOD = 2.1), and a site on chromosome 2 between 194 and 216 cM with a peak at 211 cM (LOD = 2.6). These findings are presented in Figure 1 and Table 2.
Figure 1.
Multipoint Linkage Analysis for the Cannabis dependence (upper graph), Cannabis craving (middle graph) and Cannabis withdrawal (lower graph) phenotypes for the entire genome. Results for each chromosome are aligned end to end with the p terminus on the left. Log of the Odds (LOD) score is plotted on the Y-axis. Vertical lines indicate the boundaries between the chromosomes. The numbers above on the X-axis indicate the chromosome number.
Table 2. Linkage findings for cannabis phenotypes in the UCSF Family study.
| Chr | Phenotype | LOD | Chr loc | Nearest Marker(s) | Supporting reference, phenotype |
|---|---|---|---|---|---|
| 1 | Cannabis cravin | 3.6 | 43cM (36-48cM) | D1S199-D1S2864 | |
| Cannabisdependence | 2.1 | 154cM (142-159cM) | D1S498 | ||
| 2 | Cannabis dependence | 2.6 | 211cM (194-216cM) | D2S2361 | Nurnberger et al., (2001) alcohol dep and depression; Schuckit et al., (2001) subjective response to alcohol; Hill et al., (2004) alcohol dep; Gelernter et al., (2005, 2006) cocaine, opiod dep; Agrawal et al., (2008d), alc dep; Gizer et al.,(unpublished work), nicotine dep |
| 3 | Cannabis withdrawal | 2.5 | 96cM (89-102cM) | D3S1566-D3S3681 |
Stallings et al., (2003), substance dep; Ehlers et al., (2008), ASPD/CD; Hopfer et al., (2007) cannabis dep |
| Cannabis craving | 4.4 | 171cM (164-176cM) | D3S1279-D3S3668 | ||
| 6 | Cannabis craving | 3.2 | 202cM (195-202cM) | D6S281 | |
| 7 | Cannabis withdrawal | 2.2 | 71cM (65-90cM) | D7S506 |
Reich et al., (1998) alcohol dep; Agrawal et al., (2007) alcohol consumption; Hill et al., (2004) alcohol dep |
| Cannabis craving | 5.7 | 79cM (70-83cM) | D7S502-D7S2476 | ||
| 9 | Cannabis craving | 2.6 | 35cM (25-44cM) | D9S157 | Gizer et al., (unpublished work) alcohol dep; Stallings et al., (2003), substance dep; Hopfer et al., (2007) cannabis dep |
| Cannabis withdrawal | 3.6 | 174cM (167-177cM) | D9S1838 | ||
| 15 | Cannabis craving | 2.3 | 81cM (75-84cM) | D15S127 |
Dick et al., (2002) alcohol dep subtype; Ehlers et al., (2004b) alcohol withdrawal |
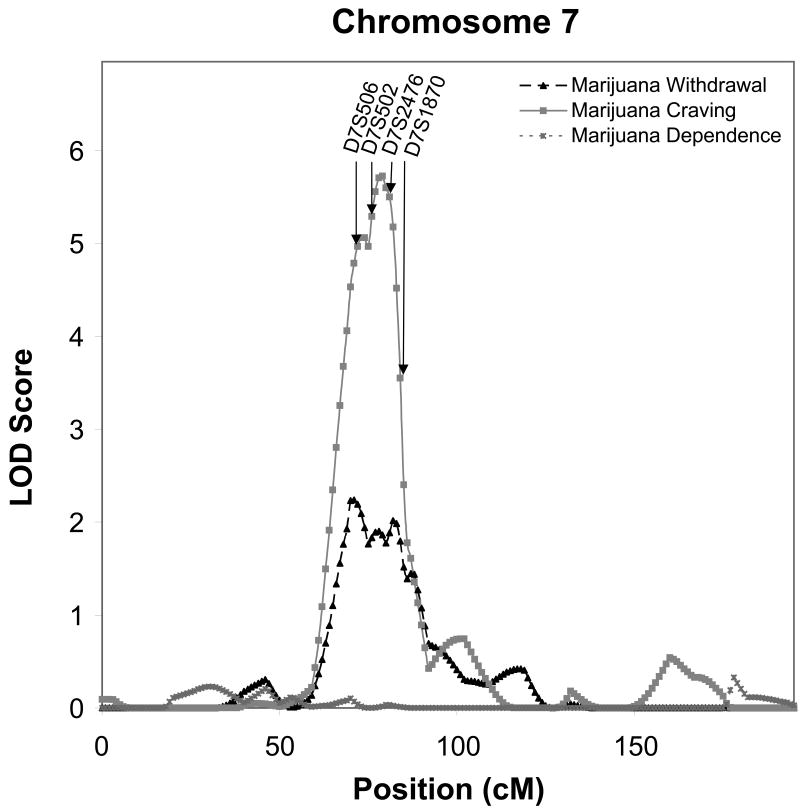
Linkage analyses of cannabis “craving” e.g. endorsing: “In situations where you couldn't use marijuana, did you ever have such a strong desire for it that you couldn't think of anything else” and the cannabis withdrawal symptom of: “Did stopping or cutting down after a period of regular use of marijuana ever cause you to feel nervous, tense, restless or irritable” revealed 5 sites with LOD scores over 3.0. The symptom of cannabis craving yielded the strongest evidence for linkage (LOD = 5.7) on chromosome 7 between 70 and 83 cM with a peak at 79 cM. At that same site there was suggestive evidence of linkage within that same support interval with a peak at 71 cM for the withdrawal phenotype (LOD = 2.2) as illustrated in Figure 2.
Figure 2.
Multipoint Linkage Analysis for Cannabis dependence (dotted line), Cannabis craving (solid gray line) and Cannabis withdrawal (solid black line) phenotype for chromosome 7. The analysis assumes a latent normally distributed variable with a threshold above which an individual is affected. Log of the Odds (LOD) score (Y-axis) is plotted for the chromosome location map (in centimorgans (cM), X-axis). Nearest markers to the peak are presented within the support interval.
The symptom of cannabis craving also yielded strong evidence for linkage on chromosome 3 between 164 and 176 cM with a peak at 171 cM (LOD = 4.4). There was additional evidence for linkage for cannabis craving on chromosome 1 between 36 and 48 cM with a peak at 43 cM (LOD = 3.6), and on chromosome 6 between 195 an 202 cM with a peak at 202 cM (LOD = 3.2). There was suggestive evidence on chromosome 9 between 25 an 44 cM with a peak at 35 cM (LOD = 2.6), and on chromosome 15 between 75 and 84 cM with a peak at 81 cM (LOD = 2.3).
The symptom “nervous, tense, restless or irritable” during cannabis withdrawal also demonstrated evidence for linkage on chromosome 9 between 167 and 177 cM with a peak at 174 cM (LOD = 3.6). Additionally, suggestive evidence for linkage for this withdrawal phenotype was found on chromosome 3 between 89 and 102 cM with a peak at 96 cM (LOD = 2.5) and on chromosome 7 between 65 and 90 cM with a peak at 71cM (LOD = 2.2) (see Table 2 for complete results). Nominal p-values generated by simulation analyses are presented for all three cannabis phenotypes in table 3.
Table 3. Nominal p-values for cannabis phenotypes generated by simulation analyses.
| Chr | Loc | Phenotype | LOD | Nominal p-value |
|---|---|---|---|---|
| 1 | 43cM | Cannabis craving | 3.6 | 0.0077 |
| 154cM | Cannabis Dependence | 2.1 | 0.004 | |
| 2 | 211cM | Cannabis Dependence | 2.6 | 0.0016 |
| 3 | 96cM | Cannabis | 2.5 | 0.0041 |
| 171cM | Withdrawal Cannabis craving | 4.4 | 0.0031 | |
| 6 | 202cM | Cannabis craving | 3.2 | 0.0109 |
| 7 | 71cM | Cannabis | 2.2 | 0.0062 |
| 79cM | Withdrawal Cannabis craving | 5.7 | 0.0007 | |
| 9 | 35cM | Cannabis craving | 3.6 | 0.0077 |
| 174cM | Cannabis Withdrawal | 3.6 | 0.0007 | |
| 15 | 81cM | Cannabis craving | 2.3 | 0.0249 |
Secondary analyses were conducted in order to determine whether these genome locations were specific only to the “nervous, tense, restless or irritable” symptom of withdrawal or whether the “sleeplessness” and a “number of withdrawal symptoms count” phenotype could also produce similar LOD scores in those locations. Evidence for linkage on chromosome 9 was found for the “sleeplessness” phenotype at 158 cM (LOD = 4.2) and the symptom count phenotype (LOD = 3.2). Additionally, suggestive evidence for linkage for the sleeplessness (LOD = 2.6) and symptom count (LOD = 3.0) phenotype was found on chromosome 3 with a peak at 95 cM. Thus it appears that similar regions in the genome may harbor genes for several withdrawal symptoms.
In order to determine whether the sites identified for the cannabis phenotypes were mainly due to comorbid alcohol dependence a separate linkage analysis was conducted using alcohol dependence as a covariate and few differences were found. The results of those analyses are presented in table 4. The linkage results for cannabis dependence, craving and withdrawal were also compared to the results obtained from previous linkage scans for alcohol and nicotine dependence that were also conducted in the UCSF family study population. None of the linkage regions reported in a previous linkage scan for alcohol dependence [Gizer et al., 2009] yielded neither significant nor suggestive linkage in a subsequent scan for nicotine dependence [Gizer et al., unpublished work] in the UCSF family study population. However, in the present set of analyses, suggestive evidence for linkage to cannabis dependence was found at a site on chromosome 2 previously identified in a scan for nicotine dependence [Gizer et al., unpublished work], and another site identified for cannabis craving on chromosome 9 in the present study was also identified in a previous scan for alcohol dependence [Gizer et al., 2009] in the UCSF family study population. These results provide evidence suggesting that two of the regions identified in the present set of analyses may also confer risk towards nicotine and alcohol dependence in this population.
Table 4. Linkage results for MJ phenotypes with and without Alcohol Dependence included as a covariate.
| Chr | Phenotype | Chr loc (cM) | LOD | LOD w/AlcDep as covar | Difference |
|---|---|---|---|---|---|
| 1 | Craving | 43 | 3.6 | 3.5 | -0.1 |
| Dependence | 154 | 2.1 | 2.6 | +0.5 | |
| 2 | Dependence | 211 | 2.6 | 2.4 | -0.2 |
| 3 | Withdrawal | 96 | 2.5 | 2.0 | -0.5 |
| Craving | 171 | 4.4 | 4.4 | 0 | |
| 6 | Craving | 202 | 3.2 | 3.3 | +0.1 |
| 7 | Withdrawal | 71 | 2.2 | 2.6 | +0.4 |
| Craving | 79 | 5.7 | 6.1 | +0.4 | |
| 9 | Craving | 35 | 3.6 | 2.4 | -1.2 |
| Withdrawal | 174 | 3.6 | 3.7 | +0.1 | |
| 15 | Craving | 81 | 2.3 | 2.1 | -0.2 |
Discussion
Several studies suggest that there is a moderate genetic influence on cannabis dependence, and recently there have been additional reports that have identified regions in the genome that may be linked to cannabis dependence [see Kendler and Prescott, 1998; Tsuang et al., 1998; Maes et al., 1999; Miles et al., 2001; Lynskey et al., 2002; Kendler et al., 2003; Rhee et al., 2003; Wilhelmsen and Ehlers, 2005; Agrawal and Lynskey, 2006; Hopfer et al., 2007; Agrawal et al., 2008a,b]. Only two sites, one on chromosome 1 and one on chromosome 2, were identified in the present study for DSM-IV cannabis dependence. The site on chromosome 1 at 154 cM (LOD = 2.1) does not appear to be within 30 cM of other sites identified using substance dependence phenotypes. However, this peak was between two sites; one at around 197 cM that was identified by Dick et al. [2002] in the Collaborative Study for the Genetics of Alcoholism (COGA) dataset for a quantitative alcohol-related phenotype and one at around 102 cM for a cannabis problems factor score in an autosomal linkage scan for cannabis use disorders in the nicotine addiction genetics project [Agrawal et al, 2008a].
The site identified on chromosome 2 near 211 cM (LOD = 2.6) for DSM-IV cannabis dependence in the UCSF Family Alcoholism Study appears to be in the same general region (within 30 cM) as a site identified in the COGA dataset for comorbid alcohol dependence and major depression [Nurnberger et al., 2001], and for subjective response to alcohol [Schuckit et al., 2001] using microsatellite markers and for alcohol dependence using SNP genotyping [Agrawal et al., 2008d]. It also appears to be near a site for alcohol dependence identified in multiplex families in Pittsburgh [Hill et al., 2004], and for heavy cocaine use [Gelernter et al., 2005] and opioid dependence [Gelernter et al., 2006] in African Americans. Additionally, this site was identified by a previous genome scan for nicotine dependence in the UCSF family study [Gizer et al., unpublished work]. Thus, this site appears to harbor genes that confer risk for a number of different substance-related phenotypes.
It has been suggested that the effort to identify genetic factors and the mechanisms whereby they influence addiction may be aided by the use of phenotypes that may be more closely related to the biological processes underlying risk for use disorders [Gottesman and Gould, 2003]. One phenotype that most DSM-IV substance dependence syndromes have in common is a withdrawal syndrome defined as a combination of physiologic and psychological processes that occurs after cessation of its use [Edwards, 1986; Weisbeck et al., 1996; Hasin et al., 2006, 2008]. Although withdrawal is not required to meet DSM-IV criteria for drug/alcohol dependence it is associated with a more severe clinical course and a poorer prognosis [Schuckit et al., 1999; Budney et al., 1999, 2003, 2004, 2008; Budney and Hughes, 2006; Hasin et al., 2000, 2008; Ehlers et al., 2004a]; and as such, has been shown to be more heritable than the clinical diagnosis itself in some populations [Ehlers et al., 2004b]. Another phenotype that is common to many drugs of abuse is craving. Human and animal studies have demonstrated that craving is an important element in the addictive process and that control of craving may improve efforts at abstinence [see Wise, 1988; Robinson and Berridge, 1993; Anton, 1999; Sinha and O'Malley, 1999; Field et al., 2004; Heishman and Singleton, 2006; Haughey et al., 2008]. Evaluation of the heritability of cannabis withdrawal (h2 =0.28, p<0.001) and craving (h2 =0.36, p<0.0001) demonstrated that these two phenotypes were more heritable than the DSM-IV diagnosis of cannabis dependence (h2 =0.20, p<0.0001) in the UCSF family study population [Ehlers et al, unpublished work]. Additionally, five sites in the genome that provided evidence for linkage to these phenotypes were identified.
Two of the chromosome sites that were identified in the UCSF family study population for cannabis craving and cannabis withdrawal were on chromosomes 3q and 9q. These locations have been identified previously in genome scans for cannabis and other drug dependence phenotypes [see Stallings et al., 2003, 2005; Hopfer et al., 2007]. Hopfer and colleagues [2007] conducted a genome wide scan for loci influencing adolescent cannabis dependence symptoms in 324 sibling pairs from 192 families. In that study, probands (52.1% of whom were EuroAmerican, 36.5% of whom were Hispanic, and 7.8% of whom were African-American) were identified from consecutive admissions to substance abuse treatment facilities. The authors found evidence for suggestive linkage on chromosomes 3q21 (LOD = 2.61) and 9q34 (LOD = 2.57). Using the same population of adolescents, Stallings et al. [2003, 2005], conducted linkage scans for the average number of dependence symptoms, conduct disorder symptoms and a composite index of antisocial substance dependence, and also found evidence for linkage to the chromosome 9q34 region (LOD = 3.15) and 3q24-3q25 (LOD = 3.27) for the composite index. The site on chromosome 3q was within 30 cM of a site identified for antisocial personality disorder/conduct disorder in an American Indian community [Ehlers et al., 2008]. The site identified on chromosome 3q for cannabis craving in the UCSF family study (LOD = 4.5) was also within 30 cM of the site identified on that chromosome by Stallings et al. [2005]. Additionally, a site on chromosome 9q (LOD=3.5) for cannabis withdrawal in the UCSF family study was also within 30 cM of the site identified on that chromosome by Stallings et al. [2005]. Additionally, a site on chromosome 9q (LOD=3.5) for cannabis withdrawal in the UCSF family study was also within 30 cM of the site identified on that chromosome by Stallings et al., [2005]. Taken together these findings suggest that these sites on chromosome 3q and 9q may harbor genes that confer risk for severe phenotypes associated with drug dependence including antisocial symptoms.
The region of the genome that provided the best evidence for linkage in the UCSF family study was on chromosome 7q (LOD = 5.6) for the cannabis craving phenotype. This region has consistently emerged with significant evidence of linkage in the COGA project. In the initial COGA sample this region on chromosome 7q provided the strongest evidence of linkage to alcohol dependence with a multipoint LOD score of 3.49 [Reich et al., 1998]. An additional linkage analysis with expanded microsatellite markers in the full COGA sample revealed a LOD score of 2.9 [Wang et al., 2004], and a set of SNPs genotyped in the region that were not in linkage disequilibrium with adjacent markers generated a LOD score at the peak of 4.1 [Dunn et al., 2005]. Other population samples have also found evidence for linkage on 7q. A report of linkage to this region on 7q has been made by the Nicotine Addiction Genetics project for alcohol consumption phenotypes [Agrawal et al., 2008a], and for alcohol dependence in a sample of multiplex families ascertained at Pittsburgh [Hill et al., 2004]. A systematic SNP screen to fine map this region on chromosome 7 using the COGA dataset identified an association with ACN9 a gene involved in gluconeogenesis and the assimilation of ethanol or acetate into carbohydrate [Dick et al., 2008].
There were several areas of the genome that were reported previously as being linked to cannabis use that were not identified in the current linkage scan for cannabis dependence phenotypes in the UCSF Family study. For instance, Agrawal and colleagues [2008d] conducted a linkage scan in a sample from the COGA study for six DSM-IV cannabis dependence criteria considered as a continuous variable (0-6) and found a maximum LOD score of 1.9 at 95 cM on chromosome 14. A site on chromosome 14 at 122 cM was also found for a severe cannabis subtype generated by a cluster analysis in an American Indian community [Ehlers et al., 2009]. This region on chromosome 14 was also identified previously in a genome-wide linkage analysis for ASPD/CD in that American Indian community [Ehlers et al., 2008]. Two areas of the genome were also identified in that American Indian community study with LOD scores that provide evidence for linkage for a severe cannabis use/antisocial subtype on chromosome 16 (@139 cM, LOD score = 4.4), and on chromosome 19 (@74 cM, LOD score = 6.4) [Ehlers et al., 2009]. There have also been some findings in genome scans for cannabis dependence and other drug use related phenotypes on chromosome 19. A region has been identified on chromosome 19 at 17 cM for early-onset cannabis use, and for frequency of use on chromosome 18, in a study of 2314 Australian families [Agrawal et al., 2008b]. A site on chromosome 4 for a cannabis problems factor score has been identified on chromosome 4 in the region of the gamma aminobutyric acid type A gene cluser with a LOD score of 2.2 in the Nicotine Addiction Genetics Project [Agrawal et al., 2008a].
In summary, data from the UCSF Family study did not reveal evidence for genome-wide significance for linkage (LOD > 3.0) to the DSM-IV cannabis dependence diagnosis, however, linkage analyses of cannabis “craving” and the cannabis withdrawal symptom of “nervous, tense, restless or irritable” revealed five sites with LOD scores over 3.0 on chromosomes 1, 3, 6, 7, 9. These results have identified new regions of the genome associated with cannabis use phenotypes as well as corroborated the importance of several chromosome regions highlighted in previous linkage analyses for other substance dependence phenotypes. The results of this study should, however, be interpreted in the context of several limitations. First, multiple phenotypes were tested in the present study, thus increasing the possibility that a false positive result was interpreted as substantive. To protect against this possibility, empirically-derived p-values were reported for each linkage signal allowing for the objective evaluation of the linkage evidence. Second, the findings may not generalize to other large studies that were not ascertained for alcoholic probands. Third, only retrospective and cross-sectional data on cannabis use and use disorders were assessed. Fourth, comparisons to other large samples may be limited by differences in a number of genetic and environmentally determined variables. The exclusion criteria for this study eliminated participants reporting serious drug dependence on stimulants, cocaine, or opiates, current diagnosis of schizophrenia, bipolar disorder, or other psychiatric illness involving psychotic symptoms. This exclusion was included to create a more homogeneous sample for genetic analyses. However, this limitation reduces external validity and clinical applicability since the co-occurrence of mental health disorders and substance abuse is common. Additionally, this sample was comprised of almost entirely Caucasians that also limits applicability to other ethnic groups.
Acknowledgments
Supported in part by funds from the State of California for medical research on alcohol and substance abuse through the University of California at San Francisco, the Ernest Gallo Clinic and Research Center (KCW), the National Institute of Alcohol Abuse and Alcoholism grants T32 AA007573 (KCW) and AA010201 and Clinical Translational Studies U54 RR0250204. (CLE).
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. The genetic epidemiology of cannabis use, abuse and dependence. Addiction. 2006;101(6):801–812. doi: 10.1111/j.1360-0443.2006.01399.x. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Pergadia ML, Saccone SF, Lynskey MT, Wang JC, Martin NG, Statham D, Henders A, Campbell M, Garcia R, et al. An autosomal linkage scan for cannabis use disorders in the nicotine addiction genetics project. Arch Gen Psychiatry. 2008a;65(6):713–721. doi: 10.1001/archpsyc.65.6.713. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Morley KI, Hansell NK, Pergadia ML, Montgomery GW, Statham DJ, Todd RD, Madden PA, Heath AC, Whitfield J, et al. Autosomal linkage analysis for cannabis use behaviors in Australian adults. Drug Alcohol Depend. 2008b;98(3):185–190. doi: 10.1016/j.drugalcdep.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Pergadia ML, Lynskey MT. Is there evidence for symptoms of cannabis withdrawal in the national epidemiologic survey of alcohol and related conditions? Am J Addict. 2008c;17(3):199–208. doi: 10.1080/10550490802019519. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, Saccone SF, Saccone NL, Grucza RA, Wang JC, Cloninger CR, et al. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Drug Alcohol Depend. 2008d;93(1-2):12–20. doi: 10.1016/j.drugalcdep.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony JC, Warner LA, Kessler RC. Comparative epidemiology of dependence on tobacco, alcohol, controlled substances, and inhalants: basic findings from the National Comorbididty Survey. Exp Clin Psychopharmacol. 1994;2(3):244–268. [Google Scholar]
- Anton RF. What is craving? Models and implications for treatment. Alcohol Res Health. 1999;23(3):165–173. [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Budney AJ, Novy PL, Hughes JR. Marijuana withdrawal among adults seeking treatment for marijuana dependence. Addiction. 1999;94(9):1311–1322. doi: 10.1046/j.1360-0443.1999.94913114.x. [DOI] [PubMed] [Google Scholar]
- Budney AJ, Moore BA, Vandrey RG, Hughes JR. The time course and significance of cannabis withdrawal. J Abnorm Psychol. 2003;112(3):393–402. doi: 10.1037/0021-843x.112.3.393. [DOI] [PubMed] [Google Scholar]
- Budney AJ, Hughes JR, Moore BA, Vandrey R. Review of the validity and significance of cannabis withdrawal syndrome. Am J Psychiatry. 2004;161(11):1967–1977. doi: 10.1176/appi.ajp.161.11.1967. [DOI] [PubMed] [Google Scholar]
- Budney AJ, Hughes JR. The cannabis withdrawal syndrome. Curr Opin Psychiatry. 2006;19(3):233–238. doi: 10.1097/01.yco.0000218592.00689.e5. [DOI] [PubMed] [Google Scholar]
- Budney AJ, Vandrey RG, Hughes JR, Thostenson JD, Bursac Z. Comparison of cannabis and tobacco withdrawal: severity and contribution to relapse. J Subst Abuse Treat. 2008;35(4):362–368. doi: 10.1016/j.jsat.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton WM, Grant BF, Colliver JD, Glantz MD, Stinson FS. Prevalence of marijuana use disorders in the United States: 1991-1992 and 2001-2002. JAMA. 2004;291(17):2114–2121. doi: 10.1001/jama.291.17.2114. [DOI] [PubMed] [Google Scholar]
- De Bruijn C, Korzec A, Koerselman F, van den Brink W. Craving and Withdrawal as Core Symptoms of Alcohol Dependence. J Nerv Ment Dis. 2004;192(7):494–502. doi: 10.1097/01.nmd.0000131912.71344.e4. [DOI] [PubMed] [Google Scholar]
- Degenhardt L, Hall W, Lynskey M. Exploring the association between cannabis use and depression. Addiction. 2003;98(11):1493–1504. doi: 10.1046/j.1360-0443.2003.00437.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Nurnberger J, Jr, Edenberg HJ, Goate A, Crowe R, Rice J, Bucholz KK, Kramer J, Schuckit MA, Smith TL, et al. Suggestive linkage on chromosome 1 for a quantitative alcohol-related phenotype. Alcohol Clin Exp Res. 2002;26(10):1453–1460. doi: 10.1097/01.ALC.0000034037.10333.FD. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Wang JC, Saccone S, Hinrichs A, Bertelsen S, Budde J, Saccone N, Foroud T, Nurnberger J, Jr, et al. A systematic single nucleotide polymorphism screen to fine-map alcohol dependence genes on chromosome 7 identifies association with a novel susceptibility gene ACN9. Biol Psychiatry. 2008;63(11):1047–1053. doi: 10.1016/j.biopsych.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn G, Hinrichs AL, Bertelsen S, Jin CH, Kauwe JS, Suarez BK, Bierut LJ. Microsatellites versus single-nucleotide polymorphisms in linkage analysis for quantitative and qualitative measures. BMC Genet. 2005;6 1:S122. doi: 10.1186/1471-2156-6-S1-S122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G. The alcohol dependence syndrome: a concept as stimulus to enquiry. Br J Addict. 1986;81(2):171–183. doi: 10.1111/j.1360-0443.1986.tb00313.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Betancourt M, Gilder DA. The clinical course of alcoholism in 243 Mission Indians. Am J Psychiatry. 2004a;161(7):1204–1210. doi: 10.1176/appi.ajp.161.7.1204. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004b;129B(1):110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Slutske WS, Lind PA, Wilhelmsen KC. Externalizing disorders in American Indians: Comorbidity and a genome wide linkage analysis. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(6):690–698. doi: 10.1002/ajmg.b.30666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Gizer IR, Wilhelmsen KC. Heritability and a genome-wide linkage analysis of a Type II/B cluster construct for cannabis dependence in an American Indian community. AddictBiol. 2009;14(3):338–348. doi: 10.1111/j.1369-1600.2009.00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergusson DM, Horwood LJ. Does cannabis use encourage other forms of illicit drug use? Addiction. 2000;95(4):505–520. doi: 10.1046/j.1360-0443.2000.9545053.x. [DOI] [PubMed] [Google Scholar]
- Field M, Mogg K, Bradley BP. Cognitive bias and drug craving in recreational cannabis users. Drug Alcohol Depend. 2004;74(1):105–111. doi: 10.1016/j.drugalcdep.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Fisher BA, Ghuran A, Vadamalai V, Antonios TF. Cardiovascular complications induced by cannabis smoking: a case report and review of the literature. Emerg Med J. 2005;22(9):679–680. doi: 10.1136/emj.2004.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136B(1):45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, Weiss R, Sonne S, Zhao H, Farrer L, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78(5):759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizer IR, Ehlers CL, Vietan C, Seaton-Smith KL, Feiler HS, Lee JV, Segall SK, Wilhelmsen KC. Linkage scan of alcohol dependence in the UCSF family alcoholism study. Alc Clin Exp Res. 2009;33(6):14A. doi: 10.1016/j.drugalcdep.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Hall WD, Degenhardt L, Teesson M. Cannabis use and psychotic disorders: an update. Drug Alcohol Rev. 2004;23(4):433–443. doi: 10.1080/09595230412331324554. [DOI] [PubMed] [Google Scholar]
- Hashibe M, Straif K, Tashkin DP, Morgenstern H, Greenland S, Zhang ZF. Epidemiologic review of marijuana use and cancer risk. Alcohol. 2005;35(3):265–275. doi: 10.1016/j.alcohol.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Hasin D, Paykin A, Meydan J, Grant B. Withdrawal and tolerance: prognostic significance in DSM-IV alcohol dependence. J Stud Alcohol. 2000;61(3):431–438. doi: 10.15288/jsa.2000.61.431. [DOI] [PubMed] [Google Scholar]
- Hasin D, Hatzenbuehler ML, Keyes K, Ogburn E. Substance use disorders: Diagnostic and Statistical Manual of Mental Disorders, fourth edition (DSM-IV) and International Classification of Diseases, tenth edition (ICD-10) Addiction. 2006;101 1:59–75. doi: 10.1111/j.1360-0443.2006.01584.x. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Keyes KM, Alderson D, Wang S, Aharonovich E, Grant BF. Cannabis withdrawal in the United States: Results from NESARC. J Clin Psychiatry. 2008;69(9):1354–1363. doi: 10.4088/jcp.v69n0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughey HM, Marshall E, Schacht JP, Louis A, Hutchison KE. Marijuana withdrawal and craving: influence of the cannabinoid receptor 1 (CNR1) and fatty acid amide hydrolase (FAAH) genes. Addiction. 2008;103(10):1678–1686. doi: 10.1111/j.1360-0443.2008.02292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heishman SJ, Singleton EG. Assessment of cannabis craving using the Marijuana Craving Questionnaire. Methods Mol Med. 2006;123:209–216. doi: 10.1385/1-59259-999-0:209. [DOI] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Med Genet B Neuropsychiatr Genet. 2004;128B(1):102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer CJ, Lessem JM, Hartman CA, Stallings MC, Cherny SS, Corley RP, Hewitt JK, Krauter KS, Mikulich-Gilbertson SK, Rhee SH, et al. A genome-wide scan for loci influencing adolescent cannabis dependence symptoms: evidence for linkage on chromosomes 3 and 9. Drug Alcohol Depend. 2007;89(1):34–41. doi: 10.1016/j.drugalcdep.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel DB, Chen K. Types of marijuana users by longitudinal course. J Stud Alcohol. 2000;61(3):367–378. doi: 10.15288/jsa.2000.61.367. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA. Cannabis use, abuse, and dependence in a population-based sample of female twins. Am J Psychiatry. 1998;155(8):1016–1022. doi: 10.1176/ajp.155.8.1016. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Neale MC, Prescott CA. Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Arch Gen Psychiatry. 2000;57(3):261–269. doi: 10.1001/archpsyc.57.3.261. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160(4):687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Koob GF. Animal models of craving for ethanol. Addiction. 2000;95 2:S73–S81. doi: 10.1080/09652140050111663. [DOI] [PubMed] [Google Scholar]
- Lynskey M, Hall W. The effects of adolescent cannabis use on educational attainment: a review. Addiction. 2000;95(11):1621–1630. doi: 10.1046/j.1360-0443.2000.951116213.x. [DOI] [PubMed] [Google Scholar]
- Lynskey MT, Heath AC, Nelson EC, Bucholz KK, Madden PA, Slutske WS, Statham DJ, Martin NG. Genetic and environmental contributions to cannabis dependence in a national young adult twin sample. Psychol Med. 2002;32(2):195–207. doi: 10.1017/s0033291701005062. [DOI] [PubMed] [Google Scholar]
- Lynskey MT, Heath AC, Bucholz KK, Slutske WS, Madden PA, Nelson EC, Statham DJ, Martin NG. Escalation of drug use in early-onset cannabis users vs co-twin controls. JAMA. 2003;289(4):427–433. doi: 10.1001/jama.289.4.427. [DOI] [PubMed] [Google Scholar]
- Maes HH, Woodard CE, Murrelle L, Meyer JM, Silberg JL, Hewitt JK, Rutter M, Simonoff E, Pickles A, Carbonneau R, et al. Tobacco, alcohol and drug use in eight- to sixteen-year-old twins: the Virginia Twin Study of Adolescent Behavioral Development. J Stud Alcohol. 1999;60(3):293–305. doi: 10.15288/jsa.1999.60.293. [DOI] [PubMed] [Google Scholar]
- McGue M, Elkins I, Iacono WG. Genetic and environmental influences on adolescent substance use and abuse. Am J Med Genet, B Neuropsychiatric Genet. 2000;96(5):671–677. doi: 10.1002/1096-8628(20001009)96:5<671::aid-ajmg14>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66(3):1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles DR, van den Bree MB, Gupman AE, Newlin DB, Glantz MD, Pickens RW. A twin study on sensation seeking, risk taking behavior and marijuana use. Drug Alcohol Depend. 2001;62(1):57–68. doi: 10.1016/s0376-8716(00)00165-4. [DOI] [PubMed] [Google Scholar]
- Mittleman MA, Lewis RA, Maclure M, Sherwood JB, Muller JE. Triggering myocardial infarction by marijuana. Circulation. 2001;103(23):2805–2809. doi: 10.1161/01.cir.103.23.2805. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, et al. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158(5):718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope HG, Jr, Gruber AJ, Hudson JI, Huestis MA, Yurgelun-Todd D. Neuropsychological performance in long-term cannabis users. Arch Gen Psychiatry. 2001;58(10):909–915. doi: 10.1001/archpsyc.58.10.909. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Hum Genet. 1998;81(3):207–215. [PubMed] [Google Scholar]
- Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch Gen Psychiatry. 2003;60(12):1256–1264. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Berridge KC. The neural basis of drug craving: an incentive-sensitization theory of addiction. Brain Res Brain Res Rev. 1993;18(3):247–291. doi: 10.1016/0165-0173(93)90013-p. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Drug and alcohol abuse: a clinical guide to diagnosis and treatment. New York: Plenum Medical Book Co.; 1995. [Google Scholar]
- Schuckit MA, Daeppen JB, Danko GP, Tripp ML, Smith TL, Li TK, Hesselbrock VM, Bucholz KK. Clinical implications for four drugs of the DSM-IV distinction between substance dependence with and without a physiological component. Am J Psychiatry. 1999;156(1):41–49. doi: 10.1176/ajp.156.1.41. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A, Foroud T. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res. 2001;25(3):323–329. [PubMed] [Google Scholar]
- Schuckit MA. Drug and Alcohol Abuse. New York: Springer Science+Business Media, Inc.; 2006. [Google Scholar]
- Seaton KL, Cornell JL, Wilhelmsen KC, Vieten C. Effective strategies for recruiting families ascertained through alcoholic probands. Alcohol Clin Exp Res. 2004;28(1):78–84. doi: 10.1097/01.ALC.0000107200.88229.57. [DOI] [PubMed] [Google Scholar]
- Sharma BP. Cannabis and its users in Nepal. Br J Psychiatry. 1975;127:550–552. doi: 10.1192/bjp.127.6.550. [DOI] [PubMed] [Google Scholar]
- Sinha R, O'Malley SS. Craving for alcohol: findings from the clinic and the laboratory. Alcohol Alcohol. 1999;34(2):223–230. doi: 10.1093/alcalc/34.2.223. [DOI] [PubMed] [Google Scholar]
- Solowij N, Stephens RS, Roffman RA, Babor T, Kadden R, Miller M, Christiansen K, McRee B, Vendetti J. Cognitive functioning of long-term heavy cannabis users seeking treatment. JAMA. 2002;287(9):1123–1131. doi: 10.1001/jama.287.9.1123. [DOI] [PubMed] [Google Scholar]
- Southwest Foundation for Biomedical Research [S.F.B.R] San Antonio, TX: Southwest Foundation for Biomedical Research; 2008. Sequential Oligogenic Linkage Analysis Routines. Available at: [ http://solar.sfbrgenetics.org/] [Google Scholar]
- Stallings MC, Corley RP, Hewitt JK, Krauter KS, Lessem JM, Mikulich SK, Rhee SH, Smolen A, Young SE, Crowley TJ. A genome-wide search for quantitative trait loci influencing substance dependence vulnerability in adolescence. Drug Alcohol Depend. 2003;70(3):295–307. doi: 10.1016/s0376-8716(03)00031-0. [DOI] [PubMed] [Google Scholar]
- Stallings MC, Corley RP, Dennehey B, Hewitt JK, Krauter KS, Lessem JM, Mikulich-Gilbertson SK, Rhee SH, Smolen A, Young SE, et al. A genome-wide search for quantitative trait Loci that influence antisocial drug dependence in adolescence. Arch Gen Psychiatry. 2005;62(9):1042–1051. doi: 10.1001/archpsyc.62.9.1042. [DOI] [PubMed] [Google Scholar]
- Stinson FS, Grant BF, Dawson DA, Ruan WJ, Huang B, Saha T. Comorbidity between DSM-IV alcohol and specific drug use disorders in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Drug Alcohol Depend. 2005;80(1):105–116. doi: 10.1016/j.drugalcdep.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Stinson FS, Ruan WJ, Pickering R, Grant BF. Cannabis use disorders in the USA: prevalence, correlates and co-morbidity. Psychol Med. 2006;36(10):1447–1460. doi: 10.1017/S0033291706008361. [DOI] [PubMed] [Google Scholar]
- Swift W, Hall W, Teesson M. Cannabis use and dependence among Australian adults: results from the National Survey of Mental Health and Wellbeing. Addiction. 2001;96(5):737–748. doi: 10.1046/j.1360-0443.2001.9657379.x. [DOI] [PubMed] [Google Scholar]
- Tashkin DP. Smoked marijuana as a cause of lung injury. Monaldi Arch Chest Dis. 2005;63(2):93–100. doi: 10.4081/monaldi.2005.645. [DOI] [PubMed] [Google Scholar]
- Taylor DR, Poulton R, Moffitt TE, Ramankutty P, Sears MR. The respiratory effects of cannabis dependence in young adults. Addiction. 2000;95(11):1669–1677. doi: 10.1046/j.1360-0443.2000.951116697.x. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55(11):967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- van den Bree MB, Johnson EO, Neale MC, Svikis DS, McGue M, Pickens RW. Genetic analysis of diagnostic systems of alcoholism in males. Biol Psychiatry. 1998;43(2):139–145. doi: 10.1016/S0006-3223(97)00225-4. [DOI] [PubMed] [Google Scholar]
- Vieten C, Seaton KL, Feiler HS, Wilhelmsen KC. The University of California, San Francisco Family Alcoholism Study. I. Design, methods, and demographics. Alcohol Clin Exp Res. 2004;28(10):1509–1516. doi: 10.1097/01.alc.0000142261.32980.64. [DOI] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, Kwon JM, Wu W, Dick DM, Rice J, et al. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet. 2004;13(17):1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Wiesbeck GA, Schuckit MA, Kalmijn JA, Tipp JE, Bucholz KK, Smith TL. An evaluation of the history of a marijuana withdrawal syndrome in a large population. Addiction. 1996;91(10):1469–1478. [PubMed] [Google Scholar]
- Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, Kalmijn J. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27(7):1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen KC, Ehlers C. Heritability of substance dependence in a Native American population. Psychiatr Genet. 2005;15(2):101–107. doi: 10.1097/00041444-200506000-00006. [DOI] [PubMed] [Google Scholar]
- Wise RA. The neurobiology of craving: implications for the understanding and treatment of addiction. J Abnorm Psychol. 1988;97(2):118–132. doi: 10.1037//0021-843x.97.2.118. [DOI] [PubMed] [Google Scholar]