Abstract
The chick limb bud has been used as a model system for studying pattern formation and tissue development for more than 50 years. However, the lineal relationships among the different cell types and the migrational boundaries of individual cells within the limb mesenchyme have not been explored. We have used a retroviral lineage analysis system to track the fate of single limb bud mesenchymal cells at different times in early limb development. We find that progenitor cells labeled at stage 19–22 can give rise to multiple cell types including clones containing cells of all five of the major lateral plate mesoderm-derived tissues (cartilage, perichondrium, tendon, muscle connective tissue, and dermis). There is a bias, however, such that clones are more likely to contain the cell types of spatially adjacent tissues such as cartilage/perichondrium and tendon/muscle connective tissue. It has been recently proposed that distinct proximodistal segments are established early in limb development; however our analysis suggests that there is not a strict barrier to cellular migration along the proximodistal axis in the early stage 19–22 limb buds. Finally, our data indicate the presence of a dorsal/ventral boundary established by stage 16 that is inhibitory to cellular mixing. This boundary is demarcated by the expression of the LIM-homeodomain factor lmx1b.
Keywords: Lineage, limb bud, lmx1b, chick, dorsal/ventral, cartilage, perichondrium, tendon, muscle connective tissue, dermis, CHAPOL
Introduction
The developing vertebrate limb bud has served as an important model system in which to study the mechanisms that control patterning and differentiation during embryogenesis. However, despite recent progress in understanding the genetic regulation of limb pattern, there are still critical holes in the description of the morphogenic events themselves. For example, while detailed fate maps have been derived for the limb using various labeling techniques including chick:quail chimeras, carbon particle labeling, and diI injection, none of the fate maps reported thus far provide cellular-level resolution. Hence the lineage relationships among different cell types in the developing limb bud and the times at which cells become committed to various tissue fates in the limb remain unclear. In particular, it is known that the mesenchymal cells at the distal tip of the early limb bud, referred to as the Progress Zone, are maintained in an undifferentiated state through the influence of factors produced by the overlying Apical Ectodermal Ridge (AER) (Globus and Vethamany-Globus, 1976) and that this undifferentiated population of cells gives rise to most of the cell types of the mature limb including cells of the dermis, perichondrium, cartilage, bone, muscle connective tissue, tendons, and ligaments. However, the Progress Zone cells could, in principle, be composed of a collection of distinct progenitors, each with a restricted potential for forming different cell types or alternatively a set of equipotential cells capable of giving rise to the same range of cell types. Moreover, the timing of when these cell fate decisions are made remains unknown. While it has been demonstrated that mesenchymal cells from limb buds as early as stage 21 are capable of differentiating into cartilage cells in primary cultures treated with appropriate differentiating agents (Ahrens et al., 1977), it is unclear when these cells, within the context of the developing limb, become restricted to a specific cellular fate.
The one lineage restriction that is well understood in the developing limb can be traced back to the dual origin of the cells that form the limb bud itself. Future muscle and endothelial cells of the limb originate in the somitic epithelia. These muscle and epithelial cell precursors migrate into the limb bud mesenchyme before differentiating and prior to receiving the pattern information directing them to form an elaborate network of muscles and vasculature respectively (Chevallier et al., 1977; Christ et al., 1977; Kardon, 1998). Most of the remaining limb bud cell types arise from the lateral plate mesoderm (Wachtler et al., 1981), a flat sheet of tissue on either side of the neural tube and somites. At the time of limb bud specification the ventral (splanchnic) and the dorsal (somatic) lateral plate mesoderm are separated from each other by the coelomic cavity. The limb of the chick is formed from an initial thickening of the somatic lateral plate mesoderm starting at developmental stage (HH) 16 (Hamburger and Hamilton, 1951). As this thickening proceeds, the lateral edges of the left and right somatopleure fold under the embryo fusing at the midline and converting the initial dorsal/ventral thickening into a medial/lateral outgrowth of limb bud tissue. It is during this outgrowth of the limb bud that the expanding population of lateral plate mesoderm derived cells is invaded by the somitic muscle and vasculature cell progenitors.
Knowing the lineage relationships among limb bud cells of the somitic and lateral plate populations is critical for understanding the mechanism by which specific cell fates are achieved in the limb. For example, a recent retroviral lineage tracing of the somitic population of limb progenitors revealed that myogenic and endothelial cells are derived from a common somitic precursor and are not specified until after they have reached their final destination within the limb bud (Kardon et al., 2002). Moreover, the future myogenic cells are not committed to forming slow or fast muscle fibers prior to entering the limb. Thus local extrinsic limb signals are responsible for determining muscle versus endothelial cell fate and fast versus slow fiber type. As indicated above, equivalent lineage information is currently lacking for the population of tissues in the limb bud derived from the lateral plate mesoderm.
Lineage analysis of the somite-derived cell population in the limb also revealed that myogenic precursors in the somites are not committed to forming particular anatomical muscles or muscles within specific proximal-distal or dorsal-ventral limb regions. Previous diI based fate mapping studies in the limb have indicated that small populations of labeled lateral plate-derived cells in the Progress Zone are also capable of spreading along the proximo-distal axis of the limb (Vargesson et al., 1997). Although in other experiments, injection of lipophilic dyes to has suggested that stage HH19 cells may not mix between future zeugopod and stylopod segments, even though they distribute quite widely within each limb segment (Dudley et al., 2002), whereas the future zeugopod/autopod boundary may not be precisely defined at this early stage (Sato et al., 2007). However, as in considering lineage restrictions, single cell resolution is required since labeling a population of cells could overestimate the amount of expansion that the descendants of any single cell in the early limb bud are capable of attaining and, conversely, following the spatial fate of a population of labeled cells could underestimate the range the descendants of individual cells can achieve by focusing on the bulk of labeled tissue. This issue is particularly important as it has implications for current models of proximodistal patterning.
The progress zone model of proximodistal patterning maintains that the cells of the distal sub-Apical Ectodermal Ridge (AER), mesenchyme (the Progress Zone), are continuously reprogrammed to a more distal patterning fate as long as they remain under the influence of the AER (Summerbell et al., 1973). As soon as cells exit the Progress Zone, due to displacement from outgrowth of the limb, they cease their progression and maintain their specific proximodistal identity. The proximodistal patterning of the limb is continued in this manner until the entire extent of proximodistal limb pattern is established. In principle, the Progress Zone model would accommodate a fate map where an individual cell labeled within the distal tip of the limb bud produced progeny cells in multiple proximodistal segments of the limb, if there were considerable cell mixing within the Progress Zone. This model could also fit with a fate map where all the descendants of any individual Progress Zone cell always ended up within the same proximodistal segment, if the cells maintained a strict spatial relationship with their neighbors within the Progress Zone.
In contrast, an alternative model postulates that the proximodistal patterning of the limb is specified very early (as opposed to progressively), followed by expansion of the patterned primordium (Dudley et al., 2002; Sun et al., 2002). In this scenario a single cell labeled in the Progress Zone would produce progeny confined to a single proximodistal segment.
The patterning of the dorsoventral limb axis is better understood. The secreted inductive signal Wnt7a, which is specifically expressed in the dorsal limb bud ectoderm, acts upstream of the LIM-homobox gene Lmx1b in the underlying mesenchyme. Lmx1b, in turn, has been shown to be necessary and sufficient for specifying dorsal pattern in the distal limb bud (Riddle et al., 1995; Vogel et al., 1995; Dreyer et al., 1998; Chen et al., 1998). The striking expression of Lmx1b in the dorsal half of the limb bud from the earliest stages of its formation (Riddle et al. 1995; Vogel et al., 1995) raises the intriguing possibility that Lmx1b expression establishes a dorsoventral compartment boundary through the limb mesenchyme, across which cells are unable to migrate. Such a mesenchymal compartment would be particularly notable as most of the previously described compartments in the vertebrate embryo are within epithelia. Indeed, in the context of the limb bud, it has been demonstrated that the ectodermal cells remain tightly confined into either dorsal, ventral or AER compartments (Michaud et al., 1997; Kimmel et al., 2000; Guo et al., 2003). However, arguing against the idea that Lmx1b specifies a distinct compartment, a previous study (Altabef et al., 1997) has suggested that there are no dorsoventral compartments in the mesodermal cells of the early limb field. This conclusion, however, was drawn from diI labeling where populations of cells, rather than individual clones, were followed. Thus, while the authors observed a low level of “mixing” when cells were labeled far from the dorsoventral border and extensive mixing distally when cells were labeled near the dorsoventral border, this could in principle reflect the simultaneous labeling of multiple limb progenitor cells with different dorsoventral fates by the diI method. Thus, once again, this issue needs to be reexamined with single cell resolution.
To address the issues of multipotency within the Progress Zone, the timing of cellular commitment to a specific tissue type, and the limitations of cellular movements along the proximodistal and dorsoventral limb axis, we have carried out a lineage analysis using a library of retroviral vectors to trace the cellular fate and location of the progeny of individually marked single cells within the context of the developing limb bud.
Results and Discussion: Marking Individual Progenitor Cells within the Limb Bud
We followed the fate of clonally related cells using the CHAPOL retroviral library (Golden et al., 1995), which consists of a replication incompetent retrovirus carrying both a histochemical tag (human PLacental Alkaline Phosphatase, PLAP) and a degenerate 24bp oligonucleotide insert (Figure 1). PLAP activity marks retrovirally infected cells which are individually dissected from labeled, cryosectioned tissue and used as a substrate for PCR amplification and subsequent sequencing. Because the retroviral genome is irreversibly integrated into the genome of the infected cell, each insert indelibly labels all clonal descendants of the initially infected cell and because the retrovirus is replication incompetent it can not spread to non-descendents. The degenerate oligonucleotide insert in our CHAPOL library has a complexity of over 2×106 unique sequence tags, allowing us to confidently classify groups of PLAP expressing cells containing identical sequence tags as being clonally related descendants from a primarily infected single cell.
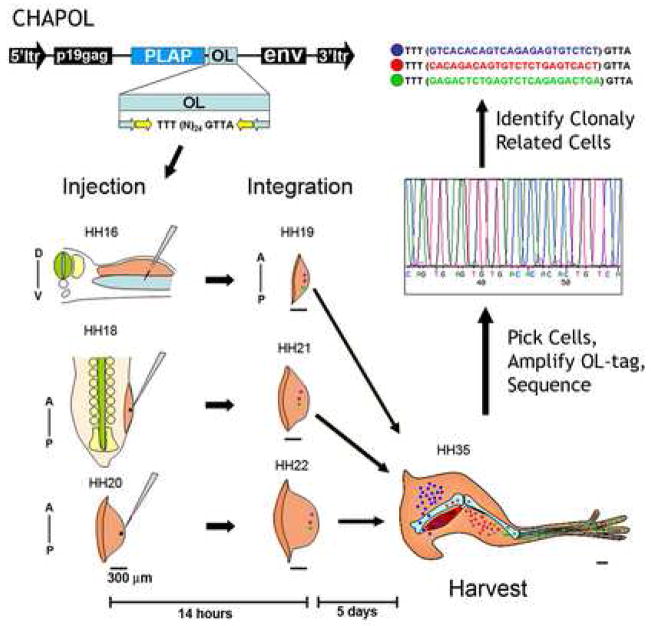
Figure 1.
CHAPOL is a replication incompetent retrovirus which carries the human PLacental Alkaline Phosphatase (PLAP), histochemical tag, allowing identification of cells that have incorporated the viral genome into their own. Additionally the viral genome contains a degenerate oligonucleotide that is flanked by nested PCR primer sequences (light blue and yellow arrows). Thus, each transduced cell contains a unique oligonucleotide tag that is incorporated into its genome and can be easily amplified by PCR. For our experiments chick embryos were injected with CHAPOL retrovirus at three stages of embryonic development (HH16, HH18, and HH20; Hamburger and Hamilton 1951). HH16 injections will integrate into the host genome and label cells by HH19. HH18 injections integrate and label cells by HH21 and HH20 injections label cells by HH22. The time difference between the earliest injected embryos (HH16) and the latest injected embryos (HH20) was 18 hours of incubation. All labeled tissues were further incubated for 5 days (to HH35) prior to harvest and analysis. At HH16 the CHAPOL retrovirus was injected into the coelomic space between the somatic and splanchnic lateral plate mesodermal layers. At HH18 and HH20 CHAPOL virus was injected into the distal most tip of the emerging limb bud to label the undifferentiated Progress Zone cells and to avoid labeling migrating somitic endothelial and myoblast cells. In the schematic blue, red, and green dots represent uniquely transduced single cells which subsequently undergo clonal expansion until the tissue is harvested. After harvesting, PLAP positive cells were manually dissected, incubated in lysis buffer, and the integrated oligonucleotide amplified using nested PCR, and sequenced. Clonal populations of cells were identified based on their sharing a sequence tag. Black bars under the schematic limbs indicate a reference measure of 300μm. A–P and D–V indicate the anterior-posterior and dorsal-ventral axes respectively.
To analyze the lineage relationship among the lateral plate mesoderm derived limb bud cells, presumptive hind limbs were infected with the CHAPOL retroviral library at stages HH16, HH18 and HH20. At HH16, the virus was injected into the coelomic cavity underneath the limb-forming lateral plate mesoderm on the right side of the embryos (Figure 1). At later stages of limb bud outgrowth, the virus was injected into the distal-most tip of the right hind limb bud to preferentially label the undifferentiated Progress Zone cells and to avoid infecting the vasculature and somitic myoblasts migrating into the limbs. It is important to realize that the different injection sites used for HH16 versus HH18–20 injected embryos result in different proximal-distal labeling of the mature tissues. The coelomic (HH16) injections label cells in all limb segments while the distal tip (HH18, HH20) injections result in progressively more distally labeled cells. However, our primary goal in this analysis is to label and trace the least differentiated tissue at the given stage of injection which corresponds to the entire lateral plate mesoderm at HH16, and the distal tip of the HH18-HH20 limbs. Infected embryos were further incubated for 5 days and harvested at stage HH35 for analysis (Figure 1). By this time, all five of the major lateral plate mesoderm derived cell types in the limb have differentiated into mature cells expressing markers characteristic of their particular cell type. Cartilage cells were identified by Collagen II staining and by morphology. Perichondrial cells were identified by their proximity to the cartilage and by morphology. Tendon cells were identified by Scleraxis staining (Schweitzer et al., 2001), and Myosin Heavy Chain protein staining marked the area around muscle connective tissue (Bader et al., 1982). Dermal cells were identified by their location and morphology (Figure 2).
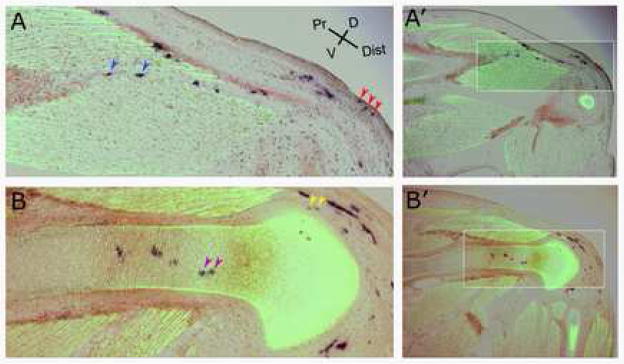
Figure 2.
Clonal descendents of a single cell labeled by the CHAPOL retrovirus by injection at stage 16 can contribute to all multiple tissue types. Panels (A) and (B) are images taken at the level of the thigh to shank junction from distinct sections through a CHAPOL infected limb. Tissues are stained for the indicated markers and are oriented with the proximal limb to the left and dorsal toward the top of the panel. The clone depicted contains cells of all five tissue types four of which are shown here (remaining images of this clone are in Supplemental figure 1). (A) A more superficial view illustrating PLAP positive Dermal, Tendon, and Muscle connective tissue cells. (B) Deeper tissues illustrating PLAP positive Cartilage and Perichondrium. Condensed cartilage is visualized by an antibody staining for Collagen II and muscle cells are stained with an antibody to Myosin Heavy Chain (both in Green and distinguished by morphology). Tendon cells are visualized by in situ hybridization for Scleraxis (brown). Cells descended from those cells primarily infected with the CHAPOL virus are visualized by staining for PLAP (dark purple). The arrowheads mark the cells determined to be clonally related by virtue of their carrying identical sequence tags. The indicated clone contained cells in dermis (red arrows), muscle connective tissue (blue arrows), cartilage (purple arrows), perichondrium (yellow arrows), and tendon (Supplemental figure 1). A′ and B′ panels are lower magnifications of A and B for orientation.
Our retroviral library only infects dividing cells. Since in principle progenitors of specific cell types might not be cycling at a given stage, it was important to establish that we were indeed capable of infecting all cell types in the limb. To test for any inherent bias in the frequency at which different cell types can be infected, or bias in cell types which can be successfully used for PCR and sequencing we injected limb buds with the CHAPOL retroviral library at stage 22, and picked cells at stage 35 (the same stage assayed in the experiments described below). Approximately 100 cells were picked from each of the five different tissues types and processed by PCR amplification and sequencing. Good amplification and sequence was obtained for 46/100 (46%) muscle connective tissue cells, 48/98 (49%) tendon cells, 44/81 (54%) dermis cells, 26/66 (40%) perichondrial cells and 43/100 (43%) cartilage cells. Thus, there is no appreciable cell-type bias inherent in our methodology or approach.
The primary data from our lineage analysis are summarized in Tables 1 and 2. From the stage HH16 injections, 1636 cells were picked and sequence was amplified from 996 cells (61%). A usable single sequence was found in 713 cells (44%), of which 377 cells (23%) were grouped into 62 multi-cell clones. In the remaining 336 cells, the sequences were identified and were all unique to that single dissection (Table 1). These rates of amplification are comparable to previous studies using the CHAPOL retroviral library (Golden and Cepko, 1996; Kardon et al., 2002).
Table 1.
Primary lineage analysis data.
Summary of CHAPOL lineage analysis in chick hind limbs injected at stages HH16, HH18, and HH20 and harvested after 5 days of incubation. Percentages in parentheses represent the percentage relative to the total number of cells picked at that stage. Numbers presented for 3–5 tissue types clones are formatted as [number of clones: smallest clone-largest clone (average clone size)].
| HH | Total Cells Picked |
PCR Amplified (%) |
Good Sequence (%) |
Multi-Cell Clones |
No. Cells in Clones |
Avg. Clone Size |
1 Tissue Type |
2 Tissue Types |
3 Tissue Types |
4 Tissue Types |
5 Tissue Types |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 1636 | 996 (61%) | 713 (44%) | 62 | 377 (23%) | 6.1 | 20 | 28 | 6: 7-26 (13) | 6: 6-33 (21) | 2:13-25 (19) |
| 18 | 1245 | 723 (58%) | 527 (42%) | 73 | 322 (28%) | 4.4 | 39 | 25 | 7: 6-34 (13) | 2: 10-14 (12) | 0 |
| 20 | 549 | 361 (69%) | 208 (38%) | 29 | 78 (14%) | 2.7 | 22 | 7 | 0 | 0 | 0 |
Table 2.
Tissue composition of CHAPOL Clones.
Lineage analysis data of one and two tissue type clones. (A) Each of the five tissue types is listed at the top of a column with the stage of injection listed at the left. Values indicate the number of multi-cell clones that were restricted to the indicated tissue type at the indicated stage of injection. (B) Each of the ten possible two tissue-type combinations is represented by the intersection of a row and column. Values in each intersection indicate the number of clones containing exactly two tissue-types that are composed of the indicated tissue types from HH16 (top), HH18 (middle), and HH20 (bottom), injected embryos. The numbers listed in A and B are formatted as [number of clones in category: smallest clone-largest clone (average clone size)].
| A | |||||
|---|---|---|---|---|---|
| Inj. Stage (HH) | Size:Range (Avg.) Cartilage | Perichondrium | Tendon | Muscle Connective Tissue | Dermis |
| 16 | 10: 2-11(4) | 1:2(2) | 5:2-3(2) | 0 | 4:2-7(4) |
| 18 | 19: 2-5(2) | 7:2-4(3) | 4:2-4(4) | 8:2-7(4) | 1:2(2) |
| 20 | 13: 2-4(2) | 1:2(2) | 5:2(3) | 1:2(2) | 2:2(2) |
| B | ||||
|---|---|---|---|---|
| PC | Tendon | MCT | Dermis | |
| 16 | 7:2-24(7) | 3:7-12(10) | 2:2-6(4) | 1:3(3) |
| Cartilage 18 | 5:2-5(3) | 1:3(3) | 0 | 0 |
| 20 | 5:2-6(4) | 0 | 0 | 0 |
|
| ||||
| 16 | 1:10(10) | 0 | 0 | |
| PC 18 | 4:2-9(5) | 2:5-8(7) | 1:2(2) | |
| 20 | 1:2(2) | 0 | 0 | |
|
|
||||
| 16 | 11:3-20(11) | 3:5-11(8) | ||
| Tendon 18 | 8:3-12(6) | 0 | ||
| 20 | 1:10(10) | 0 | ||
|
|
||||
| 16 | 0 | |||
| MCT 18 | 4:3-4(4) | |||
| 20 | 0 | |||
|
|
||||
From the stage HH18 injected embryos, 1245 cells were analyzed and sequence was amplified for 723 cells (58%). Usable single sequences were found in 527 cells (42%), again giving rates comparable to previous studies. At this stage, 322 cells (28%) were placed into 73 multi-cell clones (Table 1).
Because of the decreased number of infected cells per injected embryo in stage HH20 injections, 549 cells were picked of which 361 (69%) gave amplified sequence, and 208 (38%) had a usable single sequence. Seventy-eight (14%) of these cells were placed into 29 multi-cell clones (Table 1).
The average clone size decreased from 6.1 cells for HH16 injections to 4.4 cells for HH18 injections to 2.7 cells for HH20 injections.
In interpreting these results, it is critical to bear in mind that type-C retroviruses (like the avian leucosis virus-based vectors used here) do not integrate into the host chromosome until the infected cell divides and the nuclear envelope breaks down. Moreover, integration occurs behind the DNA replication fork with the consequence that only one daughter cell carries the retroviral DNA. Thus one does not actually tag all the descendants of the first cell infected; but rather all the descendants of one daughter of the cell initially infected. We have empirically determined how quickly this takes place during limb development. Our findings indicate a 14 hour time lag from the time of injection of the virus to fully marking early limb cells with an integrated provirus. For the current studies, this means, for example, that injecting our retroviral library at stage 16 is actually testing the fate determination of limb bud mesenchymal cells at stage 19 (Figure 1).
Lineage relationship among cell types
The first question we addressed was that of the lineage relationship among cartilage, perichondrium, tendon, muscle connective tissue, and dermis, specifically asking whether individually labeled progenitors ever became restricted to a single cell type and, if so, at what stage this occurred. Absolute lineage restrictions among the five cell types could not be found at any of the three stages analyzed. Two clones containing cells of all five tissue types were found in the stage 16 injected limbs (Table 1; Figure 2; Supplemental Figure 1), indicating that they resulted from the infection of completely uncommitted progenitor cells at HH16. We also saw six clones containing cells of four tissue types. There, in fact, were more clones containing multiple cell types at this stage than there were clones restricted to a single cell type. Furthermore, clone size corresponded closely to the number of cell types in a clone. Clones of just one cell type contained an average of 4.8 cells, whereas those with two cell types averaged 8.7 cells, and those with three and four cell types averaged 21 and 19 cells, respectively (Table 1). Thus, limb bud mesenchymal cells at HH19 (injected at HH16) are not specified to any single lineage and at least a subset of progenitor cells is capable of generating all of the five mature tissue types.
While we no longer observed clones spanning all five tissues in limbs injected at HH18, the clones were smaller and two clones contained cells of four tissue types (Table 1, Figure 3A). Again, larger clones generally consisted of more cell types. The average size of a clone with only one cell type was 2.5 cells and for two cell types was 4.7 cells, whereas for clones with three or four cell types it was 13 and 12 cells, respectively. We observed many more clones (52) containing only one cell type than we saw that contained more than one cell type (34). Thus, it seems that at stage 21 (when cells injected at HH18 are labeled), progenitors have less of a chance to become all possible cell types but are still fairly plastic as to their tissue type commitments.
Figure 3.
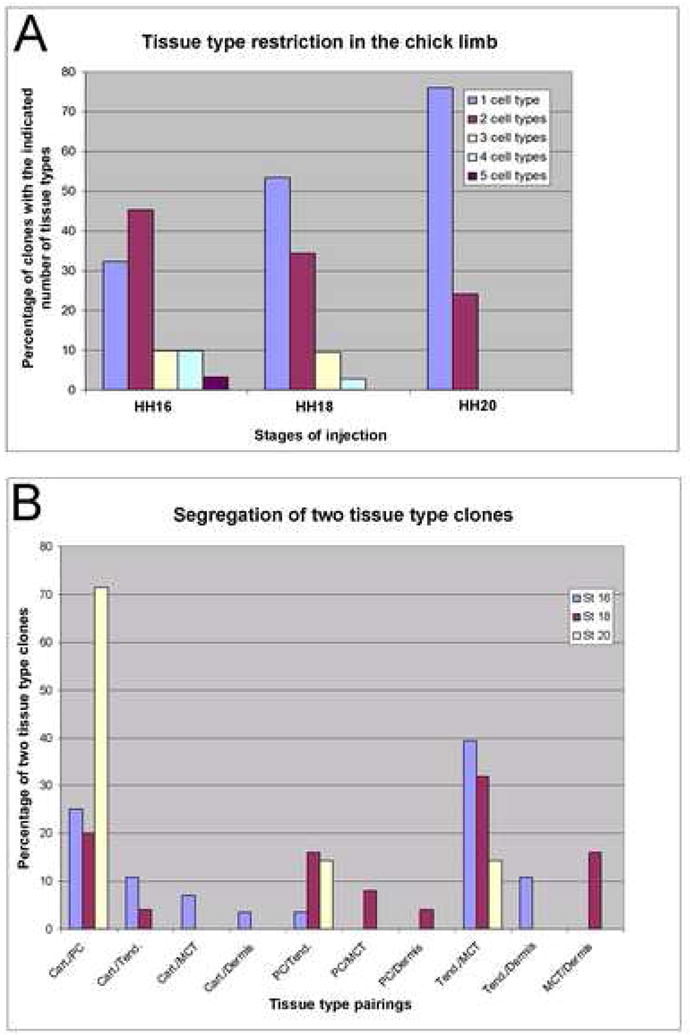
Graphical analysis of CHAPOL lineage data. (A) Graph of the clonal complexity of the progeny of a single labeled cell at each stage. Bars represent the percentage of all clones at the indicated stage that contain the indicated number of tissue types. (B) Graph of two tissue type clone segregation. Bars represent the percentage of all clones containing exactly two tissue types from embryos injected at the indicated stage represented by the indicated tissue combination.
In the limbs injected at stage HH20, more than two-thirds of the sequence tags identified were unique to a single cell pick and most of the multi-cell clones contributed to only a single tissue type, with only 7 of 35 clones containing two cell types (Table 1, Figure 3A). Clones containing only one tissue type averaged 2.2 cells while clones containing two tissue types averaged 4.6 cells, almost the same average pick numbers for these classes as in the stage HH18 injected limbs.
In the data set as a whole we can identify clones containing every possible pairing of tissue type. While we see a progressive decrease in clonal complexity in the progeny of cells labeled from HH19 to HH22 (Figure 3A, Table 1), the lower clonal complexity is accompanied by a correspondingly dramatic decrease in clone size making it difficult to discern whether the reduction of complexity is due to an actual restriction of cellular potency or simply a bias due to sampling errors of smaller clone sizes. Nonetheless, since the developmental time between injection stages (HH16-HH20, 18 hours) is very short compared to the amount of time of incubation after injection (5 days for all stages of injections), it is striking that there is such a dramatic decrease in average clone size resulting from single cells labeled at these stages. This could imply that the rate of proliferation of the lateral plate mesoderm derived cells may be considerably slower at later stages of limb development, perhaps correlating with progressive differentiation.
It should be noted that, as in previous studies using retroviral libraries for lineage analysis, there is a less than 50% success rate in amplifying and sequencing the tag from the dissected PLAP-labeled cells. As a consequence, the complexity of each clone can only give a lower bound of the range of cell types in a clone and at times will under-represent the number of cell types actually descended from a given progenitor. If we had, in fact, observed highly restricted clones at some stage of limb development we would have needed to utilize statistical methods to confirm that our inability to identify clonally related cells in different tissues was not due to sampling errors. Our data, however, show no restriction in spite of this technical limitation. Indeed some of our 2-, 3- and 4-tissue type clones would certainly have been scored as 3-, 4- and 5-tissue type clones if all tags were identified in our analysis, which would only reinforce our conclusions in this regard. In fact, our statistical simulation analyzing of the probability of losing tissue type representation indicated that about 47% of 5 tissue type clones would be mis-identified as 4 tissue type clones while about 5% would be mis-identified as 3 tissue type clones giving us a loss of perfect representation in 52% of actual five tissue type clones (Supplemental text 1). However since we only identified 2 five tissue type clones, even if 66% of actual 5 tissue type clones were not identified (4 out of 6 actual clones lost), our analysis still indicates that a majority of our primarily infected cells did not give rise to clones containing all five tissue types.
Developmental relationship of limb tissue types
While we observed no strict restriction of tissue types, our observations imply that some tissue types are more closely related than others developmentally. This is more obvious if we focus on only the clones containing two tissue types. If progenitors are indeed multipotent, and their ultimate fate is random, then a progenitor cell undergoing a terminal commitment will be as likely to produce (for example) a cartilage/tendon two tissue type clone as it is to form a cartilage/dermis clone. Thus, since there are 10 possible combinations of two tissue type clones, we would then expect each combination to show up in about 10% of the clones. In contrast, we actually observe that three of the two tissue type pairings make up 72% of all two tissue type clones (averaged between the 3 stages). Specifically, we see cartilage/perichondrium in 28%, tendon/muscle connective tissue in 33%, and perichondrium/tendon in 10% of all two tissue type clones (Figure 3B, Table 2B). Two pairings, cartilage/dermis and perichondrium/dermis were dramatically under-represented relative to other pairings, each case being represented by only one small clone (3 cells and 2 cells, respectively; Table 2B). Thus, in our two tissue type clones containing cartilage cells as one of the tissues, 71% had perichondrium, 17% had tendon, 8% had muscle connective tissue, and 4% of clones had dermis as their partner tissue type. Likewise, in our two tissue type clones containing muscle connective tissue as one of the cell types, we saw 71% containing tendon, 14% containing dermis, 7% containing perichondrium, and 7% containing cartilage as the second cell type. Importantly, our analysis identified a similar total number of two tissue type clones containing cartilage, perichondrium, muscle connective tissue, and tendon (24, 26, 28 and 33, respectively). Yet 71% of the cartilage-containing two tissue type clones contain perichondrium as their partner and only 8% contain muscle connective tissue, while 61% of the tendon-containing two tissue type clones contain muscle connective tissue as their partner and only 12% contain cartilage.
Interestingly, our observations indicate that the degree of relationship between two limb tissues, as indicated by an increased representation in the two tissue type clone group, is perfectly correlated with the ultimate spatial distribution of those tissues. That is, a cartilage cell is more closely related to a perichondrium cell than it is to a tendon, muscle connective tissue, or dermal cell; and a dermal cell is more closely related to a muscle connective tissue cell than it is to a tendon, perichondrium or cartilage cell. This implies that tissue specific differentiation occurs with regard to the spatial distribution of plastic limb mesenchymal tissue. Perhaps the most likely explanation is that there is little mixing of the progenitor population between those closer to the surface and those closer to the core of the expanding limb bud. As a consequence, related cells see similar, albeit not always identical, extracellular environments and intracellular signals and hence differentiate into tissue types that are the same or adjacent to one another in the mature limb. It is important to emphasize that our lineage analysis can only address the fate that labeled cells will adopt under normal in vivo conditions without perturbation, not what fates they could possibly adopt if transferred to a different spatial environment. Studies of recombinant limbs have demonstrated that limb bud mesenchyme that is removed disaggregated and repacked into a donor ectodermal hull is still competent to recapitulate normal patterning and differentiation even though there is little cellular re-organization (Ros et al., 1994; Wada et al., 1998).
Although we cannot exclude the possibility that there are subsets of progenitors committed to a single tissue, the most parsimonious interpretation of our data is that the cells within the Progress Zone have not yet received signals directing them along any particular cell differentiation pathway and are multipotent.
Proximodistal lineage restriction
Of equal importance to determining whether clones in the limb display cell type restriction is determining whether clones are restricted spatially in the limb. As discussed above, this question is particularly important to answer along the proximodistal axis as the outcome could potentially affect the way we view various models for proximodistal patterning. We therefore analyzed the cells of each clone in terms of location of the limb girdle, stylopod, zeugopod, or autopod of the leg. In the stage HH16 injections, most clones localize to a single segment, but 9 of 80 extend between the stylopod/zeugopod (thigh/shank) segments and 12 of 80 clones spanned the zeugopod/autopod (shank/foot) segments. There was a size difference between clones located in a single segment and clones spanning two segments. Single segment clones averaged 7 cells, whereas two segment clones averaged 11 cells. No clone spanned all three segments. For stage 18 injections, three of the 86 clones spanned two segments. At this stage single segment clones averaged 4.1 cells and two segments clones average 5 cells, so there is no great size difference between these categories of clones, and one 34 cell clone remained contained entirely within the stylopod. At stage HH20 injections, only one of 35 clones spanned two segments. Recalling that, due to the time required for integration and cell division, injections done at stage HH16 label clones at stage HH19, and injections at stage HH18 label clones at stage HH21, this suggests that at least a significant percentage of the progenitor cells in the early limb bud are not restricted to a particular proximodistal cell fate.
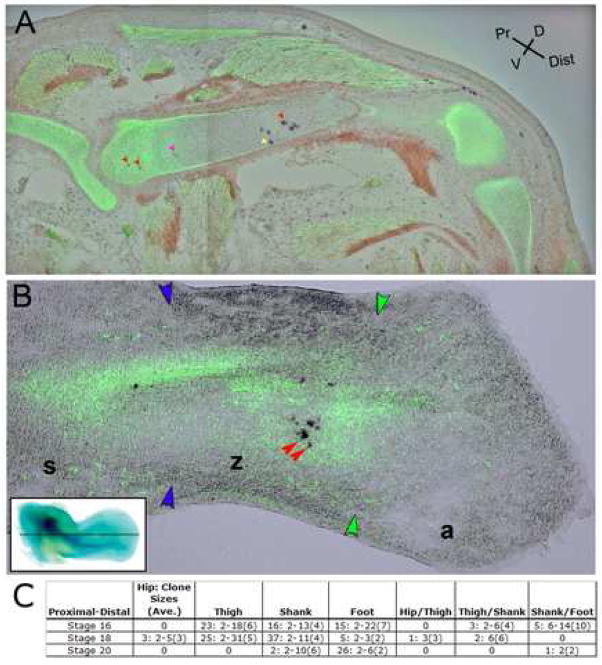
Quite surprisingly, however, when we evaluated the proximodistal fate of cells in cartilage separately, we found that no clones extended beyond a single cartilage element. None of the 44 clones containing only cartilage cells contained cells in multiple segments. Likewise, all of the cartilage cells in multi-cell type clones were confined to one proximodistal segment, although other members of the same clones often were found in different limb segments in the soft tissue, indicating a greater proximodistal restriction on cartilage limb tissue than on surrounding tissues. Additionally, cells in cartilage clones were, in some cases quite widely dispersed, separated by 66% or more of the length of the cartilage element in which they reside, without having clonal cohorts in adjacent elements (Figure 4A; Supplemental Figure 2). Taken at face value, this would seem to indicate that cells in the early limb bud are already restricted to form skeletal elements of a particular segment, although their descendents that differentiate into other cell types can traverse between the forming segments. An alternative explanation, however, would be that the cartilage condensations form before the cells have had a chance to migrate widely along the proximodistal axis. And once a condensation does form, the cells are simply trapped within the deposited matrix preventing further migration. Thus, our clonal populations may appear restricted to individual segments without having true proximodistal positional information at this stage.
Figure 4.
Cartilage clones marked by CHAPOL in embryos injected at stage HH16. (A) Clones from embryos harvested at stage HH35 spread across large portions of proximodistal elements. Clonally related cells are marked by the same color arrow. Cells without arrows were picked but remain unidentified due to the technical attrition discussed in the text. The red arrows demonstrate a clonally related cell population which spreads over half the femur but never crosses the proximodistal segment boundaries. Non-clonally related PLAP positive cells were identified in all segments of this embryo (Supplemental figure 2). (B) Clonally related cells from embryos injected at stage HH16 and harvested prior to cartilage segmentation (HH26), show insignificant spread along the proximodistal axis of the condensing cartilage elements. Inset in (B) demonstrates the plane of section shown in (B) on a HH26 whole mount chick hind limb stained with Alcian Blue to reveal branching cartilage condensations. The presumptive femur is the single branch in the proximal limb while the distal bifurcated cartilage marks the presumptive radius and ulna. Digital cartilage condensations have not yet formed at this stage. Boundaries between stylopod (s), zeugopod (z), and autopod (a) are demarcated by blue and green arrowheads.
This issue can be resolved because the skeletal condensations do not initially form as separate, discrete units isolating cells from one another. Rather they start as a single, continuous branching element (Shubin and Alberch, 1986). As such, at stage HH26 there is no more of a barrier between the tibia and the femur than there is between different regions of the femur. Moreover, by this stage there has not been time for cell division after cartilage condensation, hence these early clones represent proliferation prior to condensation. If, at this early stage of analysis, clones are dispersed across large portions of a single cartilage element, but do not cross between cartilage elements, it would provide strong evidence for early proximodistal specification in cartilage progenitors. However, if clones analyzed at stage HH26 are not widely spaced within any given cartilage element, it would imply that the absence clonal cohorts in multiple proximodistal cartilage segments has the more trivial explanation of a single progenitor cell being trapped in the extra-cellular matrix of a discrete element and it’s spread along the long bones is likely the result of post-condensation interstitial growth.
We stained limbs that were injected at HH16 and harvested at HH26 with Collagen II antibodies to mark the cartilage and by in situ hybridization for HoxA11 which marks the zeugopod allowing us to identify the segment in which a cell lies. We picked 175 cartilage cells, 78 of which (44%) gave good sequence information. This is on the same order as the number of cartilage picks that we analyzed in the stage 18 and stage 20 injections. Sixteen other embryos were processed and then excluded from further analysis because they had only a small number of PLAP labeled cartilage cells lying too close together to provide significant information. Of the informative picks, 42 could be arranged into fifteen multi-cell cartilage clones, giving an overall average of 2.8 cells per clone, suggesting that not many cell divisions occurred between labeling and harvesting. The largest clone contained six cartilage cells. In contrast to the pattern seen when limbs injected at the same stage were analyzed at HH35, at HH26 none of the clones exhibited significant proximodistal spread within the cartilage elements (Figure 4B). The cells in a clone tended to lie very close to one another, spanning only a small fraction of a segment, not the half to two-thirds of a segment seen in many later cartilage clones. This indicates that there is not much cell mixing along the proximodistal axis prior to the time of cartilage condensation. Since, without significant cell mixing, cells do not have the ability to switch segments, it is impossible to determine by this method whether there is a molecular restriction of proximodistal fate during early limb development in the cells fated to form cartilage. However, the soft tissue clones are not restricted to particular limb segments suggesting that, in all likelihood, there are no strict proximodistal boundaries at the stages we examined. Thus the data argue against the early specification model for proximodistal limb patterning. Similar conclusions have recently been obtained by others studying mouse and chick limb proximo-diatal patterning (Arques et al., 2007; Sato et al., 2007).
In previous experiments from our lab, analyzing the positional fate of cells in the distal limb mesenchyme using diI to mark small cell populations, we focused on labeled populations incorporated into skeletal elements and found that the bulk of labeled cells were restricted to a single proximodistal segment (Dudley et al., 2002). Results from these current studies indicate that this result was likely the result of technical limitations resulting from the labeling of cartilage progenitors that underwent minimal migration prior to segmentation.
Dorsal ventral compartmentalization of the limb mesoderm
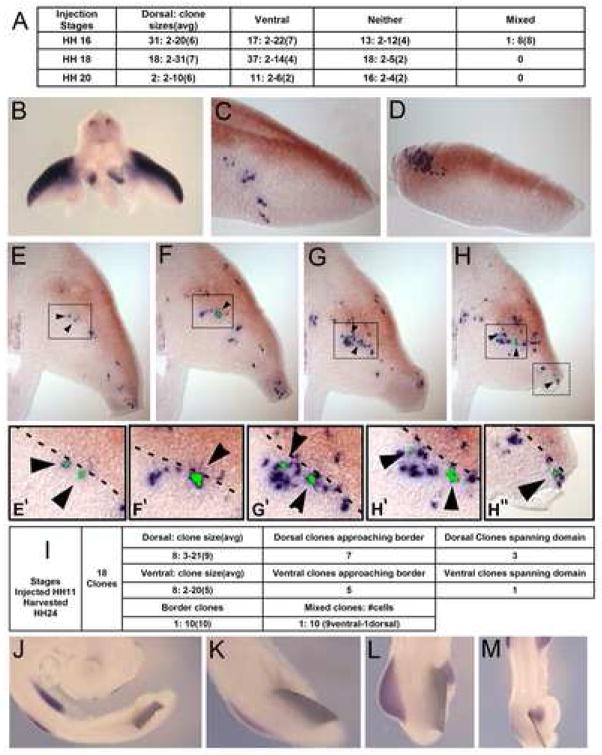
While we saw no evidence of strict cellular compartmentalization along the proximal distal axis at the stages analyzed, we did observe a strong tendency for clonally related cells to segregate into either the dorsal or ventral regions of the mature limbs. Out of a total of 164 identified clonal cell populations from injections at stages HH16, HH18, and HH20, only one contained a single cell that crossed the dorsal-ventral boundary based on using the skeletal elements as the dorsoventral midline. Of the remaining clonal populations, 116 were completely restricted to either the dorsal or ventral regions of the limb (Figure 5). However, at this stage of limb development, it is impossible to define a clear dorsal ventral boundary within the condensed cartilage and developing perichondrium. Because of this, clones that contained cartilage or perichondrium cells could not be classified as either dorsally or ventrally restricted. That is, in an otherwise dorsally restricted clone, cartilage cells belonging to that clone could be found at the ventral-most extent of the cartilage tissue and vice versa for otherwise ventrally-restricted clones. Despite the ambiguity in assignment of cells in the cartilage and perichondrium to either a dorsal or ventral identity, this observation that clonal descendants of single cells infected at stages 16–20 have a strong predilection toward dorsal or ventral restriction suggested to us that earlier in development there may exist a boundary in the limb bud mesenchyme that is restrictive to cellular mixing.
Figure 5.
A dorsal/ventral boundary to cellular migration in the limb bud mesenchyme. (A) Summary of dorsal/ventral CHAPOL analysis of chicks harvested at HH35. Stages of embryo injections are listed on the left. Clones are classified as dorsally restricted (Dorsal), Ventrally restricted (Ventral), Unassignable due to the presence of cartilage and/or perichondrium cells in clone (Neither), or as boundary crossing (Mixed). Numbers presented are formatted as [number of clones: smallest clone-largest clone (average clone size)]. (B) Whole mount in situ hybridization for Lmx1b, shown by blue staining in cross-section at limb bud level, demonstrates the dorsally restricted expression domain. (C, D) Embryos injected with CHAPOL library at stage HH11 (labeling lateral plate mesoderm at stage HH16) produced clonally related cell populations predominantly localized to either the dorsal (Lmx1b positive), or ventral (Lmx1b negative) domains. Examples examples shown are clusters of PLAP positive cells containing ventrally restricted (C) and dorsally restricted (D) clones that span their respective domains but fail to cross the lmx1b demarcated boundary. Lmx1b expression is in brown to distinguish from retroviral PLAP staining. (E–H) show a separate clone that extends along the proximal distal axis of the limb skirting the edge of the Lmx1b demarcated D/V boundary but remaining firmly in the ventral Lmx1b negative domain. Clonally related cells are labeled with black arrowheads and green pseudocoloring of the dissected regions. (E′–H″) show magnified views of the boxes in E–H. The D/V boundary is illustrated with a dashed line. (I) Summary of dorsal/ventral CHAPOL data analysis of chick embryos injected at HH11 and harvested at HH24. (J–M) Tantalum foil barriers implanted between HH15 somites and lateral plate mesoderm block lmx1b induction. Embryos shown were surgically manipulated to either completely block the limb field (J,K) or partially block (L,M) the midline originating signals specifying the dorsal right hind limb mesenchyme. Partial blockages allow induction of lmx1b only in the unblocked region of the limb bud. All embryos in J–M were hybridized with probes detecting lmx1b transcript.
The LIM-homeobox gene Lmx1b marks a dorsal domain in the mesenchyme of the early limb bud in both chick (Figure 5B) and mouse and it has been demonstrated that Lmx1b expression is both necessary (Dreyer et al., 1998; Chen et al., 1998) and sufficient (Riddle et al., 1995; Vogel et al., 1995) to direct dorsal pattern formation. Because lmx1b is expressed in the dorsal mesenchyme from the earliest stage of limb development, it allows us to more accurately mark the dorsoventral location of cells within the early limb bud and to test whether an observed dorsoventral restriction obeys this molecular boundary.
We repeated the lineage analysis injecting at a stage HH11 which would generate two cell clones in the budding lateral plate mesoderm just as Lmx1b expression is first seen (early HH16). We harvested these samples at stage HH24 and analyzed the expansion of clonal populations relative to the Lmx1b expression boundary.
We identified a total of 18 clones in our analysis, 16 of which were restricted to either the dorsal Lmx1b positive compartment or to the ventral Lmx1b negative compartment (Figure 5I). One of the clones was classified as a border clone, all of its cells being right at the dorsal/ventral boundary. Finally, one clone was predominantly ventral, but contained a single cell that was unambiguously in the Lmx1b positive dorsal domain. Thus, as in the original analysis, we observe a strong predilection toward restriction along the D/V axis, in this case with the Lmx1b expression domain marking the boundary. These observations could arise from three possible scenarios. The first is that the limb bud mesenchymal cells are actually restricted in their movements along the dorsal ventral axis during limb outgrowth, second we may see an apparent restriction due to a lack of dorsal to ventral spread of clonally related cells, and third we may have misidentified unrestricted clones as being restricted because of a combination of attrition during processing of the picked cells and of small clone sizes. The second possibility is unlikely since among the 16 restricted clones 12 (7 dorsal and 5 ventral) were large clones that approached the dorsal/ventral boundary, but failed to cross and 4 of the clones (3 dorsal and 1 ventral), spanned the entire domain in which they were restricted, still failing to cross (Figure 5C–H). Finally, it is unlikely that the observed restriction is due to a sampling error resulting from attrition and small clone sizes since many of the clones in this analysis were quite large compared to the original lineage analysis at stage HH35 (averaging 10 and going as high as 20 cells per clone).
The observation that Lmx1b marks a dorsal/ventral boundary that is resistant to cellular mixing raises the possibility that Lmx1b plays a causal role in preventing dorsal/ventral mixing. We attempted to test this possibility, re-engineering the CHAPOL library to misexpress Lmx1b in each infected cell and retesting the restriction of clones along the dorsal/ventral axis of the limb relative to endogenous Lmx1b. If Lmx1b indeed acts as a dorsal selector gene establishing the dorsal-ventral compartment boundary, then ventral clones of cells in which Lmx1b is misexpressed should be capable of crossing that boundary. However, in our analysis we were unable to identify Lmx1b misexpressing clones crossing the endogenous Lmx1b expression boundary (data not shown).
Recent work in two other laboratories using the mouse limb bud as their model system has suggested the existence of a dorsoventral compartment border defined by Lmx1b expression (Arques et al. 2007, and Qui et al., 2007) and one of those additionally obtained genetic evidence that Lmx1b itself is responsible for establishing the compartment (Qui et al., 2007). Thus, it is likely that technical limitations, such as levels of Lmx1b expression from our retroviral system, have prevented us from identifying boundary crossing clones with the Lmx1b-CHAPOL vector.
Our data also indicate that, since the D/V restricted clones in our experiment are the progeny of cells labeled in the lateral plate mesoderm, the dorsal ventral boundary is established very early and even prior to definitive limb bud outgrowth. This would predict that the dorsoventral axis of the limb is established when the limb field of the lateral plate mesoderm is still a relatively flat sheet of tissue and thus that the ultimate dorsal/ventral axis of the limb bud originates as the medial/lateral axis of the lateral plate mesoderm. A prediction of this model would be that early midline signals are responsible for establishing the future D/V axis of the limb bud. It has, in fact, been beautifully demonstrated that a signal from the developing somites initiates a program that establishes the dorsal limb identity in the lateral plate mesoderm (Michaud et al., 1997; Ohuchi et al. 1999). We also see that implanting a barrier between the somites and the lateral plate mesoderm to block signals from the midline at HH15, just prior to lmx1b induction, will prevent the primary induction of lmx1b in the nascent limb bud (Figure 5J–M). Taken together, the preponderance of data generated from multiple labs on the establishment if the limb D/V axis leads us to the conclusion that this axis of the limb is established in two distinct phases. The early phase consists of a signal originating from the HH13–14 somites and effecting events that lead to the primary induction of lmx1b in the medial lateral plate mesoderm (Michaud et al., 1997; Ohuchi et al. 1999; and our data). This signal would appear to be independent of Wnt7a in the ectoderm as ectodermal reversals prior to HH15 have no effect on D/V patterning and Wnt7a mutants still show Lmx1b induction in the proximal limb bud (Geduspan and MacCabe 1987; Cygan et al. 1997). The second phase, starting at HH15 (Geduspan and MacCabe 1987; Michaud et al. 1997; Altebef et al. 1997), is regulated by Wnt7a secreted from the dorsal ectoderm and is responsible for D/V patterning in the most distal limb tissues (autopod). Ectodermal reversals and loss of ectodermal Wnt7a result in alterations of D/V patterning in distal (autopodial), tissues (Geduspan and MacCabe 1987; Geduspan and MacCabe 1989; Riddle et al. 1995; Cygan et al. 1997). Thus, the early midline lmx1b induction seems to be responsible for D/V polarity of the proximal (stylopod, zeugopod), elements of the limb while the later ectodermally maintained signal is responsible for distal (autopodial) D/V patterning.
We have shown that undifferentiated progenitor cells in the early limb bud are not committed to individual cell fates nor do they appear to be restricted to any particular proximodistal segment along the length of the limb bud. In contrast, the limb progenitors are specified at a very early stage to either a dorsal or ventral fate, although they are not restricted from moving freely within the dorsal or ventral compartments. Elucidating the mode by which fates are determined in the limb provides a deeper understanding of how limb morphogenesis is organized and will help pave the way for future studies directed towards identifying the factors and cell interactions that direct these fate choices.
Materials and Methods
Viral construction
CHAPOL virus generation has been described in Golden et al. (1995). Briefly, the avian retroviral vector CHAP which expresses the human PLacental Alkaline Phosphatase (PLAP) gene was modified to carry a degenerate sequence of [(G or C)(A or T)]12 just down stream of PLAP. This degenerate region was flanked by nested PCR sequences to allow for efficient amplification and sequencing. The stretch of degeneracy within CHAPOL allows for a theoretical library complexity of >107 unique inserts. Two steps in the production of virus have the potential to reduce the complexity of the library, the ligation of the degenerate oligonucleotide into the CHAP vector and subsequent transformation, and the transfection of the CHAPOL plasmid library for viral production. The CHAPOL stocks used in our experiments have a complexity of >2×106 after processing.
ROLmx virus (modified CHAPOL misexpressing lmx1b), was generated by subcloning the same degenerate sequence from CHAPOL into a replication incompetent (RISAP) avian retrovirus that expresses chick Lmx1b. The complexity of the original construct was maintained by PCR amplification of 10ngs CHAPOL plasmid library (>1012 plasmids). This amplified, degenerate fragment was ligated and batch transformed to retain a complexity of >106.
Chick Manipulation
Fertilized Gallus gallus eggs were purchased from SPAFAS and incubated in a humidified 37°C incubator until reaching the desired stage. Embryos were staged according to the methods of Hamburger and Hamilton (1951). Embryos at the appropriate stage were windowed and injected with either CHAPOL or ROLmx virus, resealed with cellophane tape and re-incubated until they reached the desired harvesting stage. For barrier experiments we implanted tantalum foil squares perpendicular to the plane of the embryo, at the hind limb level, lateral to the somites and medial to the lateral plate mesoderm in HH15 embryos. The barriers were implanted to only disrupt the somatic and not the splanchnic lateral plate mesoderm. Embryos were harvested 12, 24, 36 and 48 hours after implantation and hybridized with a probe for cLmx1b.
Marker detection
In situ hybridization was performed as described in our lab protocol (http://genepath.med.harvard.edu/~cepko/protocol/). For Scleraxis detection we used probes corresponding to a 450bp BglII-EcoRV fragment of Scx containing mostly 3′UTR (Schweitzer et al., 2001). For cLmx1b we generated antisense probes to either the entire ORF or the 3′UTR (in embryos infected with lmx1b misexpressing virus). All in situ probes were labeled with Digoxigenin. Myosin Heavy Chain was detected by immunohistochemistry (after in situ hybridization), using the MF20 monoclonal antibody from the Developmental Studies Hybridoma Bank which recognizes primary myotubes. CollagenII was detected using the goat polyclonal C-19 antibody from Santa Cruz Biotechnology.
Lineage Analysis
Chick embryos injected with the CHAPOL virus were harvested, rinsed in PBS and fixed in 4%PFA at 4°C overnight. Fixed embryos were rinsed in PBS and heated to 70°C for 1.5 hours to inactivate endogenous alkaline phosphatase activity then stained with NBT and BCIP according to standard protocols. Embryos were then photographed, cryo-sectioned at 30μm, rinsed and re-stained with NBT and BCIP to better visualize individual cells. Samples were then processed for in situ hybridization for either Scleraxis or Lmx1b. Scleraxis stained embryos were further processed by staining with antibodies for Myosin Heavy Chain and Collagen II both of which were detected with a FITC-conjugated secondary antibody. Each processed section was then cover slipped under Gelvatol and photographed. Sections with PLAP positive cells were soaked in water to remove the cover slips and individual cells were picked using a single clean pulled glass needle for each cell. Needles were discarded after each cell dissection to prevent nuclear cross contamination between two cell picks. Each picked cell was annotated on a photograph of the section and on a spreadsheet indicating the number of the pick, the cell type picked and the position of the cell in the limb. Whenever possible, only a single positive cell was picked. When this was impossible due to the close proximity of the cells it was ensured that only cells of a single tissue were picked in an individual dissection. When this was impossible, or if a cell type could not be unambiguously identified, that pick was discarded from analysis. While an individual dissection includes negative tissue along with the positive cell and occasionally contains more than one positive cell, we refer to each pick as a cell. Thus, a clonally related population containing 10 picks is referred to as a 10 cell clone. Because of this we don’t conclude that a 10 cell clone contains only 10 cells, but do conclude that it contains at least 10 cells and is comparable in size to a 10 cell clone of another cell type. Picked cells were lysed in 0.2mg/ml Proteinase K (50mM KCl, 2.25mM MgCl2, 10mM Tris-HCl, pH8, 10% Tween-20, and 0.1μM of each Oligo-0 and Oligo-5) under oil at 60°C for 2 hours, 85°C for 20 mins, and 95°C for 10 mins. Lysed cells were then amplified using olig-0 (TGTGGCTGCCTGCACCCCAG GAAAG) and oligo-5 (GTGTGCTGTCGAGCCGCCTTCAATG), resulting in a 251 bp PCR product. One microliter of this reaction was transferred to a clean tube and further amplified using nested oligos, oligo-2 (GCCACCACCTACAGCCCAGTGG) and oligo-3 (GAGAGAGTGCCGCGG TAATGGG) resulting in a 121 bp amplified product with the sequence 5′-GCCACCACCTACAGCCCAGTGGGGTCGATGGCGCGCCTTT[(G/C)(A/T)]12GTTACGCGTTAATTAACTCGAGATCTTCGACAGATCCCATTACCGCGGCACTCTCTC-3′. The amplified fragments were sequenced and assigned to clones based on the presence of identical sequence information in multiple cell picks.
Supplementary Material
All remaining cells belonging to the clone depicted in figure 2 are shown here. The embryo shown was injected with CHAPOL lineage tracer at stage HH16 and harvested at HH35. All clonally related cells are indicated with red arrowheads. The clone depicted contained 25 cells, four of which were shown in figure 2 and the remaining 21 are shown here. Sections are all oriented with dorsal to the top of the frame and proximal to the left (as in figure 2). The section series proceeds from the more posterior region of the hind limb (panel A), toward the anterior (panel I). Only sections containing cells of this clone are shown, thus not all sections in the progression are adjacent. Sections are numbered on the bottom of each panel corresponding to their A–P position in the section series with panel (A) being defined as section 1. Thus, C, D, E are adjacent sections as are F–I. Unshown sections either did not contain cells in this clone, or are already shown in figure 2 (figure 2A is section 4, 2B is section 10 in this series). All sections are 30μm thick.
Demonstration of PLAP positive cells in all proximal-distal segments of the embryo shown in fig 4A. Sections shown are progressively more anterior in the specimen. Panels A, B are oriented with Dorsal to the top, and Proximal to the left as demonstrated in the schematic (modified from Hudson et al., 1959). Panel C is an anterior section that is out of the plane of the thigh and shank and only shows the distal phalanges of the foot and is oriented according to the marker inset. Groups of PLAP positive cells are marked with arrowheads colored according to their P–D segment identity. Blue = Thigh, Red = Shank, Yellow = Foot as indicated in the schematic.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahrens PB, Solursh M, Reiter RS. Stage-related capacity of limb chondrogenesis in cell culture. Dev Biol. 1977;60:69–82. doi: 10.1016/0012-1606(77)90110-5. [DOI] [PubMed] [Google Scholar]
- Altabef M, Clarke JD, Tickle C. Dorso-ventral ectodermal compartments and origin of apical ectodermal ridge in developing chick limb. Development. 1997;124(22):4547–4556. doi: 10.1242/dev.124.22.4547. [DOI] [PubMed] [Google Scholar]
- Arques CG, Doohan R, Sharpe J, Torres M. Lineage compartmentalization of the mouse limb mesenchyme along the dorso-ventral but not the proximo-distal or anterior-posterior axes. 2007 Submitted for publication. [Google Scholar]
- Bader D, Masaki T, Fischman DA. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–70. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lun Y, Ovchinnikov D, Kokubo H, Oberg KC, Pepicelli CV, Gan L, Lee B, Johnson RL. Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat Genet. 1998;19:51–55. doi: 10.1038/ng0598-51. [DOI] [PubMed] [Google Scholar]
- Chevallier A, Kieny M, Mauger A. Limb-somite relationship: origin of the limb musculature. J Emb Exp Morph. 1977;41:245–58. [PubMed] [Google Scholar]
- Christ B, Jacob HJ, Jacob M. Experimental analysis of the origin of the wing musculature in avian embryos. Anat Embryol (Berl) 1977;150(2):171–86. doi: 10.1007/BF00316649. [DOI] [PubMed] [Google Scholar]
- Cygan JA, Johnson RL, McMahon AP. Novel regulatory interactions revealed by studies of murine limb pattern in Wnt-7a and En-1 mutants. Development. 1997;124:5021–5032. doi: 10.1242/dev.124.24.5021. [DOI] [PubMed] [Google Scholar]
- Dreyer SD, Zhou G, Baldini A, Winterpacht A, Zabel B, Cole W, Johnson RL, Lee B. Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genetics. 1998;19(1):47–50. doi: 10.1038/ng0598-47. [DOI] [PubMed] [Google Scholar]
- Dudley AT, Ros MA, Tabin CJ. A re-examination of proximodistal patterning during vertebrate limb development. Nature. 2002;418:539–544. doi: 10.1038/nature00945. [DOI] [PubMed] [Google Scholar]
- Geduspan JS, MacCabe JA. The Ectodermal control of mesodermal patterns of differentiation in the developing chick wing. Dev Biol. 1987;124:398–408. doi: 10.1016/0012-1606(87)90492-1. [DOI] [PubMed] [Google Scholar]
- Geduspan JS, MacCabe JA. Transfer of dorsoventral information from mesoderm to ectoderm at the onset of limb development. The Anat Rec. 1989;224:79–87. doi: 10.1002/ar.1092240110. [DOI] [PubMed] [Google Scholar]
- Globus M, Vethamany-Globus S. An in vitro analogue of early chick limb bud outgrowth. Differentiation. 1976;6(2):91–96. doi: 10.1111/j.1432-0436.1976.tb01474.x. [DOI] [PubMed] [Google Scholar]
- Golden JA, Fields-Berry SC, Cepko CL. Construction and characterization of a highly complex retroviral library for lineage analysis. Proc Natl Acad Sci USA. 1995;92:5704–8. doi: 10.1073/pnas.92.12.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JA, Cepko CL. Clones in the chick diencephalon contain multiple cell types and siblings are widely dispersed. Development. 1996;122(1):65–78. doi: 10.1242/dev.122.1.65. [DOI] [PubMed] [Google Scholar]
- Guo Q, Loomis C, Joyner AL. Fate map of mouse ventral limb ectoderm and the apical ectodermal ridge. Dev Biol. 2003;264:166–178. doi: 10.1016/j.ydbio.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton H. A series of normal stages in the development of the chick embryo. J Embryol Exp Morphol. 1951;8:241–245. [PubMed] [Google Scholar]
- Hudson GE, Lanzillotti PJ, Edwards GD. Muscles of the pelvic limb in galliform birds. Am Midland Nat. 1959;61:1–67. [Google Scholar]
- Kardon G. Muscle and tendon morphogenesis in the avian hind limb. Development. 1998;125:4019–4032. doi: 10.1242/dev.125.20.4019. [DOI] [PubMed] [Google Scholar]
- Kardon G, Campbell JK, Tabin CJ. Local extrinsic signals determine muscle and endothelial cell fate and patterning in the vertebrate limb. Dev Cell. 2002;3:533–45. doi: 10.1016/s1534-5807(02)00291-5. [DOI] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Michaud JL, Lapointe F, Le Douarin NM. The dorsoventral polarity of the presumptive limb is determined by signals produced by the somites and by the lateral somatopleure. Development. 1997;124:1453–1463. doi: 10.1242/dev.124.8.1453. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Nakagawa T, Itoh N, Noji S. FGF10 can induce Fgf8 expression concomitantly with En1 and R-fng expression in chick limb ectoderm, independent of its dorsoventral specification. Dev Growth Diff. 1999;41:665–673. doi: 10.1046/j.1440-169x.1999.00466.x. [DOI] [PubMed] [Google Scholar]
- Qui Q, Chen H, Johnson RL. Lmx1b-expressing cells in the mouse limb bud define a dorsal mesenchymal lineage compartment. 2007 doi: 10.1002/dvg.20430. Submitted for publication. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Ensini M, Nelson C, Tsuchida T, Jessell TM, Tabin CJ. Induction of the LIM homeobox gene Lmx1 by WNT7a establishes dorsoventral pattern in the vertebrate limb. Cell. 1995;83:631–640. doi: 10.1016/0092-8674(95)90103-5. [DOI] [PubMed] [Google Scholar]
- Ros MA, Lyons GE, Mackem S, Fallon JF. Recombinant limbs as a model to study homeobox gene regulation during limb development. Dev Biol. 1994;166:59–72. doi: 10.1006/dbio.1994.1296. [DOI] [PubMed] [Google Scholar]
- Sato K, Koizumi Y, Takahashi M, Kuroiwa A, Tamura K. Specification of cell fate along the proximal-distal axis in the developing chick limb bud. Development. 2007;134:1397–1406. doi: 10.1242/dev.02822. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3566. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- Shubin N, Alberch P. A morphogenetic approach to the origin and basic organization of the tetrapod limb. Evol Biol. 1986;20:319–387. [Google Scholar]
- Summerbell D, Lewis JH, Wolpert L. Positional information in chick limb morphogenesis. Nature. 1973;244:492–496. doi: 10.1038/244492a0. [DOI] [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. Functions of FGF signaling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–509. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- Vargesson N, Clarke JDW, Vincent K, Coles C, Wolpert L, Tickle C. Cell fate in the chick limb bud and relationship to gene expression. Development. 1997;124:1909–1918. doi: 10.1242/dev.124.10.1909. [DOI] [PubMed] [Google Scholar]
- Vogel A, Rodriguez C, Warnken W, Izpisua-Belmonte JC. Dorsal cell fate specified by chick Lmx1 during vertebrate limb development. Nature. 1995;387:716–720. doi: 10.1038/378716a0. [DOI] [PubMed] [Google Scholar]
- Wachtler F, Christ B, Jacob HJ. On the determination of mesodermal tissues in the avian embryonic wing bud. Anat Embryol (Berl) 1981;161(3):283–289. doi: 10.1007/BF00301826. [DOI] [PubMed] [Google Scholar]
- Wada N, Ohsugi K, Yokouchi Y, Kuroiwa A, Ide H. Cell sorting and chondrogenesis aggregate formation in limb bud recombinants and in culture. Dev Growth Diff. 1998;35:421–430. doi: 10.1111/j.1440-169X.1993.00421.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All remaining cells belonging to the clone depicted in figure 2 are shown here. The embryo shown was injected with CHAPOL lineage tracer at stage HH16 and harvested at HH35. All clonally related cells are indicated with red arrowheads. The clone depicted contained 25 cells, four of which were shown in figure 2 and the remaining 21 are shown here. Sections are all oriented with dorsal to the top of the frame and proximal to the left (as in figure 2). The section series proceeds from the more posterior region of the hind limb (panel A), toward the anterior (panel I). Only sections containing cells of this clone are shown, thus not all sections in the progression are adjacent. Sections are numbered on the bottom of each panel corresponding to their A–P position in the section series with panel (A) being defined as section 1. Thus, C, D, E are adjacent sections as are F–I. Unshown sections either did not contain cells in this clone, or are already shown in figure 2 (figure 2A is section 4, 2B is section 10 in this series). All sections are 30μm thick.
Demonstration of PLAP positive cells in all proximal-distal segments of the embryo shown in fig 4A. Sections shown are progressively more anterior in the specimen. Panels A, B are oriented with Dorsal to the top, and Proximal to the left as demonstrated in the schematic (modified from Hudson et al., 1959). Panel C is an anterior section that is out of the plane of the thigh and shank and only shows the distal phalanges of the foot and is oriented according to the marker inset. Groups of PLAP positive cells are marked with arrowheads colored according to their P–D segment identity. Blue = Thigh, Red = Shank, Yellow = Foot as indicated in the schematic.