Abstract
Rice NH1 (NPR1 homolog 1) is a key mediator of innate immunity. In both plants and animals, the innate immune response is often accompanied by rapid cell death at the site of pathogen infection. Over-expression of NH1 in rice results in resistance to the bacterial pathogen, Xanthomonas oryzae pv. oryzae (Xoo), constitutive expression of defense related genes and enhanced benzothiadiazole (BTH)- mediated cell death. Here we describe a forward genetic screen that identified a suppressor of NH1-mediated lesion formation and resistance, snl6. Comparative genome hybridization and fine mapping rapidly identified the genomic location of the Snl6 gene. Snl6 is a member of the cinnamoyl-CoA reductase (CCR)-like gene family. We show that Snl6 is required for NH1-mediated resistance to Xoo. Further, we show that Snl6 is required for pathogenesis-related gene expression. In contrast to previously described CCR family members, disruption of Snl6 does not result in an obvious morphologic phenotype. Snl6 mutants have reduced lignin content and increased sugar extractability, an important trait for the production of cellulosic biofuels. These results suggest the existence of a conserved group of CCR-like genes involved in the defense response, and with the potential to alter lignin content without affecting development.
Author Summary
Plants possess potent and effective endogenous methods for responding to pathogen attacks, referred to as plant innate immunity. In this report we further our understanding of rice innate immunity through characterization of the Snl6 gene. The snl6 mutant was identified from a mutant screen for positive regulators of immunity. While innate immunity represents a powerful agronomic tool, identification of desirable genes from crop species is limited by the slow and laborious nature of map-based cloning. Here we describe our methodology of combining comparative genome hybridization and fine mapping to rapidly identify the Snl6 gene. Snl6 is distantly related to members of the cinnamoyl-CoA reductase gene family, is required for pathogenesis gene expression and resistance to the bacterial pathogen Xanthomonas oryzae pv. oryzae. Snl6 mutants have reduced lignin content and increased sugar extractability, an important trait for the production of cellulosic biofuels.
Introduction
Plant innate immunity is governed by a complex signaling network [1]. Hallmark events during the immune response include localized cell death at the site of pathogen recognition (known as the hypersensitive response (HR)), release of reactive oxygen species (ROS), pathogen related (PR) gene induction, callose deposition and cell wall lignification [2], [3], [4]. The HR, ROS and PR gene production are thought to be physically harmful towards the invading pathogen while callose and lignin deposition create a physical barrier to limit pathogen entry and spread [4], [5], [6], [7], [8].
NPR1 (Non-expressor of PR genes-1) is a central regulator of the disease response in Arabidopsis and has been studied extensively [9], [10], [11], [12], [13]. Plants deficient in NPR1 expression lack PR gene accumulation after pathogen treatment, display increased susceptibility to pathogens and fail to initiate systemic acquired resistance (SAR) [11]. Over-expression of the rice NPR1 ortholog, NH1 (NPR1 homologue 1) (NH1ox) in rice results in enhanced resistance to the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo), constitutive expression of PR genes and a BTH (benzothiadiazole)-mediated cell death phenotype [14]. Enhanced cell death (also called lesion mimic phenotypes) has been correlated with resistance [15], [16].
Here we describe a mutant screen to identify additional components of the rice innate immune response. NH1ox seeds (M0) were treated with fast neutron mutagenesis to generate an M1 population segregating for genomic deletions. Sixty thousand M1 lines were screened for alterations in the immune response after treatment with BTH. Plants that did not develop BTH-induced lesions were collected as putative suppressors of NH1-mediated lesion formation (snl) mutants. In this report we focus on one mutant, snl6. To expedite the cloning of Snl6 we employed comparative genome hybridization (CGH) [17], [18], [19], [20], [21], [22], [23], [24] to identify five deletions, ranging in size from 3 kb to 100.5 kb, in the snl6 mutant line. Only the deletion on rice chromosome 1 segregated with the snl6 mutant phenotype. Subsequent RNAi and allelic complementation confirmed that Snl6 encodes a member of the cinnamoyl-CoA reductase-like gene family. snl6 mutant lines fail to develop the NH1-mediated lesions, PR gene activation and resistance phenotypes. Further we show that Snl6 is not required for resistance mediated by the rice pattern recognition receptor, Xa21 [25], [26]. Finally, snl6 mutant lines show decreased phloroglucinol staining without an obvious morphologic phenotype.
Results
Identification of the Snl6 Mutant That Suppresses NH1-Mediated Lesion Formation
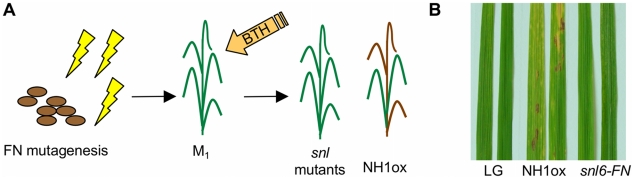
We have previously reported that over expression of the rice NPR1 homologue, NH1, in a LiaoGeng (LG) background, confers resistance to the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo) and leads to a spontaneous cell death phenotype [14]. The cell death phenotype is enhanced by application of the salicyclic acid functional analog BTH [27]. We designed a screen to identify mutants incapable of developing this cell death phenotype (Figure 1A). NH1ox seeds were treated with fast neutron (FN) mutagenesis (see Materials and Methods ). M1 progeny from 4,000 M0 were harvested in the rice field. Approximately 60,000 M1 individuals were grown in the rice field and sprayed with 10 mM BTH at 5 weeks and 7 weeks and scored for cell death at 10 weeks. BTH treatment of NH1ox plants in the field resulted in cell death, visualized by discrete lesions on the leaves. While most plants showed the typical NH1ox cell death phenotype, approximately 20 plants showed little to no cell death and were therefore named suppressor of BTH-induced, NH1-mediated lesion mimic (snl) mutants. The suppressed cell death phenotype of snl6-FN (fast neutron allele) was confirmed in the greenhouse (Figure 1B).
Figure 1. Identification of the snl6-FN mutant.
(A) Cartoon of mutant screen used to identify suppressors of BTH-induced, NH1-mediated lesion mimic (snl) mutants. NH1ox seeds were treated with Fast Neutron (FN) mutagenesis (M1). M2 plants were treated with BTH and then screened for the appearance of the lesion mimic phenotype. snl6-FN (line 11-3) did not develop lesions after BTH treatment. (B) 8-week old plants were treated with BTH. Plants were assessed for the presence of the lesion mimic phenotype. Image was taken two weeks after BTH treatment. LG, NH1ox and snl6-FN (line: 11-3-2) were compared.
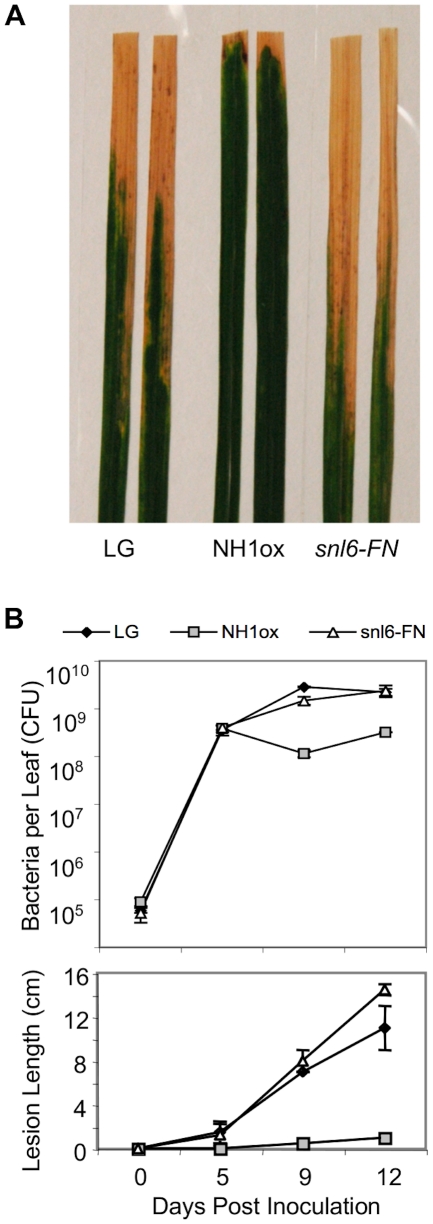
As lesion mimic mutants often have an accompanying resistance phenotype [15], [16] we hypothesized that snl6-FN plants may show enhanced susceptibility to Xoo. Indeed, snl6-FN lines show longer water-soaked lesions after infection with Xoo and support greater populations of Xoo cells than NH1ox plants (Figure 2A and 2B), indicating that Snl6 is required for NH1-mediated resistance. To ensure that the snl6-FN phenotype did not represent a mutation in the NH1 over-expression transgene, we confirmed the over-expression of NH1 in the snl6-FN line (Figure S1).
Figure 2. Snl6 is required for NH1ox-mediated resistance to Xoo.
8-week old plants were challenged with Xoo. (A) Lesion length development after 12 days. (B) Total bacterial populations per leaf (top) and lesion lengths (bottom) were measured at 0, 5, 9 and 12 days post inoculation. Mean, ± range (n = 2), are displayed. The growth curve experiment was repeated twice with similar results. LG, NH1ox and snl6-FN (line: 11-3-2) were compared.
Comparative Genome Hybridization Combined with Fine Mapping Locate Snl6 on Rice Chromosome 1
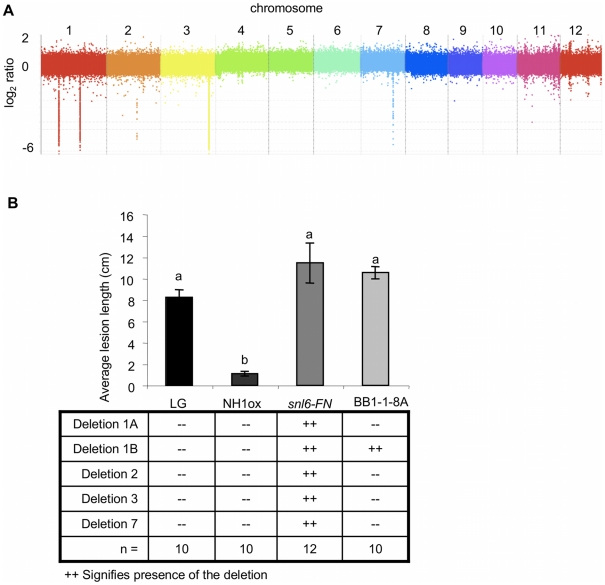
Conventional map-based cloning, while effective, is slow and laborious [25], [28]. With the availability of fully sequenced genomes, an alternative method for gene cloning has emerged called comparative genome hybridization (CGH) [29]. CGH uses tiling arrays to physically compare two genomes. In CGH, genomic DNA from each plant is fragmented and differentially labeled. The labeled DNA is then hybridized to a tiling array, composed of probes where each probe corresponds to a known location on the reference genome. Probes that show strong hybridization with the parent but not the mutant, indicate deleted regions on the mutant genome. Because the snl6-FN mutant was created with fast neutron mutagenesis, which generally induces deletions on chromosomes [30], we employed CGH to expedite the cloning of Snl6. In collaboration with Nimblegen, we designed a full genome tiling array for rice (japonica cultivar) with an average probe spacing of one 50-mer probe every 146 bp. NH1ox and snl6-FN genomic DNA was prepared, fragmented and labeled with Cy3 or Cy5, respectively. Relative hybridization intensities for each probe are reported as: log2(snl6-FN/NH1ox). A negative log2 ratio represents a deletion in the genome of the snl6-FN mutant. CGH revealed 5 deletions (Figure 3A), encompassing 35 annotated genes (Table S1), in the genome of snl6-FN and each was confirmed through PCR.
Figure 3. Snl6 segregates with deletion 1B.
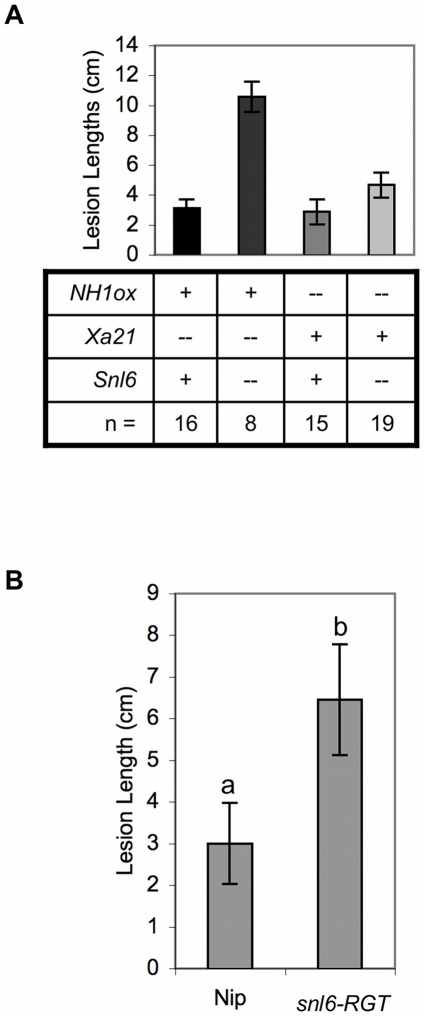
(A) Comparative Genome Hybridization (CGH) between NH1ox and snl6-FN (line: 11-3-2). Each spot represents an average of all probes in a 1.5 kb region. Identified deletions were named based on chromosome number as follows Deletion 1A, 1B, 2, 3, 7. (B) Plants were scored for resistance after challenge with Xoo. mean ± s.e.m, Different letters represent significant difference (p<0.05). LG, NH1ox, snl6-FN (line 11-3-2), BB1-1-8A (F2 mapping population individual) progeny.
Next, we created an F2 mapping population (line: BB1-1) and individuals were genotyped for the NH1ox transgene, each deletion and then scored for resistance to Xoo. Only the second deletion on chromosome 1 (Deletion 1B) cosegregated completely with susceptibility. One individual showed a recombination event between the two deletions on chromosome 1. We analyzed the next generation of this line (BB1-1-8A) and confirmed the susceptibility (Figure 3B). Deletion 1B contains 3 annotated, non-transposon genes: LOC_Os01g45160, LOC_OS01g45190 and LOC_Os01g45200 (Table S1). The former does not have any associated expression data (based on EST libraries and publicly available microarray data) and thus we de-prioritized this gene as an unlikely candidate for Snl6. LOC_Os01g45200 is annotated as a cinnamoyl CoA-reductase (CCR)-like gene (TIGR v6.1). Because CCRs catalyze the first committed step in the lignin biosynthetic pathway and have previously been linked to the defense response [31], [32], LOC_Os01g45200 became our top candidate for Snl6.
Snl6 Encodes a Member of the Cinnamoyl CoA-Reductase (CCR)-Like Gene Family
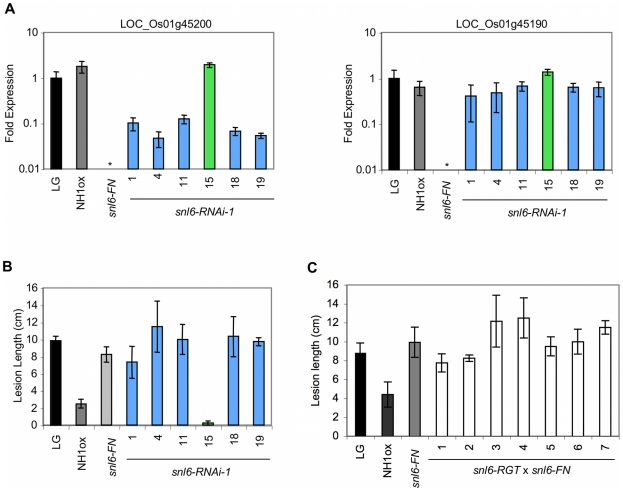
To determine if LOC_Os01g45200 is Snl6, we generated transgenic plants expressing an inverted repeat RNAi construct (snl6-RNAi) to silence LOC_Os01g45200 in an NH1ox background (Figure S2). snl6-RNAi targets 425 bp of the 3′ end of LOC_Os01g45200. This region is 74% identical to the closest homolog, LOC_Os05g50250, however this gene is intact in the snl6-FN and snl6-RGT (described below) alleles. Consequently we know that LOC_Os05g50250 does not contribute to the snl6 mutant phenotype. Four independent RNAi lines (primary transgenics, T0) challenged with Xoo displayed enhanced susceptibility as compared to the NH1ox control (Figure S3). Among six T1 progeny of snl6-RNAi-1, the enhanced susceptibility phenotype segregated perfectly with the presence of the transgene and with reduced expression of LOC_Os01g45200 (Figure 4A and 4B). LOC_Os01g45190 showed wild-type expression levels in all snl6-RNAi-1 progeny indicating that this gene does not contribute to the snl6 susceptibility phenotype. While snl6-RNAi-1 shows higher expression of Snl6 than the deletion allele, snl6-FN, the resulting lesion lengths for these different lines are comparable suggesting that Snl6 may not function in a dosage dependent manner. Next, we identified an insertion line from the Rice Transposon Flanking Sequence Tag (FST) Database [33]. We designated this line, snl6-RGT (Figure S2) and performed an allelic complementation test by crossing snl6-RGT with snl6-FN (recessive) and analyzing the progeny. Successful crosses were confirmed through PCR. All seven F1 individuals challenged with Xoo showed levels of enhanced susceptibility similar to that of snl6-FN (Figure 4C). These lines carry one copy of the NH1ox transgene, which we have previously shown is sufficient to confer high levels of resistance to Xoo [14]. Thus we conclude that snl6-RGT and snl6-FN are allelic. Taken together these data confirm that LOC_Os01g45200 is Snl6, a cinnamoyl-CoA reductase (CCR)-like gene.
Figure 4. RNAi and allelic complementation for Snl6.
(A) Expression of LOC_Os01g45190 and LOC_Os01g45200 in snl6-RNAi-1 progeny, ± s.d., n = 3 (B) Lesion lengths of snl6-RNAi-1 T1 progeny, 14 days after inoculation with Xoo. ± s.d., n = 3; * = expression level below background (C) Allelic complementation test. Lesion lengths 11 days after challenge with Xoo. ± s.d., n = 3. LG, NH1ox, snl6-FN (line 11-3-2), snl6-RNAi-1 progeny (T1), snl6-RGT (RGT6140B_5.1) x snl6-FN (line: 11-3-2) F1 individuals.
Snl6 encodes a predicted protein of 364 amino acids (39.5 kDa). CCRs exist as multi-gene families with at least 7 and 14 annotated members in Arabidopsis and rice, respectively. Previously described CCR genes show limited identity with Snl6 [31], [32], [34], [35] (Figure S4). The closest predicted rice paralog is LOC_Os05g50250, sharing 73% identity at the amino acid level. Sorghum, Brachypodium and maize all have predicted orthologs (EES03334.1, 2g44800.1 and ACR34585.1 with 86%, 81% and 82%, amino acid identity, respectively). The closest predicted ortholog in Arabidopsis thaliana is AT5G14700 with 44% amino acid identity. None of these closest predicted orthologs have been functionally characterized. In Arabidopsis, the distantly related genes, AtCCR1 (25% identity) and AtCCR2 (27% identity) have been attributed a primary role in development or pathogen attack, respectively. They appear to be able to partially compensate for each other [32].
Snl6 Is Not Required for XA21-Mediated Resistance
Both NH1ox and XA21-mediated resistance are regulated by NRR (negative regulator of resistance) suggesting a possible overlap of these pathways [36]. To determine if Snl6 is required for resistance mediated by XA21, we examined individual F2 progeny derived from a cross between snl6-FN line and a transgenic line expressing Xa21 under control of the ubiquitin promoter [37]. These lines were segregating for NH1ox, Xa21 and Snl6. Plants containing Xa21 were resistant to Xoo independent of the presence of Snl6 (Figure 5A). Thus we conclude that Snl6 is required for NH1-mediated resistance, but not XA21-mediated resistance in these lines. Our results are consistent with studies from Arabidopsis showing that the PRR, FLS2, does not require NPR1 to initiate an immune response [38].
Figure 5. Snl6 contributes to resistance in the absence of NH1ox.
(A) Individuals from line BB1-1 (snl6-FN (line:11-3-2) x Ubi-Xa21-kitaake (line 7A-8)) were genotyped for the presence of NH1ox, Xa21 and Snl6. 8-week old plants were moved to the growth chamber and challenged with Xoo. Lesion lengths were measured after 12 days. Mean ± s.e.m. and number of individuals (n) are reported. (B) Average lesion lengths 11 days after Xoo inoculation. Nip (wildtype cultivar nipponbare); snl6-RGT (T-DNA insertion in Snl6). ± s.e.m., n = 12 (nip) or 18 (snl6-RGT). Letters (a, b) indicate significant difference (p<0.05).
Snl6 Contributes to Resistance in the Absence of NH1ox
We have shown that Snl6 contributes to NH1ox–mediated resistance. To further investigate the role of Snl6 in the absence of NH1ox we compared inoculation data for snl6-RGT (lacking the NH1ox transgene) and in the genetic background of cultivar, Nipponbare used as a recipient in the transformation studies. While both lines are susceptible to inoculation with Xoo, snl6-RGT plants developed longer lesions suggesting that Snl6 contributes to resistance in Nipponbare (Figure 5B).
Snl6 is Required for NH1-Mediated PR10 Gene Expression
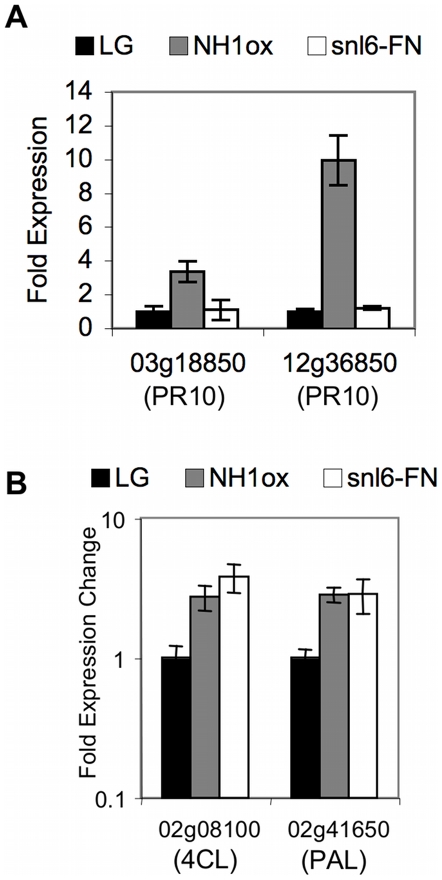
The NH1ox resistance phenotype is correlated with constitutively high expression levels of several PR genes [14]. To further elucidate the role of Snl6 in innate immunity, we examined the relative expression levels of two PR10/PBZ family members (LOC_Os03g18850 and LOC_Os12g36850), in LG, NH1ox and snl6-FN lines. Both PR10 family members show significantly higher expression levels in NH1ox lines as compared to LG. snl6-FN lines while still over-expressing NH1 (Figure S1), do not show enhanced PR10/PBZ gene expression (Figure 6A). A similar result was observed for the snl6-RNAi-1 line.
Figure 6. Characterization of the role of Snl6 in the defense response.
Relative gene expression levels in LG, NH1ox and snl6-FN (line: 11-3-2-1) lines using realtime quantitative RT-PCR, (A) two PR10 genes (B) 4CL and PAL; mean ± s.e.m, n = 3.
PAL and 4-coumarate-CoA Ligase 1 (4CL) Are Co-Expressed with Snl6
In addition to induced PR gene expression, over expression of NH1 results in constitutive activation of phenylalanine ammonia-lyase (PAL). PAL catalyzes the first step of the highly branched phenylpropanoid pathway and has been studied for its role in lignin biosynthesis as well as defense related molecules such as salicylic acid. By analyzing publicly available microarray data we found that PAL as well as a member of the lignin specific branch of the phenylpropanoid biosynthetic pathway, 4-coumarate-CoA ligase 1 (4CL), were highly co-expressed (cc >0.6) with Snl6. We hypothesized that snl6 mutant lines might contain alterations in the phenylpropanoid pathway. Both PAL and 4CL showed more than 2 fold higher expression in NH1ox and snl6-FN as compared to LG (Figure 6B). These results suggest that lignin biosynthesis is induced by over expression of NH1 and further suggest that Snl6 may function downstream of PAL and 4CL, which would be consistent with the published placement of CCR in the phenylpropanoid pathway.
Snl6 Mutants Have Reduced Lignin and Enhanced Sugar Extractability
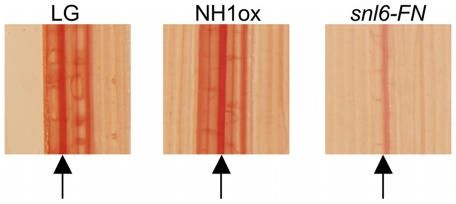
To further elucidate the function of Snl6, we used the Wiesner Test (Phloroglucinol/HCl). Phloroglucinol/HCl reacts with aromatic aldehydes to produce a pink/red color and is commonly used to detect lignin [34], [39], [40]. NH1ox and LG showed clear staining, most strongly at the midvein (arrow). While some staining is observed in snl6-FN leaves, total staining is reduced especially at the midvein (Figure 7). Among five independent experiments, the amount of staining observed in LG leaves varied, but was consistently greater than that seen for snl6 mutant line suggesting that additional environmental factors contribute to wildtype lignin accumulation. Notably, most previously characterized lignin mutants express a severe developmental phenotype [34], [40], [41], [42]. In contrast, no significant difference was found between snl6-FN and wild-type plants for plant height, seed set, tiller number or general appearance (Figure S5).
Figure 7. Characterization of the role of Snl6 in the phenylpropanoid pathway.
Tissue from 8-week old plants was cleared with lactic acid/phenol and stained with phloroglucinol/HCl. Images were taken at 2X magnification. Arrows point to the midrib of each sample. Experiment was repeated 5 times with similar results. Note: LG sample ripped during sample preparation.
Decreased expression of CCR-like genes has been correlated with increased sugar extractability from cell walls in alfalfa and poplar, an important trait for the production of cellulosic biofuels [34], [39]. We hypothesized that this correlation may also be present in snl6-FN plants. We first determined that the snl6 mutation does not affect the relative cell wall monosaccharide composition in leaves (Figure S6A). None of the individual sugars tested show a significant difference between the different genotypes (p<0.05). Next, we quantified the total sugar release after hot water pre-treatment. Compared to the LG and NH1ox controls, snl6-FN plants showed an increase in sugar release (p<0.05) (Figure S6B). These results are consistent with previous reports that correlate decreased lignin content with increased sugar extractability [43].
Discussion
The ability of a plant to recognize the presence of a pathogen and mount an effective immune response is fundamental to survival. The defense response must be tightly regulated as each of these responses present a potential fitness cost to the host. The result is a complex network of signaling cascades that govern plant innate immunity. Arabidopsis NPR1 and rice NH1 are central regulators of plant innate immunity. Our objective in the current study was to identify additional components of the rice innate immune response thus broadening our understanding of this important biological process.
In this report, we describe the identification and characterization of rice Snl6. We identified Snl6 from a screen for mutants that suppress the NH1-mediated lesion mimic phenotype. Notably, while many negative regulators of innate immunity have been identified [36], [44], [45], [46], relatively few positive regulators from rice have been characterized to date. By conducting the mutagenesis in an NH1ox background and then screening for suppressors of the lesion mimic phenotype, we were able to specifically target positive regulators, that is, genes that are required for NH1-mediated immunity. Subsequent inoculation experiments confirmed that the suppression of the NH1-mediated lesion mimic phenotype correlated with enhanced susceptibility to the bacterial pathogen Xoo.
Previous reports have demonstrated the potential usefulness of comparative genome hybridization (CGH) or similar comparative genome techniques for plant species [17], [18], [19], [20], [21], [22], [23], however routine use of this technique in crop species remains limited. In this report we have successfully combined CGH and fine mapping with RNAi silencing and allelic complementation. In this way we were able to determine that the snl6 mutant phenotype was the result of disruption of LOC_Os01g45200. In cloning Snl6 we generated a population segregating for NH1ox, Xa21 and Snl6. We were able to show that mutations in Snl6 do not compromise resistance mediated by the rice pattern recognition receptor, Xa21 driven by the ubiquitin promoter. These results are consistent with studies from Arabidopsis that have shown that the PRR, FLS2, does not require NPR1 to initiate an immune response [38]. Notably, in this population over expression of NH1 confers a similar level of resistance as XA21, highlighting the strength of NH1ox-mediated resistance as previously demonstrated [14].
Snl6 is annotated as a cinnamoyl-CoA reductase (CCR)-like gene, however overall similarity to previously characterized CCRs is low. CCRs catalyze the first committed step of the monolignol biosynthetic pathway. The first identified CCR was from Eucalyptus [47] followed a year later by an orthologue from tobacco [48]. To date, several CCRs have been characterized though their exact role in lignin biosynthesis is still unclear. Studies from Arabidopsis and tobacco indicate that down regulation of CCR results in severe developmental phenotypes, including collapsed xylem cells and dwarfism, a decrease in total lignin along with a higher S/G ratio in the lignin polymer, and the appearance of feruloyl tyramines [40], [41], [42], [49], [50]. Among the CCR family, a large amount of sequence variability exists, which may contribute to the overall uncertainty about the exact role of CCR. In Arabidopsis, two CCRs, AtCCR1 and AtCCR2 have been compared and attributed a role in lignin biosynthesis during development or pathogen attack, respectively. While AtCCR2 is primarily involved in pathogen defense, knockdown experiments have shown that it can at least partially compensate for a downregulated AtCCR1.
To the best of our knowledge there has been only one report characterizing a CCR-like gene from rice. OsCCR1 was originally identified based on its in vitro interaction with OsRac1. OsRac1 is a GTPase, important for the defense response in rice. While expression of OsCCR1 was induced by a sphingolipid elicitor in suspension cells, no mutant phenotype was observed [31]. Notably, OsCCR1 is dissimilar to Snl6, sharing only 28% identity at the amino acid level.
Here we show that mutations in Snl6 disrupt the NH1ox-mediated constitutive activation of PR genes, providing a mechanism for the role of Snl6 in NH1-mediated resistance. In addition, as Snl6 is annotated as a CCR-like gene, we hypothesized that snl6 mutant lines may contain alterations in the phenylpropanoid biosynthetic pathway. Indeed, we show that disruption of Snl6 leads to less lignin accumulation. Based on these data alone, it is unclear whether Snl6 contributes specifically to NH1ox-mediated resistance or as part of a more general resistance response. However, because Snl6 is highly co-expressed with two additional members of the phenylpropanoid biosynthetic pathway, PAL and 4CL, and because both these genes are induced in NH1 over expression plants, it seems likely that over expression of NH1 does induce lignin biosynthesis. As lignin is an important defense molecule, providing a physical barrier to pathogen entry into the plant, our results indicate that Snl6 has dual roles in the resistance response: activation of PR genes and lignin biosynthesis.
Lignin contributes to plant structure as well as pathogen defense. Most previously described lignin mutants contain a severe phenotypic detriment [34], [40], [41], [42]. Our result, that snl6 mutants contain decreased lignin, is particularly relevant to studies of bioenergy crops because snl6 mutant lines do not display an obvious morphologic phenotype. In addition, we observed a greater than 15% increase in sugar extractability from snl6 mutants as compared to controls. Our results indicate the presence of a previously uncharacterized group of CCR-like genes, alterations in which can alter lignin content without affecting development.
Materials and Methods
NH1ox Mutant Screen
NH1ox seeds were treated with fast neutron (FN) mutagenesis in three batches at 18, 20 and 22 Grays, respectively. This M0 population was grown at the UC Davis rice field and M1 seed from approximately 4,000 M0 individuals was then collected as 400 pools (10 per pool). Approximately fifteen M1 seed from each of the M0 lines were subsequently grown in the field (n = ∼60,000 M1 seed). The 60,000 M1 lines were sprayed with 10 mM BTH at 5 weeks and 7 weeks and lesion mimic severity was assessed at 10 weeks. BTH treatment of NH1ox plants in the field resulted in a lesion mimic phenotype. Plants that did not develop lesions were called, Suppressor of NH1-mediated Lesion mimic (snl) mutants. snl6-FN (originally named line 11-3) was identified in this screen.
Xoo Inoculations
Xoo (Philippine race 6, PXO99AZ) inoculations were carried out as previously described [25]. Briefly, 8-week old rice plants were transferred to the growth chamber. Scissors were dipped in a solution of Xoo cells in water (OD600 = 0.5) and then used to clip the rice leaf tip. Growth curves were performed as previously described [36].
Comparative Genome Hybridization
In collaboration with Nimblegen, we designed a full genome tiling array for rice, ssp. Japonica (average one 50-mer probe/146bp). CGH was conduced following Nimblegen's specified procedures. Briefly, genomic DNA from snl6-FN and NH1ox plants was isolated and the quality of the DNA was assessed via a spectrophotometer and agarose gel electrophoresis prior to sending to Nimblegen. At Nimblegen, the genomic DNA was sheared, labeled and then hybridized to the tilling array. A negative log2 ratio represents a deletion in the genome of snl6-FN.
Creation of Line BB1-1
snl6-FN (line: 11-3-2) was crossed to a transgenic line expressing, Xa21 (Ubi Myc-Xa21 (line 7A-8), (Park, Ronald submitted) pollen donor) under control of the ubiquitin promoter, in a kitaake background. The cross was confirmed by testing the F1 progeny (line: BB1) for the presence of Xa21. The F1 progeny was allowed to self-pollinate to create an F2 population (line: BB1-1) that segregated for Xa21, NH1ox, and all 5 deletions identified from CGH.
Fine mapping
Individuals from line BB1-1 (above) were grown in the greenhouse and genotyped for the presence of Xa21, NH1ox, and all 5 deletions identified from CGH. Individuals that did not contain Xa21 (as Xa21 also confers resistance to Xoo, Xa21+ individuals were excluded from this experiment) were moved to the growth chamber for challenge with Xoo and lesion lengths were assessed after 14 days. One individual (line: BB1-1-8A) contained NH1ox, was susceptible to Xoo, and showed a recombination event between the two deletions on chromosome 1. Phenotype and genotype were assessed in the next generation (line BB1-1-8A).
Xa21 effect on Snl6
Individual progeny from line BB1-1 (described above) were grown in the greenhouse and genotyped for the presence of Xa21, NH1ox, and Snl6. Plants were moved to the growth chamber for challenge with Xoo and lesion lengths were assessed after 14 days. Primer sequences listed in Table S3.
Creation of snl6-RNAi
A silencing construct for LOC_Os01g45200 (Genbank: Os01g0639200) was created following our previously described strategy [36]. PCR was used to amplify 425 bp from the 3′ end of LOC_Os01g45200 (Primers: GAATTCAGGCTTCGATACGAGCATGT; AGATCTGTCGAATGCGACGGAGTAG). This PCR product was confirmed through sequencing and cloned in inverse orientation (separated by ∼1000 bp of GUS intron sequence) into a gateway compatible pENTR (Invitrogen) vector. The inverted repeat sequence was then recombined into a modified version of the binary vector Ubi-pC4300 encoding a gene for mannose selection.
snl6-RGT Allele Identification
snl6-RGT (RGT6140B_5.1) was identified from the Rice Transposon Flanking Sequence Tag (FST) Database: http://sundarlab.ucdavis.edu/rice/blast/blast.html. snl6-RGT was crossed with snl6-FN (pollen donor) and the cross was confirmed through PCR with primers specific to the NH1ox transgene (not present in snl6-RGT).
Realtime Quantitative RT-PCR Analysis
Realtime reactions were prepared using Bio-RAD SsoFast EvaGreen Supermix and run on a BIO-RAD CFX96 Real-time System. Primers were designed using the Beacon Designer program and R2 and efficiency were determined for each primer pair (Tables S2 and S4). When gene expression from a single plant is displayed, error bars represent standard deviation of 3 technical replicates. When multiple plants are combined, data represent biological replicates (each with 3 technical replicates) for each genotype and the s.e.m and number of individuals (n) is reported.
Phloroglucinol Staining
Leaf tissue was collected from plants just before the emergence of the panicle and cleared in a solution of ethanol, lactic acid and phenol (2∶1∶1) as previously described [27]. Cleared tissue was then soaked in 0.6% Phloroglucinol (in 2∶1 ethanol/HCL). Tissue was vacuum infiltrated for one hour and then left in the dark over night. Note: the reported phenotype is dependent on the tissue type, age and health of the plants. Altered phenolic content in snl6 mutant plants was not observed in roots or stems, in plants showing high levels of stress or in young plants.
Sugar Analysis
Alcohol insoluble residue (AIR) was prepared from ground mature rice leaves and destarched with alpha-amylase (Megazyme). All analyses were done using five mg of destarched AIR. For hemicellulose sugar composition, AIR was treated with 2M TFA at 120C for 1 h, dried and re-suspended in water; an aliquot was taken for analysis by high performance anion exchange chromatography with pulse amperometry detection (HPAEC-PAD) using a Carbopac PA20 analytical column (Dionex) and monosaccharide standards. Cellulose content was estimated as glucose equivalents using HPAEC-PAD from AIR treated with sulfuric acid (72% w/w) for 1 h at 30C, then diluted to 4% and incubated at 120C for 1 h. For enzymatic saccharification, AIR was first pre-treated in water at 100C for 1 h, then a mixture of cellulase and beta-glucosidase (5% w/w dose, Novozyme) in 0.1 M citrate buffer, pH 5.0 was added (reaction volume of 1% total solids); samples were incubated at 50C for 8 h and an aliquot was then taken to calculate total reducing sugar amounts using the DNS assay.
Sequence Analysis
Protein sequences were obtained from NCBI, TIGR or the JGI Brachypodium resource and compared using the web-based ClustalW software (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Supporting Information
snl6-FN over-expresses NH1. Quantitative RT-PCR was used to determine the relative amounts of NH1 expression in LG, NH1ox and snl6-FN (line: 11-3-2-1) plants. Mean ± s.e.m, n = 3.
(0.21 MB TIF)
Annotated genes in Deletion 1B and schematic of snl6 RNAi and insertion lines. (Top) CGH results for Deletion 1B showing predicted gene models. Gene annotation is based on TIGR v 5. (Bottom) Schematic of snl6-RNAi and snl6-RGT. An inverted repeat RNAi construct was created to silence Os01g45200. PCR was used to amplify the 3′ end of Os01g45200 and the resulting fragment was cloned in inverse orientation, separated by an approximately 1 kb spacer. To identify the location of the insertion in snl6-RGT (line: RGT6140B_5.1), insertion specific primers (Ds5'-2a) were combined with primers specific to Os01g45200. The resulting PCR product was sequenced and revealed that the insertion is in the first intron of Os01g45200.
(0.64 MB TIF)
Four snl6-RNAi T0 lines are susceptible to Xoo. Eight-week old independently transformed lines were challenged with Xoo. Lesion length development after 12 days. Mean ± s.d., n = 3 are displayed.
(0.22 MB TIF)
Protein alignment for Snl6 and its homologues from other species. Close homologs were collected from NCBI (maize: ACR34585.1, Sorghum: EES03334.1), The Brachypodium Sequence Resource (JGI) (2g44800.1), TAIR (Arabidopsis: AT5G14700) and TIGR (rice: Os05g50250). Previously described CCRs were found through literature searches and sequences collected in NCBI as follows: Arabidopsis (AtCCR1: NP_173047, AtCCR2: NP_178197), poplar (PtCCR: AJ224986), switchgrass (PvCCR1a: GQ450296 PvCCR2a: GQ450301), and rice (OsCCR1). All sequences were compared using ClustalW2 web-based software.
(0.52 MB TIF)
Snl6 mutants display no obvious developmental defects. Plant height and tiller number were evaluated at 8 weeks and a picture was taken of each genotype. Seed set was evaluated (total panicle weight) after senescence. LG, NH1ox and snl6-FN (line 11-3-2-1) are compared. n>10; s.d. from the mean.
(0.77 MB TIF)
Sugar analysis reveals higher sugar release from snl6 mutant lines. Cell wall extraction (AIR) was performed on LG, NH1ox and snl6-FN (lines 11-3-2 or 11-3-4) adult rice leaves. Overall cell wall sugar content (A) and relative sugar release (enzymatic saccharification) after hot water pre-treatment (B), ± s.e.m. n = 3. Letters (a or b) indicate significant difference (p<0.05).
(0.38 MB TIF)
Complete list of deleted genes in snl6-FN.
(0.06 MB PDF)
Efficiency (E) and R2 values for realtime PCR primers.
(0.06 MB PDF)
Genotyping primers.
(0.06 MB PDF)
Realtime PCR primers.
(0.05 MB PDF)
Acknowledgments
We thank Venkatesan Sundaresan for the snl6-RGT line, Peijian Cao for his assistance on the gene expression analysis, Christian Gates at Nimblegen for construction and analysis of the arrays, Henrik Scheller and Chithra Manisseri for the monosaccharide analysis protocol and Brad Holmes for helpful suggestions on the saccharification experiment. We thank Laura Bartley and Sarah Hake for helpful discussions and advice.
Footnotes
The authors have declared that no competing interests exist.
RSB has been supported by a William G. and Kathleen Golden International Agriculture Fellowship, UC Davis Consortium for Women in Research Fellowship, D. Marlin Brandon Fellowship and a Henry A. Jastro and Peter J. Shields Graduate Research Scholarship. This research was supported by NIH (2-R01-GM055962-09), DOE (DE-AC02-05CH11231) and USDA (Functional Genomics of Agriculturally Important Organisms, #2004-63560416640). This work was part of the DOE Joint BioEnergy Institute (www.jbei.org) supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Panstruga R, Parker JE, Schulze-Lefert P. SnapShot: Plant immune response pathways. Cell. 2009;136:978e971–973. doi: 10.1016/j.cell.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 2.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohn J, Sessa G, Martin GB. Innate immunity in plants. Curr Opin Immunol. 2001;13:55–62. doi: 10.1016/s0952-7915(00)00182-5. [DOI] [PubMed] [Google Scholar]
- 4.Quentin M, Allasia V, Pegard A, Allais F, Ducrot PH, et al. Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog. 2009;5:e1000264. doi: 10.1371/journal.ppat.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nurnberger T, Lipka V. Non-host resistance in plants: new insights into an old phenomenon. Molecular Plant Pathology. 2005;6:335–345. doi: 10.1111/j.1364-3703.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- 6.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 7.Menden B, Kohlhoff M, Moerschbacher BM. Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry. 2007;68:513–520. doi: 10.1016/j.phytochem.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Wu G, Shortt BJ, Lawrence EB, Leon J, Fitzsimmons KC, et al. Activation of Host Defense Mechanisms by Elevated Production of H2O2 in Transgenic Plants. Plant Physiol. 1997;115:427–435. doi: 10.1104/pp.115.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis Mutant That Is Nonresponsive to Inducers of Systemic Acquired Resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 11.Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Fan W, Dong X. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell. 2002;14:1377–1389. doi: 10.1105/tpc.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rairdan GJ, Delaney TP. Role of Salicylic Acid and NIM1/NPR1 in Race-Specific Resistance in Arabidopsis. Genetics. 2002;161:803–811. doi: 10.1093/genetics/161.2.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact. 2005;18:511–520. doi: 10.1094/MPMI-18-0511. [DOI] [PubMed] [Google Scholar]
- 15.Wu C, Bordeos A, Madamba MR, Baraoidan M, Ramos M, et al. Rice lesion mimic mutants with enhanced resistance to diseases. Mol Genet Genomics. 2008;279:605–619. doi: 10.1007/s00438-008-0337-2. [DOI] [PubMed] [Google Scholar]
- 16.Lorrain S, Vailleau F, Balague C, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8:263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- 17.Rostoks N, Borevitz JO, Hedley PE, Russell J, Mudie S, et al. Single-feature polymorphism discovery in the barley transcriptome. Genome Biol. 2005;6:R54. doi: 10.1186/gb-2005-6-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mockler TC, Chan S, Sundaresan A, Chen H, Jacobsen SE, et al. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85:1–15. doi: 10.1016/j.ygeno.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Kumar R, Qiu J, Joshi T, Valliyodan B, Xu D, et al. Single feature polymorphism discovery in rice. PLoS One. 2007;2:e284. doi: 10.1371/journal.pone.0000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong JM, Waner DA, Horie T, Li SL, Horie R, et al. Microarray-based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:15404–15409. doi: 10.1073/pnas.0404780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazen SP, Borevitz JO, Harmon FG, Pruneda-Paz JL, Schultz TF, et al. Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 2005;138:990–997. doi: 10.1104/pp.105.061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruce M, Hess A, Bai J, Mauleon R, Diaz MG, et al. Detection of genomic deletions in rice using oligonucleotide microarrays. BMC Genomics. 2009;10:129. doi: 10.1186/1471-2164-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, et al. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 2003;13:513–523. doi: 10.1101/gr.541303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung DY, Kim TH, Komives EA, Mendoza-Cozatl DG, Schroeder JI. ARS5 is a component of the 26S proteasome complex, and negatively regulates thiol biosynthesis and arsenic tolerance in Arabidopsis. Plant J. 2009;59:802–813. doi: 10.1111/j.1365-313X.2009.03914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song WY, Wang GL, Chen LL, Kim HS, Pi LY, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 26.Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, et al. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science. 2009;326:850–853. doi: 10.1126/science.1173438. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald HA, Chern MS, Navarre R, Ronald PC. Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol Plant Microbe Interact. 2004;17:140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- 28.Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 29.Bignell GR, Huang J, Greshock J, Watt S, Butler A, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo DM, Wilson JW, Fogarty TN, Buck WW. A nuclear fragmentation energy deposition model. IEEE Trans Nucl Sci. 1991;38:1–8. [PubMed] [Google Scholar]
- 31.Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K, et al. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc Natl Acad Sci U S A. 2006;103:230–235. doi: 10.1073/pnas.0509875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, et al. Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry. 2001;57:1187–1195. doi: 10.1016/s0031-9422(01)00053-x. [DOI] [PubMed] [Google Scholar]
- 33.Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, et al. Establishing an efficient Ac/Ds tagging system in rice: Large-scale analysis of Ds flanking sequences. Plant Journal. 2004;37:301–314. doi: 10.1046/j.1365-313x.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 34.Leple JC, Dauwe R, Morreel K, Storme V, Lapierre C, et al. Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell. 2007;19:3669–3691. doi: 10.1105/tpc.107.054148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escamilla-Trevino LL, Shen H, Uppalapati SR, Ray T, Tang Y, et al. Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol. 2009 doi: 10.1111/j.1469-8137.2009.03018.x. [DOI] [PubMed] [Google Scholar]
- 36.Chern M, Canlas PE, Fitzgerald HA, Ronald PC. Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J. 2005;43:623–635. doi: 10.1111/j.1365-313X.2005.02485.x. [DOI] [PubMed] [Google Scholar]
- 37.Park CJ, Bart R, Chern M, Canlas PE, Bai W, et al. Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS One. 2010;5:e9262. doi: 10.1371/journal.pone.0009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 39.Jackson LA, Shadle GL, Zhou R, Nakashima J, Chen F, et al. Improving Saccharification Efficiency of Alfalfa Stems Through Modification of the Terminal Stages of Monolignol BIosynthesis. Bioenergy Research. 2008;1:180–192. [Google Scholar]
- 40.Jones L, Ennos AR, Turner SR. Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J. 2001;26:205–216. doi: 10.1046/j.1365-313x.2001.01021.x. [DOI] [PubMed] [Google Scholar]
- 41.Mir Derikvand M, Sierra JB, Ruel K, Pollet B, Do CT, et al. Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta. 2008;227:943–956. doi: 10.1007/s00425-007-0669-x. [DOI] [PubMed] [Google Scholar]
- 42.Ruel K, Berrio-Sierra J, Derikvand MM, Pollet B, Thevenin J, et al. Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol. 2009;184:99–113. doi: 10.1111/j.1469-8137.2009.02951.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Zhang Y, Clarke JD, Li Y, Dong X. Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell. 1999;98:329–339. doi: 10.1016/s0092-8674(00)81962-5. [DOI] [PubMed] [Google Scholar]
- 45.Peng Y, Bartley LE, Chen X, Dardick C, Chern M, et al. OsWRKY62 is a Negative Regulator of Basal and Xa21-Mediated Defense against Xanthomonas oryzae pv. oryzae in Rice. Molecular Plant. 2008;1:446–458. doi: 10.1093/mp/ssn024. [DOI] [PubMed] [Google Scholar]
- 46.Park CJ, Peng Y, Chen X, Dardick C, Ruan D, et al. Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol. 2008;6:e231. doi: 10.1371/journal.pbio.0060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lacombe E, Hawkins S, Van Doorsselaere J, Piquemal J, Goffner D, et al. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 1997;11:429–441. doi: 10.1046/j.1365-313x.1997.11030429.x. [DOI] [PubMed] [Google Scholar]
- 48.Ralph J, Hatfield RD, Piquemal J, Yahiaoui N, Pean M, et al. NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamylalcohol dehydrogenase and cinnamoyl-CoA reductase. Proc Natl Acad Sci U S A. 1998;95:12803–12808. doi: 10.1073/pnas.95.22.12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chabannes M, Barakate A, Lapierre C, Marita JM, Ralph J, et al. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 2001;28:257–270. doi: 10.1046/j.1365-313x.2001.01140.x. [DOI] [PubMed] [Google Scholar]
- 50.Goujon T, Ferret V, Mila I, Pollet B, Ruel K, et al. Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: effects on phenotype, lignins and cell wall degradability. Planta. 2003;217:218–228. doi: 10.1007/s00425-003-0987-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
snl6-FN over-expresses NH1. Quantitative RT-PCR was used to determine the relative amounts of NH1 expression in LG, NH1ox and snl6-FN (line: 11-3-2-1) plants. Mean ± s.e.m, n = 3.
(0.21 MB TIF)
Annotated genes in Deletion 1B and schematic of snl6 RNAi and insertion lines. (Top) CGH results for Deletion 1B showing predicted gene models. Gene annotation is based on TIGR v 5. (Bottom) Schematic of snl6-RNAi and snl6-RGT. An inverted repeat RNAi construct was created to silence Os01g45200. PCR was used to amplify the 3′ end of Os01g45200 and the resulting fragment was cloned in inverse orientation, separated by an approximately 1 kb spacer. To identify the location of the insertion in snl6-RGT (line: RGT6140B_5.1), insertion specific primers (Ds5'-2a) were combined with primers specific to Os01g45200. The resulting PCR product was sequenced and revealed that the insertion is in the first intron of Os01g45200.
(0.64 MB TIF)
Four snl6-RNAi T0 lines are susceptible to Xoo. Eight-week old independently transformed lines were challenged with Xoo. Lesion length development after 12 days. Mean ± s.d., n = 3 are displayed.
(0.22 MB TIF)
Protein alignment for Snl6 and its homologues from other species. Close homologs were collected from NCBI (maize: ACR34585.1, Sorghum: EES03334.1), The Brachypodium Sequence Resource (JGI) (2g44800.1), TAIR (Arabidopsis: AT5G14700) and TIGR (rice: Os05g50250). Previously described CCRs were found through literature searches and sequences collected in NCBI as follows: Arabidopsis (AtCCR1: NP_173047, AtCCR2: NP_178197), poplar (PtCCR: AJ224986), switchgrass (PvCCR1a: GQ450296 PvCCR2a: GQ450301), and rice (OsCCR1). All sequences were compared using ClustalW2 web-based software.
(0.52 MB TIF)
Snl6 mutants display no obvious developmental defects. Plant height and tiller number were evaluated at 8 weeks and a picture was taken of each genotype. Seed set was evaluated (total panicle weight) after senescence. LG, NH1ox and snl6-FN (line 11-3-2-1) are compared. n>10; s.d. from the mean.
(0.77 MB TIF)
Sugar analysis reveals higher sugar release from snl6 mutant lines. Cell wall extraction (AIR) was performed on LG, NH1ox and snl6-FN (lines 11-3-2 or 11-3-4) adult rice leaves. Overall cell wall sugar content (A) and relative sugar release (enzymatic saccharification) after hot water pre-treatment (B), ± s.e.m. n = 3. Letters (a or b) indicate significant difference (p<0.05).
(0.38 MB TIF)
Complete list of deleted genes in snl6-FN.
(0.06 MB PDF)
Efficiency (E) and R2 values for realtime PCR primers.
(0.06 MB PDF)
Genotyping primers.
(0.06 MB PDF)
Realtime PCR primers.
(0.05 MB PDF)