Abstract
Glyconanomaterials, nanomaterials carrying surface-tethered carbohydrate ligands, have emerged and demonstrated increasing potential in biomedical imaging, therapeutics, and diagnostics. These materials combine the unique properties of nanometer-scale objects with the ability to present multiple copies of carbohydrate ligands, greatly enhancing the weak affinity of individual ligands to their binding partners. Critical to the performance of glyconanomaterials is the proper display of carbohydrate ligands, taking into consideration of the coupling chemistry, the type and length of the spacer linkage, and the ligand density. This article provides an overview of the coupling chemistry for attaching carbohydrate ligands to nanomaterials, and discusses the need for thorough characterization of glyconanomaterials, especially quantitative analyses of the ligand density and binding affinities. Using glyconanoparticles synthesized by a versatile photocoupling chemistry, methods for determining the ligand density by colorimetry and the binding affinity with lectins by a fluorescence competition assay are determined. The results show that the multivalent presentation of carbohydrate ligands significantly enhances the binding affinity by several orders of magnitude in comparison to the free ligands in solution. The effect is sizeable even at low surface ligand density. The type and length of the spacer linkage also affect the binding affinity, with the longer linkage promoting the association of bound ligands with the corresponding lectins.
1. Introduction
Carbohydrates are the most abundant biomolecules in nature and essential elements in a wide range of processes in living systems. Besides their uses as structural materials and energy sources, they are to large extents mediating recognition events through their interactions with proteins and other biological entities. Complex carbohydrate structures are thus involved in, for example, cell communication and trafficking, tumor genesis and progression, immune responses, fertilization, apoptosis, and infection.[1–7] Many challenges are, however, associated with the study of these processes, and the development of glycoscience has been largely hampered by the complexity and low abundance of the glycan structures involved and by the weak affinities often associated with carbohydrate–protein interactions. The field has recently experienced a dramatic upsurge, much on account of the very strong developments in carbohydrate synthesis, glycan analysis methods, and nanotechnology.[8] New synthetic methods, such as automated strategies and enzyme-mediated protocols, have resulted in increased availability of complex carbohydrate structures promoting advances in the entire field.[9–11]
An important development in the field of glycoscience is the discovery that in biological systems, carbohydrates bind lectins, i.e., carbohydrate-binding proteins, in a highly cooperative manner to improve the weak affinity of individual carbohydrate ligands to the lectin.[12–15] This cluster or multivalency effect involves multiple carbohydrate ligands and lectins interacting with each other, enhancing the binding affinity by several orders of magnitude. For example, oligosaccharides exhibit higher binding affinity than monosaccharides towards the same lectin. In biological systems, lectins associate with cells by interacting with the multiple copies of carbohydrate ligands on the cell surface, exhibiting binding affinities significantly higher than those of the interactions between the lectins and the isolated carbohydrate ligands. Although the quantitative aspect of the multivalency effect is yet to be established, the fact that multivalency can significantly enhance binding affinity has helped fuel a renewed interest in fundamental glycoscience and glycomaterial development. Extensive work has been conducted for conjugating carbohydrates to the scaffolds of proteins, peptides, lipids, and synthetic polymers.[15,16] Synthetic strategies are applied to control the number of ligands on the scaffold, the spatial display of the ligands, and the structure of the scaffold, which, in turn, impact the binding affinity of the resulting glycoconjugates with their binding partners. These synthetic multivalent glycoconjugates could bind to receptors competitively, with the potential to serve as inhibitors displacing natural ligands in the applications of carbohydrate-based drug design and therapeutics.[17] When the scaffold is a flat solid surface, efficient glycan microarrays can be generated, facilitating the development of high-throughput analysis of ligand–protein interactions in applications of ligand screening and diagnostics.[18–25]
Nanomaterials as scaffolds for carbohydrate ligand display have recently emerged, and glyconanomaterials have thus been synthesized, demonstrating great potential in biomedical imaging, diagnostics, and therapeutics. Compared with molecular scaffolds, nanomaterials as ligand carriers offer a number of attractive features. Nanomaterials, being small in size, have high specific surface areas and can therefore accommodate highdensity ligands promoting multivalent interactions with their binding partners. The ligand density can be modulated by the size and shape of the nanomaterial, and multiple epitopes of the same ligand can be exposed and presented in a three-dimensional format. Nanomaterials possess unique optical, electronic, magnetic, and mechanical properties as well as chemical reactivities. These properties, together with their nanosized dimensions, allow for their incorporation into cells for in vitro and in vivo imaging, drug-delivery, and targeting tumor cells. This opens up a wide range of possibilities, the potential of which is just emerging.[26–31]
A critical step in the preparation of glyconanomaterials is the surface coupling chemistry for attaching carbohydrates to the nanomaterial.[32] Nanomaterials come in different forms, sizes, and shapes. Conjugation chemistry should therefore be designed by taking into consideration the chemical nature of the nanomaterial to afford efficient ligand coupling and to provide optimal ligand presentation. In this Research News article, we begin by discussing surface conjugation chemistries for the synthesis of glyconanomaterials, followed by the photocoupling method developed in our laboratory including the process of surface functionalization and conjugation of carbohydrates to gold nanoparticles. A competition assay was developed to determine the apparent dissociation constants of the resulting glyconanomaterials with lectins. Our results show that the binding affinity of the resulting glyconanoparticles (GNPs) is profoundly affected by how the carbohydrate ligands are presented on the nanoparticle surface.
2. Conjugation of Carbohydrates to Nanomaterials
Two general strategies for nanomaterial functionalization can be discerned, based on either noncovalent or covalent protocols. Both approaches are associated with advantages and drawbacks, although covalent protocols are generally preferred due to the considerably higher stabilities of the constructs.
2.1. Noncovalent Attachment
A variety of glyconanomaterials based on physisorption of carbohydrate ligands to the material surface has been reported. The attachment relies on noncovalent interactions, including, for example, hydrogen bonding, Coulombic interactions, and hydrophobic effects. A method for producing metallic glyconanoparticles through electrostatic adsorption was reported by Yang and co-workers, in which metal/chitosan nanocomposites were prepared on a range of different metals, including Au, Ag, Pt, and Pd.[33] The nanoparticles were synthesized by reducing metal salts in the presence of chitosan, resulting in simultaneous ligand adsorption. Rosenzweig et al. synthesized dextran-coated quantum dots (QDs) where negatively charged carboxymethyldextran was adsorbed onto QDs by mixing with positively charged polylysine via electrostatic interactions.[34] Khiar et al. functionalized carbon nanotubes (CNTs) with pyrene-modified neoglycolipids.[35] Carbohydrate-conjugated, self-assembled CNT bundles could be exfoliated, yielding individual functionalized nanotubes. As noticed from these examples, a notable advantage of the physisorption strategy is that the reaction conditions are relatively mild, and minimal chemical derivatization is required for the nanomaterial substrates and the carbohydrate ligands. Nevertheless, the physical adsorption is relatively random and disordered compared to covalent linkages. In addition, the association is not sufficiently strong, which may lead to potential bond breakage during interactions, as well as increased nonspecific or unexpected interactions with the target molecules. This can significantly affect the specificity and sensitivity in applications such as biological sensing and recognition. However, as demonstrated in the mentioned examples, oligomer/polymer-based ligands can to some extent circumvent the stability problems.
2.2. Covalent Attachment
The most commonly used method for conjugating carbohydrate structures to nanomaterials is based on covalent attachment. Among the various nanomaterials, gold nanoparticles (Au NPs) are the most extensively used scaffold materials especially in fundamental studies due to their ease of preparation, exceptional stability, and high reproducibility.[36] Au NPs of different sizes, shapes, and controlled dispersity can now be synthesized using simple solution-based methods. The well-established thiol– and disulfide–Au chemistry, first applied to nanoparticles using a two-phase system by Brust et al., allows the preparation of Au NPs with well-defined surfaces.[37] These surface ligands serve as a protective layer to provide high stability for the nanomaterials in media ranging from organic solvents to biological milieus. The chemistry has been widely adopted to prepare Au NPs modified with various functional groups, and biological molecules including DNA, proteins, peptides, and carbohydrates have all been successfully introduced into the system.[38–40] Penadés and co-workers reported the first synthesis of carbohydrate-functionalized Au NPs.[41,42] The trisaccharide determinant of the Lewisx (Lex) antigen was derivatized with an alkylthiol, and Lex-coated Au NPs were prepared by reducing HAuCl4 with NaBH4 in presence of the thiol-derivatized Lex. Based on this strategy, Au NPs functionalized with monosaccharides (glucose), disaccharides (maltose), and tetrasaccharides (Ley) were prepared and applied to the studies of various biological interactions.[43,44] Later, several other research groups utilized a similar strategy to produce Au and Ag glyconanoparticles using thiolated carbohydrates.[45–51] Furthermore, thiolated carbohydrate derivatives have been adopted in the preparation of glyco-quantum dots (GQDs).[52–54] Additional coupling methods based on the reaction of complementary functional groups have also been developed to facilitate the conjugation of carbohydrates other than the thiolated derivatives. Examples include coupling N-hydroxysuccinimide (NHS)-functionalized dextran to amine-functionalized Ag NPs,[55] and amine-derivatized carbohydrates to aldehyde-functionalized Au NPs.[56]
Current methods for the preparation of carbohydrate-conjugated nanomaterials generally require the use of derivatized carbohydrates, amenable to coupling to the chosen nanomaterial surface. Un-derivatized carbohydrate structures present a considerable challenge. A few reported examples apply to flat substrates in microarray construction. One approach used hydrazide-modified gold films, where the hydrazide reacted with the terminal aldehyde group of the carbohydrates.[57,58] A similar approach employed amine-functionalized surfaces and the coupling of the carbohydrates took place by reductive amination.[59,60] In both cases, reducing carbohydrates are necessary and, for monosaccharides, the coupled products often became acyclic and lost their binding affinities.
2.3. Photoinitiated Coupling of Un-derivatized Carbohydrates to Nanomaterials
We have developed a simple method for attaching un-derivatized carbohydrates to gold[61] and iron oxide nanoparticles.[62] The coupling chemistry is based on the well-established procedure for the covalent attachment of molecules and materials to solid substrates using functionalized perfluorophenylazides (PFPAs).[63–65] The azide moiety on PFPAs can be activated by UV light, converting into the highly active nitrene that undergoes insertion reaction into diverse CH bonds and addition reaction to C=C bonds. Polymers,[66–70] carbon nanotubes,[71] graphene,[72] and small organic molecules[73] have been successfully immobilized onto PFPA-modified flat substrates and nanoparticles, providing highly robust and stable linkages. Carbohydrates are another category of substances that are well-suited for this photoinitiated immobilization chemistry. Carbohydrates have a number of CH bonds that can be used for the insertion reaction with PFPA, while leaving OH groups intact for the binding interactions with lectins. More importantly, the coupling chemistry does not require chemical derivatization of the carbohydrates. This is especially attractive for higher carbohydrate structures, the syntheses of which are often complex and time-consuming due to the stereochemistry control and multiple protection/deprotection steps involved in the site-specific glycosylation and derivatization reactions. The photocoupling reaction is also facile and efficient, taking place in a few minutes at room temperature in the ambient environment. Photochemical methods for carbohydrate attachment have been explored and reported in the literature. Sprenger and co-workers employed carbenes to attach glycans and glycoconjugates in the fabrication of microarrays.[74] The photoactive aryltrifluoromethyldiazirine was conjugated to dextran and was then applied to glass slides to form a photoactive coating. Activation by UV light produced highly reactive carbenes, which attached glycans via insertion reactions. In the method of Wang et al., the photoactive species was a phthalimide chromophore that induces H abstraction and subsequent recombination reaction with neighboring molecules.[75,76] This photochemistry was adopted to covalently attach unmodified mono-, oligo-, and polysaccharides on glass slides. PFPA as the photocoupling agent was used by Addadi and co-workers in the preparation of hyaluronan-coated polystyrene beads.[77] PFPA was first coupled onto amino-capped polystyrene beads, and hyaluronan was subsequently immobilized by UV irradiation.
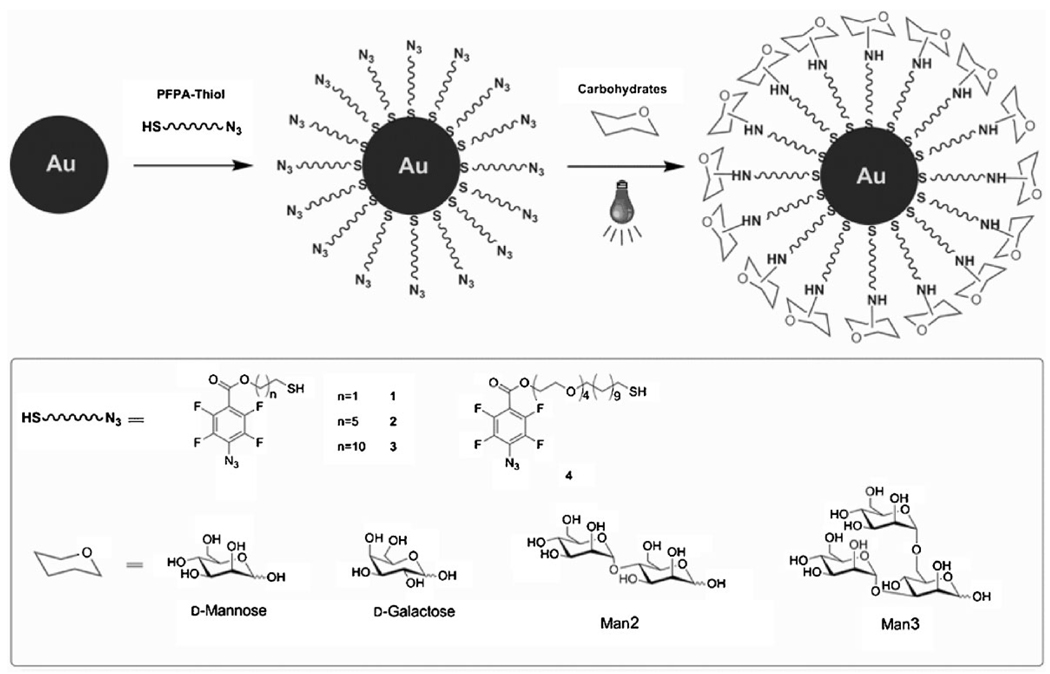
We now demonstrate that PFPAs can be employed to conjugate monosaccharides and oligosaccharides to nanomaterials. Compared with polysaccharides, mono- and oligo-saccharides are smaller carbohydrate structures and more challenging for this coupling chemistry. In principle, only one covalent bond is needed to attach the entire molecule to the surface. The probability of bond formation increases with the number of CH bonds, or the size of the carbohydrate structure. Indeed, our results showed that the coupling yield increased from 57% for d-mannose, to 74% and 81% for 2-O-α-d-mannopyranosyl-d-mannopyranose (Man2) and 3,6-di-O-(α-d-mannopyranosyl)-d-mannopyranose (Man3), respectively.
The key step in our photocoupling chemistry is the preparation of PFPA-functionalized nanomaterials. Depending on the chemical nature of the nanomaterial, the corresponding functionalized PFPA can be used. The synthesis of these PFPAs started with commercially available pentafluorobenzene derivatives that can be easily converted to the corresponding 4-azidotetrafluorobenzene derivatives via a facile nucleophilic substitution reaction using NaN3. Large quantities of these compounds can be prepared and they are stable when protected from light. From these precursors, a series of PFPAs carrying thiol,[78] disulfide,[61] phosphate,[62] and silane[68,70] groups were synthesized. We have successfully functionalized Au, iron oxide, and silica nanoparticles using these reagents. In the case of Au and silica nanoparticles, a one-pot process was developed, whereby the synthesized nanoparticles were functionalized in situ with the corresponding thiol- and silane-functionalized PFPAs. The presence of PFPA on the NPs was confirmed by NMR, FTIR, and X-ray photoelectron spectroscopy (XPS). UV–vis spectroscopy and transmission electron microscopy (TEM) images of PFPA/Au NPs showed excellent dispersibility and stability of these nanoparticles in organic solvents. To couple carbohydrates to the NPs, a solution of PFPA NPs mixed with the carbohydrate ligand was irradiated with 280-nm UV light for 5 min (Fig. 1) to yield nanoparticles that were well-dispersed and readily soluble in water.
Figure 1.
Synthesis of Au NPs functionalized with PFPA-thiol and subsequent photoinitiated coupling of carbohydrates.
The recognition ability of the surface-tethered carbohydrate ligands was evaluated using lectins. Concanavalin A (Con A), obtained from jack bean Canavalia ensiformis, is a well-studied lectin; its structure has been thoroughly characterized and established.[79] At pH above 7, Con A exists as a tetramer and each monomeric unit binds to α-d-mannopyranose with free –OH groups at the C-3, C-4, and C-6 positions, as well as α-linked oligomannosides. α-d-Glucopyranose and α-linked oligoglucosides such as maltooligosaccharides also bind Con A, although the affinity is lower than for mannopyranose.[80] The interactions of Con A with these ligands and derivatives have been investigated using various characterization methods, and the binding affinity and dissociation constants have been measured and reported.[81–83] These studies are used as the benchmark to validate our photocoupling chemistry.
The tetrameric Con A acts as a multifunctional crosslinking agent bringing together GNPs causing aggregation of mannose- or glucose-modified Au NPs. This was indeed observed in TEM as well as in UV–vis spectroscopy, where a large red shift in the plasmon resonance band occurred leading to solution color change from burgundy to blue. Au NPs functionalized with d-galactose did not induce aggregation or solution color change; neither did the PFPA-functionalized Au NPs. The method was further tested by treating nanoparticles bearing different carbohydrates with lectins including Griffonia simplicifolia lectin II (GS II), peanut agglutinin (PNA) and soybean agglutinin (SBA). Results showed the expected binding pattern of each carbohydrate ligand with its corresponding lectin, consistent with the solution binding studies reported.[84]
3. Characterization of Glyconanomaterials
Glyconanomaterials are synthesized under various conditions using specific chemistry and reagents. These materials must therefore be carefully evaluated to fully characterize the structure, composition, density of surface ligands, and biological activities in order to obtain proper correlation with their performances. Thanks to the significantly increased specific surface areas of nanomaterials, conventional chemical analytic techniques that are insensitive to flat substrates can be readily adopted for nanomaterial characterization. In the paper by Brust et al. on the preparation of thiol-capped Au NPs, the products were characterized by FTIR showing the presence of alkanethiol and TEM revealing the size and shape of the nanoparticles.[37] With the rapid development of advanced analytical tools, especially sensitive surface characterization techniques, nanomaterials can now be analyzed more accurately,[85] providing in-depth understanding of the chemical and physical properties of glyconanomaterials. NMR, FTIR, and surface-enhanced Raman spectroscopy (SERS) offer detailed structural analysis of nanomaterials and surface ligands. Thermogravimetric analysis (TGA) yields the amount of organic components on the nanomaterials, from which the ligand densities can be derived. Elemental analysis and XPS provide information on the elemental composition and chemical state of the bulk nanomaterials and the surface ligands. A combination of microscopy techniques, scanning probe techniques (STM, AFM), TEM, and small-angle X-ray scattering (SAXS) reveals the physical characteristics of size, shape, and assembly behavior of the nanomaterials.[86,87] Caution should be used when analyzing the results as the experimental conditions applied to each technique (vacuum, ambient, solution) can significantly impact the outcome. Microscopic techniques can also be used to directly visualize the interactions of glyconanomaterials with their binding partners. In our study, when d-mannose-functionalized iron oxide nanoparticles were treated with Escherichia coli strain ORN178, the nanoparticles selectively bound to the FimH lectin on the bacteria, which was clearly shown by TEM.[62] The surfaces can be further characterized by taking advantage of the unique properties offered by the nanomaterials. Classic examples are metal nanoparticles, which exhibit plasmon resonance that is highly sensitive to the surface constituents and can be conveniently monitored colorimetrically, as the molecular recognition event occurs at, or close to, the surface of the nanoparticles.[88]
4. Binding Affinity of Glyconanomaterials
Biomedical imaging, therapeutics, medical diagnosis, and drug delivery are among the many areas glyconanomaterials have the potential to impact. The interaction of glyconanomaterials with biological receptors and targets is a critical process involved in these applications and the binding affinity is thus an important parameter for evaluating the performance of glyconanomaterials. When a ligand is conjugated to a solid substrate, the structure of the ligand is in a sense altered. The binding affinity of the free ligand in solution can no longer be used as the substitute for the glyconanomaterial with the corresponding binding partner. In glyconanomaterials, multiple ligands are assembled on a single solid entity. Multivalency effect comes in play whereby ligands can act cooperatively enhancing the overall binding affinity with the receptor. This multivalency effect is highly sensitive to the manner in which the ligands are presented on the nanomaterial surface, i.e., the number of ligands or the ligand density, the structure and length of the spacer linkage, and how the ligand is attached or the coupling chemistry. Therefore the binding affinity of the glyconanomaterials must be carefully evaluated taking into consideration these parameters.
Carbohydrate–lectin interactions of free ligands in solution have been studied by many biochemical and biophysical methods including NMR spectroscopy,[89] surface plasmon resonance (SPR),[90,91] X-ray crystallography,[92,93] isothermal titration calorimetry (ITC),[94] and fluorescence spectroscopy.[95] Quantitative analysis of glyconanomaterials is investigated to a lesser extent and only a few protocols were reported to determine the binding affinity of glyconanoparticles. Lin and coworkers used SPR to analyze the multivalent interactions between mannose-, glucose-, or galactose-encapsulated gold nanoparticles with Con A.[45] A competition binding study was carried out where equilibria were established between mannopyranoside attached on the SPR sensor, Con A, and varied concentrations of mannose-encapsulated Au NPs. The dissociation constant Kd of mannose/Au NPs with Con A was determined to be 2.3 nM, representing a binding affinity over 5 orders of magnitude higher than that of the free d-mannopyranoside with Con A in solution (Kd = 470 µm measured by ITC[96]). In the system developed by Wu and co-workers, magnetite/gold core/shell nanoparticles coated with proteins were allowed to interact with carbohydrate ligands on a glycan array.[97] A magnetic field was applied to amplify the protein–carbohydrate interactions and the signals were visualized and quantified using a silver enhancement reagent. Apparent Kd values of 66 nm, 61 nm, and 57 nm were determined for Man, Man4, and Man9 ligands with Con A, respectively.
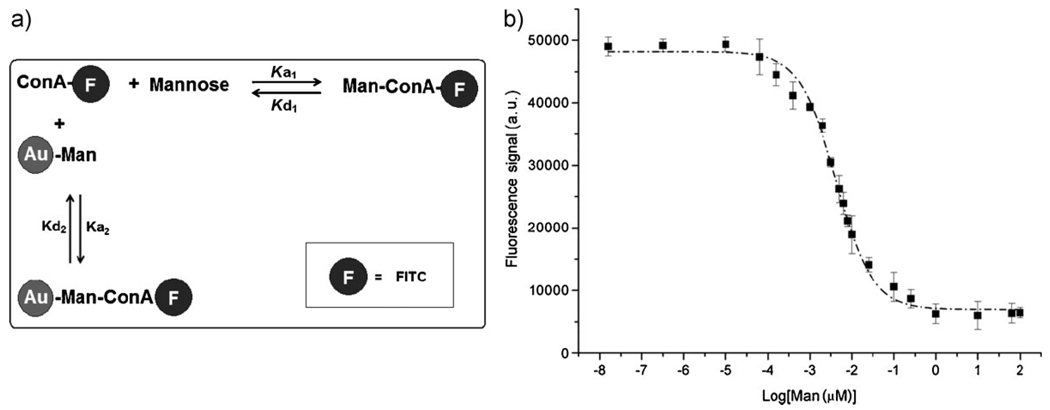
We have developed a fluorescence-based competition assay to determine the binding affinity of glyconanoparticles with lectins. In the assay, a fixed concentration of a free ligand (for example, d-mannose) and varying amounts of ligands bound to Au NPs were incubated with fluorescein isothiocyanate (FITC)-labeled Con A. The solution was then centrifuged and the fluorescence intensity of the supernatant was measured. Two equilibria co-exist in the system: FITC-Con A with free d-mannose and FITC-Con A with d-mannose bound on nanoparticles (Fig. 2). Since very low concentrations of Con A and free d-mannose were used, it was assumed that no agglomeration occurred. Both interactions are reversible, and steady equilibria are reached rapidly.
Figure 2.
a) Equilibria involved in the competition binding assay. b) Concentration dependent fluorescence intensity curve (right). [Man] is the concentration of d-mannose on NPs determined using the anthrone/H2SO4 colorimetry assay described in the text. Each data point is the average of 3 samples.
In order to calculate the binding affinity constant, the concentration of the carbohydrate ligand on Au NPs must be determined. The colorimetric assay of anthrone-sulfuric acid was adopted to measure the ligand density on the nanoparticles.[98] A calibration curve was first established using the corresponding free carbohydrate, and the amount of surface-bound ligand on the Au NPs was subsequently determined. The fluorescence intensity measured from the competition studies was plotted against the concentration of d-mannose on the Au NPs (Fig. 2). The result was a typical concentration-response curve for ligand–receptor binding, validating the assumptions made for the system. The concentration of ligands displaying 50% of specific binding (IC50) value was subsequently derived and the apparent dissociation constant (Kd2) calculated using the Cheng–Prusoff equation (Eq. (1))[99]
| (1) |
where [M] is the concentration of free ligand, i.e., d-mannose, Kd1 is dissociation constant of free ligand to Con A, and Kd2 is the apparent dissociation constant of surface bound d-mannose to Con A.
5. Ligand Presentation and Binding Affinity
Nanomaterials, being three-dimensional in shape and small in size, are capable of hosting ligands in higher densities in comparison to their flat counterparts due to greatly increased specific surface areas. This has significant implication for glyconanomaterials where the substrate configuration could dictate the ligand presentation and cooperativity and thus impact the interactions of the glyconanomaterials with their binding partners. The result is markedly enhanced affinities of these glyconanomaterials with the relevant biological targets. Data from our laboratory showed that the apparent Kd of d-mannose tethered on Au NPs with Con A can be as low as 0.43 nm (Table 1), representing a binding affinity of over six orders of magnitude higher than for free d-mannopyranoside with Con A. These results demonstrate that nanoparticles are excellent scaffolds for amplifying the weak affinities of carbohydrate ligands with lectins. Similar observations were reported by Sun and co-workers where single-walled carbon nanotubes functionalized with mannose and galactose selectively bound anthrax spores, inducing aggregation of the spores in the presence of Ca2+.[100] In contrast, carbohydrate-conjugated polystyrene beads did not exhibit the observed affinity towards anthrax spores. This was attributed to the ability of nanotubes to promote multivalent interactions of the carbohydrate ligands with receptors on the spores.
Table 1.
Binding affinity of d-mannose on Au NPs with Con A. Au NPs were functionalized with PFPA–thiol before d-mannose was coupled.
| PFPA–thiol | Spacer | Kd [nm] |
|---|---|---|
| 1 | CH2CH2 | 19 ± 2.2 |
| 2 | CH2(CH2)4CH2 | 15 ± 2.0 |
| 3 | CH2(CH2)9CH2 | 5.3 ± 0.72 |
| 4 | CH2(CH2)9CH2(OCH2CH2)4 | 0.43 ± 0.044 |
Unlike the free ligand that has the translational and rotational freedom in solution, the surface-bound ligand is no longer an unrestricted entity. Each ligand becomes a member of the nanomaterial carrier and can act cooperatively when interacting with their binding partners. The efficiency of the ligand association with the binding site, i.e., the binding affinity, is sensitive to a number of factors: how the ligand is attached, i.e., the coupling chemistry, the type and length of the spacer connecting the ligand and the nanomaterial, the flexibility/rigidity of the spacer, the density of ligands, and the distance between them. In the sections bellow, the impact of linker length and ligand density on the binding affinities of glyconanoparticles will be demonstrated using the latest results from our laboratory.
5.1. Spacer Linkage
Two types of spacer linkage were used in this study: ethylene and ethylene oxide (EO) chains. Alkyl groups promote self-organization in the monolayer formation whereas EO units are effective in improving water-solubility of nanomaterials. Using the photoinitiated coupling chemistry, we investigated the effects of linkers on the binding affinity of glyconanoparticles in carbohydrate–protein interactions. PFPA–thiols with different ethylene chain lengths (1, 2, 3) and containing additional EO units (4) were synthesized and used in the study (Fig. 1). Au NPs were first functionalized with each PFPA–thiol, and d-mannose was subsequently coupled to the NPs. It was found that the density of d-mannose coupled to Au NPs functionalized with PFPA–thiol 4 (82 nmol mg−1 NPs) was higher than those with the ethylene linkages (32, 40, and 58 nmol mg−1 NPs for 1, 2, and 3, respectively). The high coupling yield is likely due to a combination of the longer linker and enhanced water-solubility of NPs functionalized with PFPA–thiol 4. The binding affinity was subsequently measured using the fluorescence competition assay described above. Results show that the apparent Kd of Au/Man with Con A increased with the spacer length (Table 1). Similar observations were reported by Lin and co-workers, who found that the binding affinity increased with the spacer linker.[98] Remarkably, the binding affinity of glyconanoparticles prepared from PFPA–thiol 4 was more than 10 times higher than those with the ethylene linkages alone (Table 1). The longer and more flexible spacer may provide additional spatial freedom and less steric hindrance to the attached ligands for a more efficient association with their binding partners.
5.2. Ligand Density
Ligand density is another important parameter affecting the binding of surface-tethered carbohydrates. A few studies report the impact of ligand density on the binding affinity of carbohydrates immobilized on a flat surface, but the topic has not been extensively investigated for glyconanomaterials. Wong and co-workers used a fluorescence assay to analyze mannose–Con A interaction on glycan microarrays.[101] Kd decreased from 214 nM to 76.8 nM when the mannose printing concentration increased from 0.6 µm to 80 µm. However, Kd increased to 80.4 nm when the mannose printing concentration increased to 100 µm. Corn and co-workers employed the technique of SPR imaging to study carbohydrate–protein interactions using carbohydrate microarrays.[90] The binding affinity, measured by the adsorption coefficient (KADS), increased slightly from 5.0 × 106 M−1 to 5.6 × 106 M−1 when the surface mannose concentration increased from 10% to 50%, but remained unchanged up to 100% of the surface mannose density.
Using the photocoupling chemistry descried above, we synthesized glyconanoparticles and studied the relationship between ligand density and the binding affinity of the resulting glyconanoparticles. The ligand density was controlled by adding varying amounts of 1-hexanethiol to PFPA–thiol 3, and treating Au NPs with the mixed thiols before d-mannose was coupled. The ligand density was measured by the anthrone-sulfuric acid assay, and the apparent Kd values were determined by the fluorescence competition assay. Results show that the binding affinity increased with the ligand density (Table 2). Interestingly, the binding affinity of the glyconanoparticles with only 2.8% of surface coverage was over 3,800 times higher than that for free d-mannopyranoside with Con A in solution. The binding affinity increased 5.7 times from 123 nm to 21.4 nm when the ligand density increased only 2.6 times from 2.0 to 5.3 nmol mg−1 NPs. In both cases, multivalency could well be in play, demonstrating the enormous power of the ligand cooperativity.
Table 2.
Binding affinity versus ligand density. The Au NPs were functionalized with mixed thiols of 1-hexanethiol and PFPA–thiol 3.
| Percentage of PFPA–Thiol 3 |
d-Mannose Density [nmol mg−1 Au NPs] |
Surface Coverage [%] [a] |
Kd [nM] |
|---|---|---|---|
| 10% | 2.0 ± 0.54 | 2.8 | 123 ± 16 |
| 30% | 5.3 ± 0.83 | 7.4 | 21.4 ± 7.3 |
| 50% | 11.2 ± 2.32 | 16 | 16.3 ± 5.4 |
| 70% | 24.3 ± 2.78 | 34 | 14.4 ± 2.4 |
| 90% | 35.2 ± 1.29 | 49 | 12.7 ± 4.0 |
| 95% | 42.6 ± 3.81 | 59 | 9.4 ± 2.3 |
| 98% | 50.3 ± 4.17 | 70 | 6.7 ± 1.4 |
| 100% | 57.4 ± 3.21 | 80 | 5.3 ± 1.6 |
The surface coverage was defined as the percentage of ligands coupled versus the theoretical maximal number of ligands occupying the nanoparticle [61].
6. Conclusions
Merging nanotechnology with biology has seen exponential growth of research activities in functional bionanomaterials. The field of glyconanomaterials has begun to emerge, demonstrating increasing potential in applications where carbohydrate-based biological recognitions are called for. Essential to glyconanomaterial preparation is the conjugation chemistry that can efficiently attach carbohydrates to the nanomaterials. We developed a simple and versatile photocoupling chemistry that allows the covalent immobilization of a wide range of carbohydrate structures to nanomaterials. The coupling reaction was efficient and high yielding, and the resulting surface-bound carbohydrate ligands retained their binding affinities and selectivities. Nanomaterials as scaffolds are capable of presenting carbohydrate ligands such that the cooperativity greatly amplifies the affinity with the binding partners. Studies presented in this article demonstrated that surface-bound carbohydrate ligands exhibited binding affinities that were several orders of magnitude higher than those of the free ligands in solution. The strength of the interactions can furthermore be tuned by varying the ligand density, the spacer linkage between ligands and nanomaterials, and the coupling chemistry. These results highlight the importance of quantitative analysis of structural and functional properties of glyconanomaterials, especially the biorecognition properties that must be carefully analyzed in the context of ligand presentation and display.
Acknowledgements
This work was supported by the National Institute of General Medical Science (NIGMS) under the NIH award numbers R01GM080295, R01GM080295S1, and 2R15GM066279 and in part by the European Commission (MRTN-CT-19561). This article is part of a Special Issue on USTC Materials Science.
Contributor Information
Xin Wang, Department of Chemistry, Portland State University, P.O. Box 751, Portland, Oregon, 97207-0751 (USA).
Olof Ramström, Department of Chemistry, Portland State University, P.O. Box 751, Portland, Oregon, 97207-0751 (USA) Department of Chemistry, KTH - Royal Institute of Technology, Teknikringen 30, Stockholm, S-10044 (Sweden).
Mingdi Yan, Email: yanm@pdx.edu, Department of Chemistry Portland State University, P.O. Box 751, Portland, Oregon, 97207-0751 (USA).
References
- 1.Liu FT, Rabinovich GA. Nat. Rev. Cancer. 2005;5:29. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 2.Dube DH, Bertozzi CR. Nat. Rev. Drug Discovery. 2005;4:477. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 3.Fuster MM, Esko JD. Nat. Rev. Cancer. 2005;5:526. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 4.Szymanski CM, Wren BW. Nat. Rev. Microbiol. 2005;3:225. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 5.Ohtsubo K, Marth JD. Cell. 2006;126:855. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Sharon N. Biochim. Biophys. Acta, Gen. Subj. 2006;1760:527. doi: 10.1016/j.bbagen.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Crocker PR, Paulson JC, Varki A. Nat. Rev. Immunol. 2007;7:255. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 8.Blow N. Nature. 2009;457:617. doi: 10.1038/457617a. [DOI] [PubMed] [Google Scholar]
- 9.Sears P, Wong CH. Science. 2001;291:2344. doi: 10.1126/science.1058899. [DOI] [PubMed] [Google Scholar]
- 10.Koeller KM, Wong CH. Nature. 2001;409:232. doi: 10.1038/35051706. [DOI] [PubMed] [Google Scholar]
- 11.Seeberger PH. Chem. Soc. Rev. 2008;37:19. doi: 10.1039/b511197h. [DOI] [PubMed] [Google Scholar]
- 12.Lee YC, Lee RT. Acc. Chem. Res. 1995;28:321. [Google Scholar]
- 13.Mammen M, Choi SK, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 14.Huskens J. Curr. Opin. Chem. Biol. 2006;10:537. doi: 10.1016/j.cbpa.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Jayaraman N. Chem. Soc. Rev. 2009;38:3463. doi: 10.1039/b815961k. [DOI] [PubMed] [Google Scholar]
- 16.Drechsler U, Erdogan B, Rotello VM. Chem. Eur. J. 2004;10:5570. doi: 10.1002/chem.200306076. [DOI] [PubMed] [Google Scholar]
- 17.Ernst B, Magnani JL. Nat. Rev. Drug Discovery. 2009;8:661. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D. Proteomics. 2003;3:2167. doi: 10.1002/pmic.200300601. [DOI] [PubMed] [Google Scholar]
- 19.Feizi T, Chai W. Nat. Rev. Mol. Cell Biol. 2004;5:582. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 20.Stevens J, Blixt O, Paulson JC, Wilson IA. Nat. Rev. Microbiol. 2006;4:857. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coullerez G, Seeberger PH, Textor M. Macromol. Biosci. 2006;6:634. doi: 10.1002/mabi.200600090. [DOI] [PubMed] [Google Scholar]
- 22.Laurent N, Voglmeir J, Flitsch SL. Chem. Commun. 2008:4400. doi: 10.1039/b806983m. [DOI] [PubMed] [Google Scholar]
- 23.Park S, Lee MR, Shin I. Chem. Soc. Rev. 2008;37:1579. doi: 10.1039/b713011m. [DOI] [PubMed] [Google Scholar]
- 24.Horlacher T, Seeberger PH. Chem. Soc. Rev. 2008;37:1414. doi: 10.1039/b708016f. [DOI] [PubMed] [Google Scholar]
- 25.Liang PH, Wu CY, Greenberg WA, Wong CH. Curr. Opin. Chem. Biol. 2008;12:86. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Nat. Biotechnol. 2003;21:47. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 27.Rojo J, Díaz V, de la Fuente JM, Segura I, Barrientos AG, Riese HH, Bernade A, Penadés S. ChemBioChem. 2004;5:291. doi: 10.1002/cbic.200300726. [DOI] [PubMed] [Google Scholar]
- 28.Sihelnikova L, Tvaroska I. Chem. Pap. 2007;61:237. [Google Scholar]
- 29.De M, Ghosh PS, Rotello VM. Adv. Mater. 2008;20:4225. [Google Scholar]
- 30.Sperling R, Rivera GP, Zhang F, Zanella M, Parak W. Chem. Soc. Rev. 2008;37:1896. doi: 10.1039/b712170a. [DOI] [PubMed] [Google Scholar]
- 31.Cipolla L, Peri F, Airoldi C. Anti-Cancer Agents Med. Chem. 2008;8:92. doi: 10.2174/187152008783330815. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Liu LH, Ramström O, Yan M. Exp. Biol. Med. 2009;234:1128. doi: 10.3181/0904-MR-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H, Yuan Q, Yang X. Colloids Surf. B. 2004;39:31. doi: 10.1016/j.colsurfb.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Ji T, Rosenzweig Z. Nano Lett. 2003;3:581. [Google Scholar]
- 35.Assali M, Leal MP, Fernandez I, Baati R, Mioskowski C, Khiar N. Soft Matter. 2009;5:948. [Google Scholar]
- 36.El-Kouedi M, Keating CD. In: Nanobiotechnology. Niemeyer CM, Mirkin CA, editors. Germany: Wiley-VCH, Weinheim; 2004. [Google Scholar]
- 37.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J. Chem. Soc, Chem. Commun. 1994:801. [Google Scholar]
- 38.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 39.Niemeyer CM. Angew. Chem. Int. Ed. 2001;40:4128. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 40.Daniel MC, Astruc D. Chem. Rev. 2003;104:293. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 41.Rojas TC, de la Fuente JM, Barrientos AG, Penadés S, Ponsonnet L, Fernández A. Adv. Mater. 2002;14:585. [Google Scholar]
- 42.J. Hernáiz M, de la Fuente JM, Barrientos ÁG, Penadés S. Angew. Chem. Int. Ed. 2002;41:1554. doi: 10.1002/1521-3773(20020503)41:9<1554::aid-anie1554>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 43.de la Fuente JM, Eaton P, Barrientos AG, Menendez M, Penadés S. J. Am. Chem. Soc. 2005;127:6192. doi: 10.1021/ja0431354. [DOI] [PubMed] [Google Scholar]
- 44.de Paz JL, Ojeda R, Barrientos AG, Penadés S, Martin-Lomas M. Tetrahedron: Asymmetry. 2005;16:149. [Google Scholar]
- 45.Lin CC, Yeh YC, Yang CY, Chen GF, Chen YC, Wu YC, Chen CC. Chem. Commun. 2003;23:2920. doi: 10.1039/b308995a. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Geddes CD, Lakowicz JR. Anal. Biochem. 2004;332:253. doi: 10.1016/j.ab.2004.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halkes KM, de Souza Carvalho A, Maljaars CE, Gerwig GJ, Kamerling JP. Eur. J. Org. Chem. 2005;2005:3650. [Google Scholar]
- 48.Koen MH, Adriana Carvalho de S, Maljaars CEP, Gerrit JG, Johannis PK. Eur. J. Org. Chem. 2005;2005:3650. [Google Scholar]
- 49.Schofield CL, Field RA, Russell DA. Anal. Chem. 2007;79:1356. doi: 10.1021/ac061462j. [DOI] [PubMed] [Google Scholar]
- 50.Huang CC, Chen CT, Shiang YC, Lin ZH, Chang HT. Anal. Chem. 2009;81:875. doi: 10.1021/ac8010654. [DOI] [PubMed] [Google Scholar]
- 51.Marradi M, Alcántara D, de la Fuente JM, García-Martín ML, Cerdán S, Penadés S. Chem. Commun. 2009:3922. doi: 10.1039/b900957d. [DOI] [PubMed] [Google Scholar]
- 52.Robinson A, Fang JM, Chou PT, Liao KW, Chu RM, Lee SJ. ChemBioChem. 2005;6:1899. doi: 10.1002/cbic.200500112. [DOI] [PubMed] [Google Scholar]
- 53.Babu P, Sinha S, Surolia A. Bioconjugate Chem. 2007;18:146. doi: 10.1021/bc060204q. [DOI] [PubMed] [Google Scholar]
- 54.Svarovsky SA, Barchi JJJ. In: Frontiers in Modern Carbohydrate Chemistry. Demchenko AV, editor. Oxford, UK: Oxford University Press; 2007. [Google Scholar]
- 55.Earhart C, Jana NR, Erathodiyil N, Ying JY. Langmuir. 2008;24:6215. doi: 10.1021/la800066g. [DOI] [PubMed] [Google Scholar]
- 56.Otsuka H, Akiyama Y, Nagasaki Y, Kataoka K. J. Am. Chem. Soc. 2001;123:8226. doi: 10.1021/ja010437m. [DOI] [PubMed] [Google Scholar]
- 57.Lee MR, Shin I. Org. Lett. 2005;7:4269. doi: 10.1021/ol051753z. [DOI] [PubMed] [Google Scholar]
- 58.Zhi ZL, Powell AK, Turnbull JE. Anal. Chem. 2006;78:4786. doi: 10.1021/ac060084f. [DOI] [PubMed] [Google Scholar]
- 59.Seo JH, Adachi K, Lee BK, Kang DG, Kim YK, Kim KR, Lee HY, Kawai T, Cha HJ. Bioconjugate Chem. 2007;18:2197. doi: 10.1021/bc700288z. [DOI] [PubMed] [Google Scholar]
- 60.Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Nat. Methods. 2005;2:845. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, Ramström O, Yan M. J. Mater. Chem. 2009;19:8944. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu LH, Dietsch H, Schurtenberger P, Yan MD. Bioconjugate Chem. 2009;20:1349. doi: 10.1021/bc900110x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bartlett MA, Yan M. Adv. Mater. 2001;13:1449. [Google Scholar]
- 64.Yan M. Polym. News. 2002;27:6. [Google Scholar]
- 65.Yan M. Chem. Eur. J. 2007;13:4138. doi: 10.1002/chem.200700317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan M, Ren J. Chem. Mater. 2004;16:1627. [Google Scholar]
- 67.Yan M, Ren J. J. Mater. Chem. 2005;15:523. [Google Scholar]
- 68.Liu L, Engelhard MH, Yan M. J. Am. Chem. Soc. 2006;128:14067. doi: 10.1021/ja062802l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu L, Yan M. Angew. Chem. Int. Ed. 2006;45:6207. doi: 10.1002/anie.200602097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gann JP, Yan M. Langmuir. 2008;24:5319. doi: 10.1021/la7029592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pastine SJ, Okawa D, Kessler B, Rolandi M, Llorente M, Zettl A, Frechet JMJ. J. Am. Chem. Soc. 2008;130:4238. doi: 10.1021/ja8003446. [DOI] [PubMed] [Google Scholar]
- 72.Liu LH, Yan M. Nano Lett. 2009;9:3375. doi: 10.1021/nl901669h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Bataineh SA, Luginbuehl R, Textor M, Yan M. Langmuir. 2009;25:7432. doi: 10.1021/la900334w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H, Sprenger N. Glycobiology. 2005;15:31. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 75.Carroll GT, Wang DN, Turro NJ, Koberstein JT. Langmuir. 2006;22:2899. doi: 10.1021/la0531042. [DOI] [PubMed] [Google Scholar]
- 76.Carroll GT, Wang DN, Turro NJ, Koberstein JT. Glycoconjugate J. 2008;25:5. doi: 10.1007/s10719-007-9052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joester D, Klein E, Geiger B, Addadi L. J. Am. Chem. Soc. 2006;128:1119. doi: 10.1021/ja0537474. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, Ramström O, Yan M. unpublished. [Google Scholar]
- 79.Lis H, Sharon N. Chem. Rev. 1998;98:637. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 80.Baenziger JU, Fiete D. J. Biol. Chem. 1979;254:2400. [PubMed] [Google Scholar]
- 81.Anita L, Frank GL, Clement KB. Eur. J. Biochem. 1980;103:307. [Google Scholar]
- 82.Gupta D, Dam TK, Oscarson S, Brewer CF. J. Biol. Chem. 1997;272:6388. [PubMed] [Google Scholar]
- 83.Schwarz FP, Puri KD, Bhat RG, Surolia A. J. Biol. Chem. 1993;268:7668. [PubMed] [Google Scholar]
- 84.Pei ZC, Yu H, Theurer M, Walden A, Nilsson P, Yan M, Ramström O. ChemBioChem. 2007;8:166. doi: 10.1002/cbic.200600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grainger DW, Castner DG. Adv. Mater. 2008;20:867. [Google Scholar]
- 86.Hino K, Shingai R, Morita T, Toku K, Hirano T, Yoshikawa H, Nakano H, Nishi N. Chem. Phys. Lett. 2008;460:173. [Google Scholar]
- 87.Regev O, Backov R, Faure C. Chem. Mater. 2004;16:5280. [Google Scholar]
- 88.Sun Y, Xia Y. Analyst. 2003;128:686. doi: 10.1039/b212437h. [DOI] [PubMed] [Google Scholar]
- 89.Poveda A, Jimenez-Barbero J. Chem. Soc. Rev. 1998;27:133. [Google Scholar]
- 90.Smith EA, Thomas WD, Kiessling LL, Corn RM. J. Am. Chem. Soc. 2003;125:6140. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- 91.Beccati D, Halkes KM, Batema GD, Guillena G, de Souza AC, van Koten G, Kamerling JP. ChemBioChem. 2005;6:1196. doi: 10.1002/cbic.200400402. [DOI] [PubMed] [Google Scholar]
- 92.von Itzstein M, Colman P. Curr. Opin. Struct. Biol. 1996;6:703. doi: 10.1016/s0959-440x(96)80038-8. [DOI] [PubMed] [Google Scholar]
- 93.Fernandez MD, Canada FJ, Jimenez-Barbero J, Cuevas G. J. Am. Chem. Soc. 2005;127:7379. doi: 10.1021/ja051020+. [DOI] [PubMed] [Google Scholar]
- 94.Clarke C, Woods RJ, Gluska J, Cooper A, Nutley MA, Boons GJ. J. Am. Chem. Soc. 2001;123:12238. doi: 10.1021/ja004315q. [DOI] [PubMed] [Google Scholar]
- 95.Lee YC. J. Biochem. 1997;121:818. doi: 10.1093/oxfordjournals.jbchem.a021658. [DOI] [PubMed] [Google Scholar]
- 96.Sanders JN, Chenoweth SA, Schwarz FP. J. Inorg. Biochem. 1998;70:71. doi: 10.1016/s0162-0134(98)00016-6. [DOI] [PubMed] [Google Scholar]
- 97.Liang CH, Wang CC, Lin YC, Chen CH, Wong CH, Wu CY. Anal. Chem. 2009 doi: 10.1021/ac9012286. [DOI] [PubMed] [Google Scholar]
- 98.Chien Y, Jan M, Adak A, Tzeng H, Lin Y, Chen Y, Wang K, Chen C, Chen C, Lin C. ChemBioChem. 2008;9:1100. doi: 10.1002/cbic.200700590. [DOI] [PubMed] [Google Scholar]
- 99.Cheng YC, Prusoff WH. Biochem. Pharmacol. 1973;22:3099. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 100.Wang HF, Gu LR, Lin Y, Lu FS, Meziani MJ, Luo PGJ, Wang W, Cao L, Sun YP. J. Am. Chem. Soc. 2006;128:13364. doi: 10.1021/ja065455o. [DOI] [PubMed] [Google Scholar]
- 101.Liang PH, Wang SK, Wong CH. J. Am. Chem. Soc. 2007;129:11177. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]