Abstract
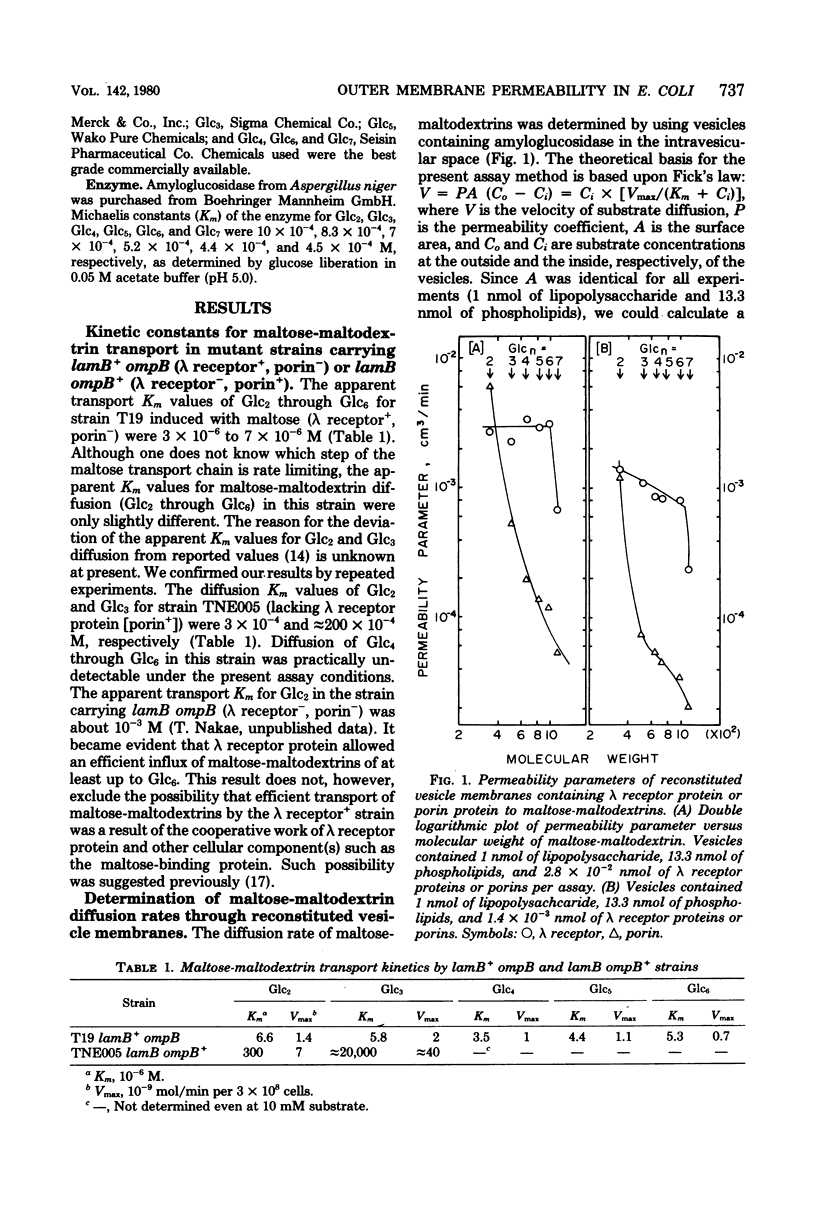
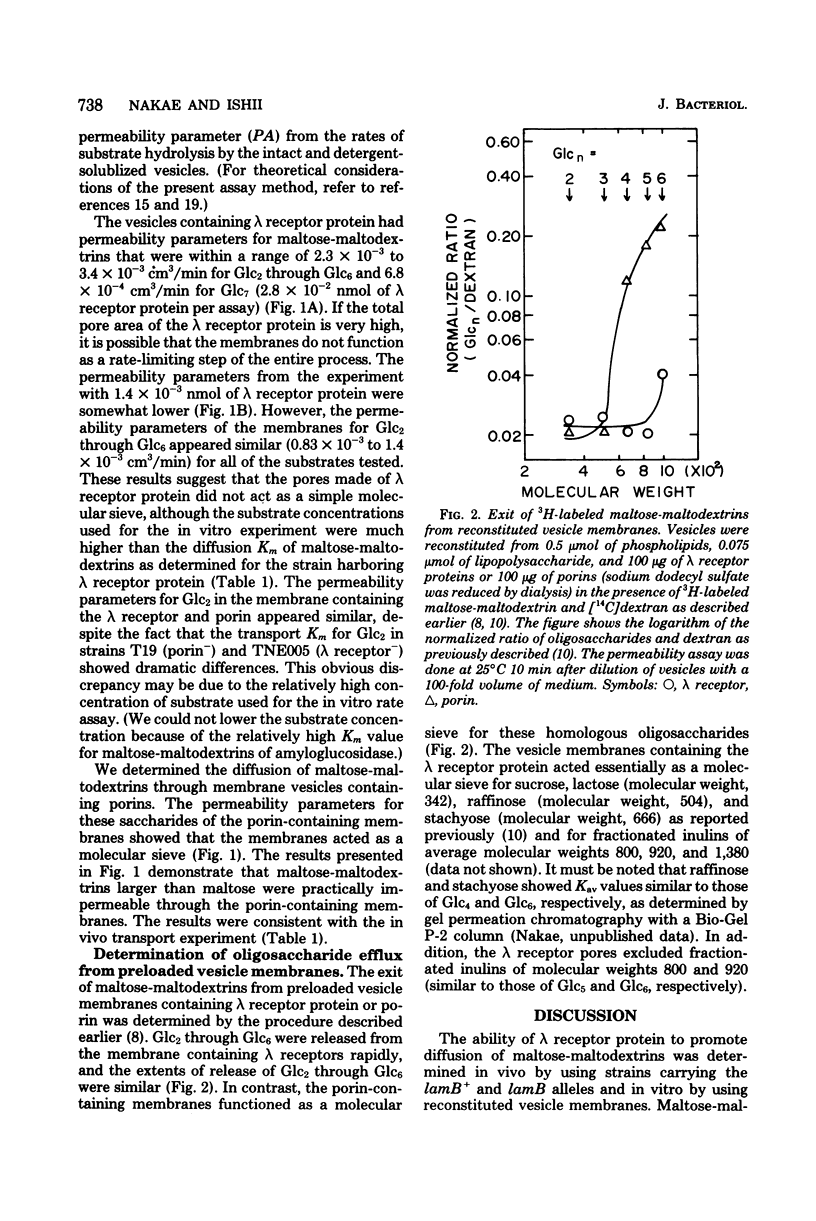
Outer membrane permeability conferred by lambda receptor protein and porins to maltose-maltodextrins and other oligosaccharides was studied in vitro with reconstituted vesicle membranes and in vivo with mutant strains lacking either one of these proteins. The vesicle membranes reconstituted from phospholipids, lipopolysaccharide, and purified lambda receptor allowed rapid diffusion of maltose and maltose-maltodextrins of up to six glucose residues, but the membranes acted essentially as a molecular sieve for sucrose, raffinose, stachyose, and inulins of molecular weights 800, 920, and 1,380. The vesicle membranes containing porins allowed rapid diffusion of maltose but not of maltose-maltodextrins larger than maltose. The apparent transport Km values for maltose-maltodextrins of up to six glucose residues from the strain carrying lamB+ ompB (lambda receptor+, porin-) were similar (about 5 X 10(-6) M), whereas the transport Km values for maltose- and maltotriose of the strain carrying lamB ompB+ (lambda receptor-, porin+) alleles appeared to be 300 and about 20,000 X 10(-6) M. These results suggest that lambda receptor protein forms permeability pores that facilitate the diffusion of maltose-maltodextrins and function as a molecular sieve for other saccharides.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J Biol Chem. 1974 Jan 25;249(2):634–644. [PubMed] [Google Scholar]
- Boehler-Kohler B. A., Boos W., Dieterle R., Benz R. Receptor for bacteriophage lambda of Escherichia coli forms larger pores in black lipid membranes than the matrix protein (porin). J Bacteriol. 1979 Apr;138(1):33–39. doi: 10.1128/jb.138.1.33-39.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edermann R., Hindennach I., Henning U. Major proteins of the Escherichia coli outer cell envelope membrane. Preliminary characterization of the phage lambda receptor protein. FEBS Lett. 1978 Apr 1;88(1):71–74. doi: 10.1016/0014-5793(78)80609-7. [DOI] [PubMed] [Google Scholar]
- Hofnung M. Divergent operons and the genetic structure of the maltose B region in Escherichia coli K12. Genetics. 1974 Feb;76(2):169–184. doi: 10.1093/genetics/76.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F. Role of a major outer membrane protein in Escherichia coli. J Bacteriol. 1977 Aug;131(2):631–637. doi: 10.1128/jb.131.2.631-637.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae T. A porin activity of purified lambda-receptor protein from Escherichia coli in reconstituted vesicle membranes. Biochem Biophys Res Commun. 1979 Jun 13;88(3):774–781. doi: 10.1016/0006-291x(79)91475-x. [DOI] [PubMed] [Google Scholar]
- Nakae T. Identification of the outer membrane protein of E. coli that produces transmembrane channels in reconstituted vesicle membranes. Biochem Biophys Res Commun. 1976 Aug 9;71(3):877–884. doi: 10.1016/0006-291x(76)90913-x. [DOI] [PubMed] [Google Scholar]
- Nakae T., Nikaido H. Outer membrane as a diffusion barrier in Salmonella typhimurium. Penetration of oligo- and polysaccharides into isolated outer membrane vesicles and cells with degraded peptidoglycan layer. J Biol Chem. 1975 Sep 25;250(18):7359–7365. [PubMed] [Google Scholar]
- Nakae T. Outer membrane of Salmonella typhimurium: reconstitution of sucrose-permeable membrane vesicles. Biochem Biophys Res Commun. 1975 Jun 16;64(4):1224–1230. doi: 10.1016/0006-291x(75)90823-2. [DOI] [PubMed] [Google Scholar]
- Nakae T. Outer membrane of Salmonella. Isolation of protein complex that produces transmembrane channels. J Biol Chem. 1976 Apr 10;251(7):2176–2178. [PubMed] [Google Scholar]
- Szmelcman S., Hofnung M. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J Bacteriol. 1975 Oct;124(1):112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmelcman S., Schwartz M., Silhavy T. J., Boos W. Maltose transport in Escherichia coli K12. A comparison of transport kinetics in wild-type and lambda-resistant mutants as measured by fluorescence quenching. Eur J Biochem. 1976 May 17;65(1):13–19. doi: 10.1111/j.1432-1033.1976.tb10383.x. [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Tokunaga H., Nakae T. The outer membrane permeability of Gram-negative bacteria: determination of permeability rate in reconstituted vesicle membranes. FEBS Lett. 1979 Oct 1;106(1):85–88. doi: 10.1016/0014-5793(79)80700-0. [DOI] [PubMed] [Google Scholar]
- WIESMEYER H., COHN M. The characterization of the pathway of maltose utilization by Escherichia coli. I. Purification and physical chemical properties of the enzyme amylomaltase. Biochim Biophys Acta. 1960 Apr 22;39:417–426. doi: 10.1016/0006-3002(60)90194-3. [DOI] [PubMed] [Google Scholar]
- Wandersman C., Schwartz M., Ferenci T. Escherichia coli mutants impaired in maltodextrin transport. J Bacteriol. 1979 Oct;140(1):1–13. doi: 10.1128/jb.140.1.1-13.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W., Rosselet A. Function of the outer membrane of Escherichia coli as a permeability barrier to beta-lactam antibiotics. Antimicrob Agents Chemother. 1977 Sep;12(3):368–372. doi: 10.1128/aac.12.3.368. [DOI] [PMC free article] [PubMed] [Google Scholar]



