Introduction
Cyclotides are fascinating micro-proteins present in plants from the Violaceae, Rubiaceae and also Cucurbitaceae and feature various biological actions such as protease inhibitory, anti-microbial, insecticidal, cytotoxic, anti-HIV or hormone-like activity.[1, 2] They share a unique head-to-tail circular knotted topology of three disulfide bridges, with one disulfide penetrating through a macrocycle formed by the two other disulfides and inter-connecting peptide backbones, forming what is called a cystine knot topology (Scheme 1A). Cyclotides have several characteristics that make them ideal drug development tools.[3, 4] The cystine knot and cyclic backbone topology makes them exceptionally resistant to thermal, chemical, and enzymatic degradation compared with other peptides of similar size.[5] Some cyclotides have been shown to be orally bioavailable. For example, the first cyclotide to be discovered, Kalata B1, was found to be an orally effective uterotonic,[6] and other cyclotides have been shown to cross the cell membrane through macropinocytosis.[7] Immunogenicity is generally considered not to be a major issue for small-sized and stable microproteins.[8, 9] Cyclotides are also amenable to substantial sequence variation and they can be considered as natural combinatorial peptide libraries structurally constrained by the cystine-knot scaffold and head-to-tail cyclization [2, 10]. Cyclotides can also be chemically synthesized thus allowing the introduction of specific chemical modifications or biophysical probes.[11–14] More importantly, cyclotides can be now biosynthesized in E. coli cells by using a biomimetic approach that involves the use of modified protein splicing units[15, 16] (Fig. 1), and therefore making them ideal scaffolds for molecular evolution strategies to enable generation and selection of compounds with optimal binding and inhibitory characteristics against particular molecular targets. Cyclotides thus appear as promising leads or frameworks for peptide drug design.[3, 4]
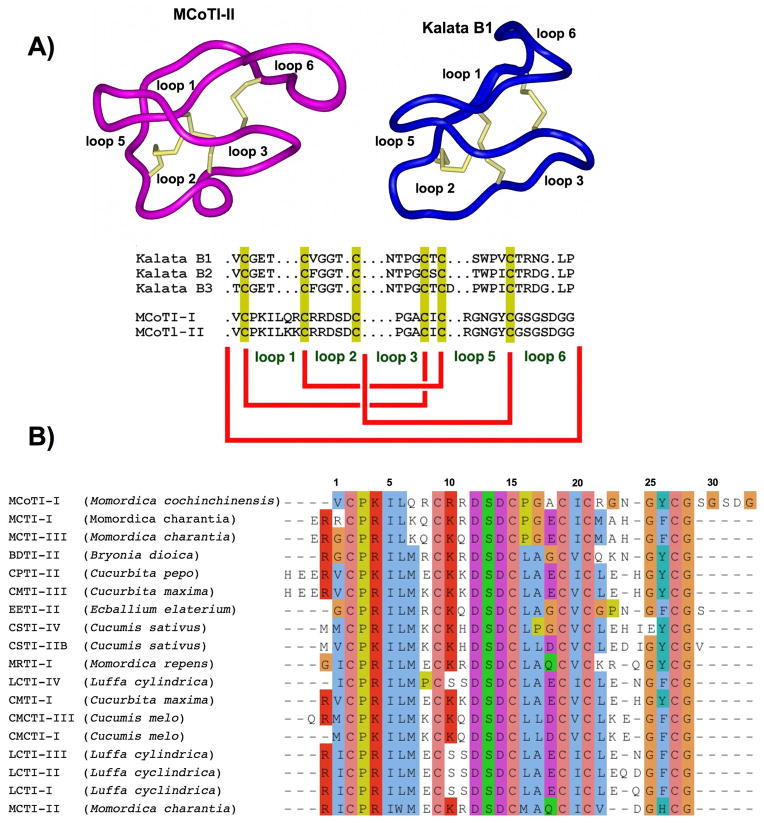
Scheme 1.
A. Primary and tertiary structure of MCoTI and Kalata cyclotides isolated from Momordica cochinchinensis and Oldenlandia affinis, respectively [6, 20, 21]. B. Multiple sequence alignment of cyclotide MCoTI-I with other squash trypsin inhibitors. Multiple sequence alignment was performed using TCoffee (http://ca.expasy.org/cgi-bin/hub) and visualized using Jalview [31].
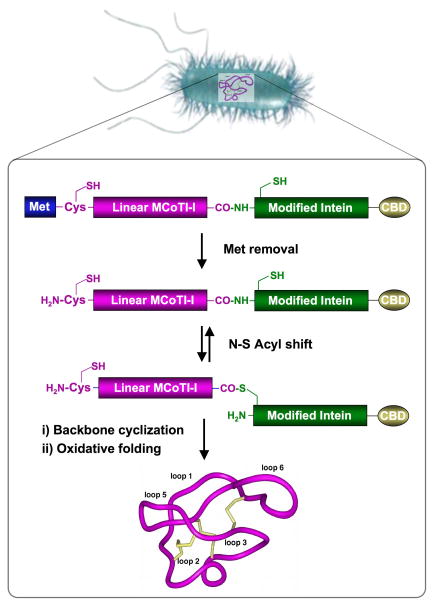
Figure 1.
Biosynthetic approach for the production of cyclotide MCoTI-I libraries inside living E. coli cells. Backbone cyclization of the linear precursor is mediated by a modified protein splicing unit or intein. The cyclized peptide then folds spontaneously in the bacterial cytoplasm.
Investigation of the contribution of individual residues to the structural integrity and biological activities of particular cyclotides is therefore crucial for their use in any potential pharmaceutical application.[17] A better understanding of the structural limitations of the cyclotide scaffold can greatly assist in the correct design of cyclotide-based libraries for molecular screening and selection of de-novo sequences with new biological activities or developing grafted analogues for use as peptide-based drugs,[14, 18] so that sequence modifications in structurally important regions are avoided. Understanding the molecular basis for bioactivity also may allow the minimization or avoidance of undesirable properties such as cytotoxic or hemolytic activity found in some cyclotides.[17]
The cyclotides MCoTI-I/II are powerful trypsin inhibitors which have been recently isolated from the dormant seeds of Momordica cochinchinensis, a plant member of cucurbitaceae family.[19] Although MCoTI cyclotides do not share significant sequence homology with other cyclotides beyond the presence of the three cystine bridges, solution NMR has shown that they adopt a similar backbone-cyclic cystine-knot topology [20, 21] (Scheme 1A). MCoTI cyclotides, however, share a high sequence homology with related cystine-knot trypsin inhibitors found in squash such as EETI, and it is likely they have a similar binding to that of the EETI-family (Scheme 1B).[19] Hence, cyclic MCoTIs represent interesting candidates for drug design, either by changing their specificity of inhibition or by using their structure as natural scaffolds possessing new binding activities.
Results and Discussion
In the current study we report the biosynthesis and screening of biological activity of libraries based on the cyclotide MCoTI-I. These libraries were designed to contain multiple MCoTI-I mutants, in which all the residues in loops 1, 2, 3, 4 and 5, except for the Cys residues involved in the cystine-knot, were replaced by different types of amino acid. These mutations included the introduction of neutral (Ala), flexible and small (Gly), hydrophilic (Ser and Thr), hydrophobic (Met and Val), constrained (Pro) and aromatic (Tyr and Trp) residues (see Table 1). The only residue in loop 6 that was mutated was Val1. This residue is a hydrophobic β-branched amino acid highly conserved in other squash trypsin inhibitors (STIs) and is in close proximity to Lys4 in loop 1, which is responsible for MCoTI ability to inhibit trypsin. The rest of the residues in loop 6 are not required for folding or biological activity in linear STIs[22] and therefore were not explored. It is believed that loop 6 acts as a very flexible linker to allow cyclization.[23] To our knowledge this is the first time that a cyclotide-based library is biosynthesized in E. coli cells and a complete amino acid scanning is carried out in the cyclotide MCoTI-I to explore the effects of individual amino acids on biological activity and structural requirements.
Table 1.
Sequences and molecular weights found for the different MCoTI-I mutants used in this work.
| Lib1 | Molecular weight (Da) | ||
|---|---|---|---|
| Name | Sequence | Expected | Found |
| wt | CGSGSDGGVCPKILQRCRRDSDCPGACICRGNGY | 3480.94 | 3480.4±0.6 |
| P3A | CGSGSDGGVCAKILQRCRRDSDCPGACICRGNGY | 3454.91 | 3454.2±0.3 |
| K4A | CGSGSDGGVCPAILQRCRRDSDCPGACICRGNGY | 3423.85 | 3423.7±0.6 |
| I5T | CGSGSDGGVCPKTLQRCRRDSDCPGACICRGNGY | 3468.89 | 3468.0±0.1 |
| L6A | CGSGSDGGVCPKIAQRCRRDSDCPGACICRGNGY | 3438.86 | 3438.7±1.0 |
| Q7G | CGSGSDGGVCPKILGRCRRDSDCPGACICRGNGY | 3409.87 | 3408.5±1.2 |
| Q7M | CGSGSDGGVCPKILMRCRRDSDCPGACICRGNGY | 3484.01 | 3483.7±0.6 |
| R8A | CGSGSDGGVCPKILQACRRDSDCPGACICRGNGY | 3395.84 | 3396.0±0.1 |
| D12A | CGSGSDGGVCPKILQRCRRASDCPGACICRGNGY | 3436.93 | 3435.7±0.6 |
| S13A | CGSGSDGGVCPKILQRCRRDADCPGACICRGNGY | 3464.95 | 3465.0±1.0 |
| G17A | CGSGSDGGVCPKILQRCRRDSDCPAACICRGNGY | 3494.97 | 3495.0±1.0 |
| A18G | CGSGSDGGVCPKILQRCRRDSDCPGGCICRGNGY | 3466.92 | 3467.0±1.7 |
| Y26A | CGSGSDGGVCPKILQRCRRDSDCPGACICRGNGA | 3388.85 | 3388.3±1.5 |
| Lib2 | Molecular weight (Da) | ||
|---|---|---|---|
| Name | Sequence | Expected | Found |
| wt | CGSGSDGGVCPKILQRCRRDSDCPGACICRGNGY | 3480.94 | 3480.4±0.6 |
| V1A | CGSGSDGGACPKILQRCRRDSDCPGACICRGNGY | 3452.89 | 3453.0±1.0 |
| V1S | CGSGSDGGSCPKILQRCRRDSDCPGACICRGNGY | 3468.89 | 3468.7±1.2 |
| R10A | CGSGSDGGVCPKILQRCARDSDCPGACICRGNGY | 3395.84 | 3396.0±0.1 |
| R10G | CGSGSDGGVCPKILQRCGRDSDCPGACICRGNGY | 3381.81 | 3380.3±0.6 |
| R11V | CGSGSDGGVCPKILQRCRVDSDCPGACICRGNGY | 3423.89 | 3423.7±0.6 |
| D14A | CGSGSDGGVCPKILQRCRRDSACPGACICRGNGY | 3436.93 | 3436.7±1.2 |
| P16A | CGSGSDGGVCPKILQRCRRDSDCAGACICRGNGY | 3454.91 | 3455.0±1.7 |
| I20G | CGSGSDGGVCPKILQRCRRDSDCPGACGCRGNGY | 3424.84 | 3423.7±0.6 |
| R22W | CGSGSDGGVCPKILQRCRRDSDCPGACICWGNGY | 3510.97 | 3510.7±1.2 |
| G23A | CGSGSDGGVCPKILQRCRRDSDCPGACICRANGY | 3494.97 | 3495.0±1.0 |
| N24W | CGSGSDGGVCPKILQRCRRDSDCPGACICRGWGY | 3553.05 | 3552.7±1.2 |
| G25P | CGSGSDGGVCPKILQRCRRDSDCPGACICRGNPY | 3521.01 | 3521.0±1.4 |
| Y26A | CGSGSDGGVCPKILQRCRRDSDCPGACICRGNGA | 3388.44 | 3388.3±1.5 |
| Y26W | CGSGSDGGVCPKILQRCRRDSDCPGACICRGNGW | 3503.98 | 3503.3±1.2 |
The biosynthesis of MCoTI-I mutants was carried out by using a protein splicing unit in combination with an in-cell intramolecular native chemical ligation reaction (NCL) (Fig. 1).[15, 16] Intramolecular NCL requires the presence of an N-terminal Cys residue and a C-terminal α-thioester group in the same linear precursor molecule.[24, 25] For this purpose the MCoTI-I linear precursors were fused in frame at their C- and N-terminus to a modified Mxe Gyrase A intein and a Met residue, respectively. This allows the generation of the required C-terminal thioester and N-terminal Cys residue after in vivo processing by endogenous Met aminopeptidase (MAP). We used the native Cys located to the beginning of loop 6 to facilitate the cyclization. This linear construct has shown to give very good expression and cyclization yields in vivo.[16]
In order to facilitate the analysis and processing of all the mutants, two libraries (Lib1 and Lib2) were produced containing 13 and 15 different MCoTI-I mutants (see Table 1). These libraries were designed to contain mutants that could be easily identified by ES-MS. In both libraries the MCoTI-I wild-type (wt) sequence was included as control. Synthetic dsDNA fragments encoding the different MCoTI-I mutants were ligated into plasmid pTXB1 in frame with Mxe Gyrase intein (Table S1). The resulting plasmid libraries were transformed into competent DH5α E. coli cells obtaining approximately 104 colonies (data not shown). All colonies were pooled and the corresponding plasmid library was transformed into E. coli Origami2(DE3) for protein overexpression.
Expression of the library in E. coli produced the corresponding MCoTI mutants - Gyrase intein linear fusion precursors with similar yields to that of the wild-type MCoTI-I wt.[16] The level of in vivo cleavage was estimated to be ≈80% following induction for 20 h at 20°C (Fig. S0). These expression conditions maximize the in vivo processing of the linear intein-fusion precursors to give natively folded MCoTI cyclotides.[16] In vivo cleavage and processing of the corresponding intein linear fusion precursor can be reduced by inducing at relatively elevated temperatures for short times (30°C for 2–4 h for example) while keeping a similar level of protein expression (Fig. S0). This allows us to vary the amounts of folded MCoTI mutants produced to access different screening methods. For in vitro screening, cyclization can be accomplished in vitro under controlled conditions, and therefore short induction times at relatively higher temperatures will yield more uncleaved linear precursor. Alternatively, in vivo cyclization yields can be easily maximized by using longer induction times and lower induction temperatures (20°C for 20 h, for example) for high throughput in vivo screening,
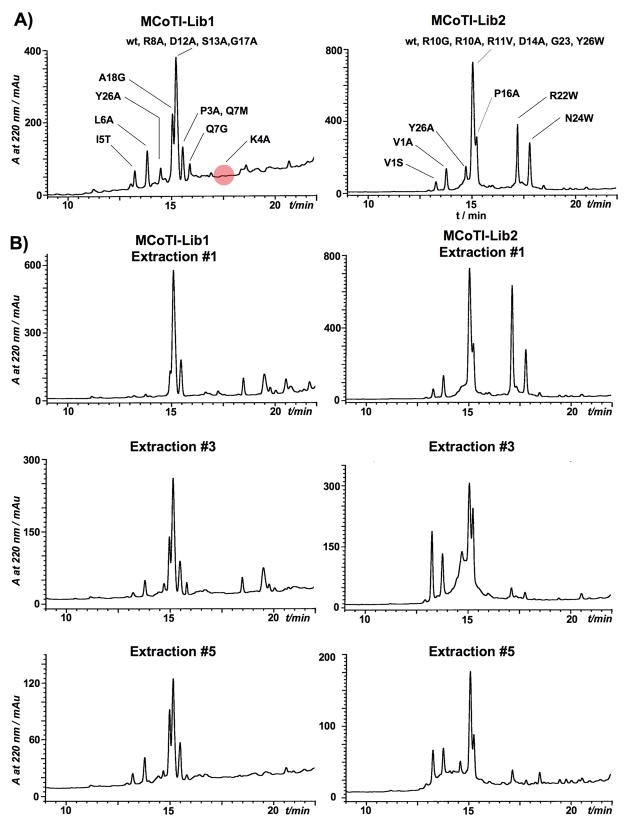
In order to characterize the MCoTI-based libraries and assess structural integrity of MCoTI mutants, we used the biological activity of MCoTI to bind trypsin. Purified MCoTI mutants-Gyrase fusion proteins were obtained from E. coli Origami2(DE3) cells which were induced at 30°C for 4 h. Under these conditions only ≈30% of the intein linear precursors were processed in vivo. The fusion precursors were cleaved and cyclized in phosphate buffer at pH 7.2 containing 50 mM GSH for 36 h. In our hands, GSH has been shown to be more effective than other thiols to promote cyclization and correct folding of cyclotides and other disulfide containing peptides in vitro[15, 16, 26]. This treatment resulted in nearly 100% cleavage of the intein precursors. The soluble fractions were purified on trypsin-sepharose beads and the bound fractions analyzed by HPLC and ES-MS to determine the presence of relative representation of the library members able to bind trypsin (Fig. 2A). As anticipated, the MCoTI-K4A mutant was not found in the trypsin-bound fraction. This residue determines binding affinity and specificity, and can only be replaced by Arg to maintain biological activity.[19] Analysis of the cyclization reaction before affinity purification confirmed the presence of this mutant in the corresponding library (Fig. S1). The K4A mutant was also individually cyclized, purified and characterized by NMR showing a native cyclotide topology when to compared to MCoTI-I wt (Fig. S2 and Table S2) therefore indicating that the lack of biological activity of this mutant was due to the replacement of Lys4 by Ala, and not to the adoption of a non-native fold. The mutant MCoTI-I G25P was also absent in the trypsin bound fraction. In vitro cyclization of the G25P revealed that the intein precursor of this cyclotide was not processed efficiently and the resulting cyclotide was not able to fold properly. Only traces of natively folded G25P were detected in the GSH-induced cyclization/folding of the corresponding intein precursor (Fig. S3). The inefficient cleavage of this mutant precursor could be explained by the proximity of a Pro residue to the MCoTI-intein junction, which may affect the ability of the Gyrase intein to produce the thioester intermediate required for the intramolecular cyclization. The Gly25 residue is located at loop 5 and it is extremely well conserved in all the STIs (Scheme 1B) thus corroborating the importance of this residue for correct folding of MCoTI cyclotides. Remarkably, the remaining mutants were identified on the trypsin bound fraction indicating that the corresponding mutants were able to adopt a native-like structure and that its ability to bind trypsin was not significantly disrupted. All the active mutants besides I20G were produced with similar yields to the MCoTI-wt (within 50% of the average value), as quantified by HPLC and ES-MS. The folded I20G mutant abundance was estimated to be ≈10% of the average. Cleavage and cyclization of I20G using GSH revealed that although the thiol-induced cleavage was very efficient, the correctly folded mutant was produced in very low yield (Fig. S4) indicating the importance of this residue for efficient folding in MCoTI cyclotides. In fact this residue, which is located between the Cys residues at the end and beginning of loops 3 and 5, respectively, is well conserved among the different linear STIs and cyclotides showing a preference for β-branched residues and hydrophilic residues. Interestingly, folded I20G mutant was able to bind trypsin beads confirming the ability to adopt a native folded structure.
Figure 2.
Analytical reversed-phase HPLC traces of trypsin-bound fractions from MCoTI Lib1 and Lib2 libraries obtained in vitro by GSH-induced cleavage and folding. A. Total trypsin-bound fractions. In red is shown the position where mutant K4A should be eluting. B. Sequential fractions extracted during competitive trypsin-binding experiments (see text for detailed description).
Next, we screened the biological activity of the MCoTI-I libraries produced in vivo. For this purpose both libraries (Lib1 and Lib2) were expressed in E. coli Origami2(DE3) cells at 20°C for 20 h in order to maximize intracellular processing and folding of the different intein precursors. After lysing the cells by sonication, the cellular supernatant was purified using trypsin-sepharose under competing binding conditions as described above. The different fractions were then analyzed and quantified by HPLC and ES-MS. The results obtained were very similar to those found with in vitro cyclized libraries (data not shown), thus indicating that the composition of the libraries obtained in vitro and in vivo were practically identical.
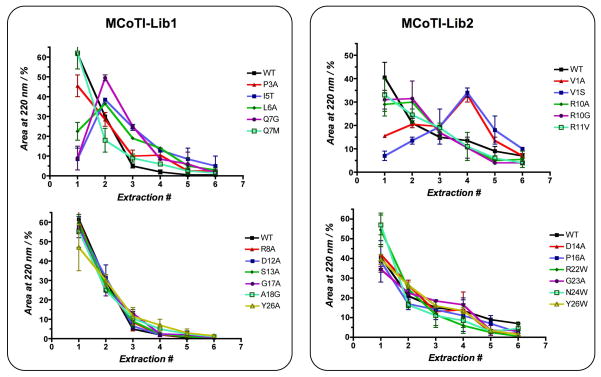
In order to establish the relative affinities of the different mutants able to bind trypsin versus MCoTI-I wild type, in vitro and in vivo cyclized libraries were incubated with trypsin-sepharose under competing conditions, i.e. using only ≈20% of the required trypsin-sepharose beads for stoichiometric binding. This process ensured that cyclotides with tighter affinities competed for binding to trypsin leaving the members of the library with weaker affinities in the superantant (i.e. unbound fraction). This supernatant was then purified again using the same approach to extract the remaining active cyclotides. This process was repeated several times until all the active cyclotides found in a particular library sample were completed extracted. This process ensured that cyclotides with tighter affinities for trypsin were extracted during the first affinity purifications leaving the library members with weaker affinities to be purified later on this sequential extraction process. All the different trypsin-bound fractions were then analyzed and quantified using HPLC and ES-MS (Fig. 2B). The results, summarized in Fig. 3, show that using MCoTI-I wt as internal reference, the mutants N24W and R22W were consistently able to compete slightly with the rest of mutants (including wt) thus indicating a somewhat tighter affinity for trypsin than the wt sequence. Most of the remaining mutants: P3A, Q7M, R8A, R10A, R10G, R11V, D12A, S13A, D14A, P16A, G17A, A18G, G23A, Y26A and Y26W showed similar elution profiles to that of wt indicating a similar affinity for trypsin. Mutants V1A, V1S, I5T, L6A and Q7G, on the other hand, were consistently extracted after MCoTI wt indicating a lower affinity for trypsin than the wt sequence. I20G was also extracted after MCoTI-I wt, however this could be due to the low abundance of folded cyclotide.
Figure 3.
Elution profiles for members of the MCoTI Lib1 and Lib2 extracted using trypsin-sepharose beads under competing conditions. The results shown are the average data obtained in vivo and in vitro (vertical bars indicate standard deviation). Quantification was done by integration of the corresponding HPLC peaks monitored at 220 nm (Fig. 2B). ES-MS was used to calculate the ratio of different cyclotides present in peaks with multiple products or not well resolved by HPLC. The area estimated for mutants with loss or gain of aromatic residues was corrected accordingly to take this into account [32].
Although there is not a structure available for the complex between MCoTI cyclotides and trypsin, the structure of several complexes formed between different STIs and trypsin have been reported so far.[27, 28] Based on the high sequence homology between these trypsin inhibitors and MCoTI cyclotides (Scheme 1), it is reasonable to assume that they possess the same binding mode to trypsin.[19] Therefore, it is not surprising that mutant K4A was not able to bind trypsin since K4 is critical for binding to the trypsin specificity pocket.[19] Other mutations in loop 1 also affected negatively trypsin binding. Hence, mutants I5T, L6A and Q7G were consistently eluted in the later fractions in our competing binding experiments, indicating a weaker affinity for trypsin. The sequence Lys/Arg-IIe-Leu in loop 1 is extremely well conserved in all linear STIs suggesting that it is required for efficient trypsin binding. Position 7, on the other hand, seems to be more promiscuous and it is able to accept hydrophobic residues (Q7M showed a similar elution pattern than MCoTI-wt) and positively charged residues (cyclotide MCoTI-II has a Lys residue in this position), but not a small and flexible residue like Gly (mutant Q7G shows weaker trypsin affinity than wt). Also in loop 1, mutation R8A did not affect significantly trypsin-binding and the corresponding mutant showed an elution pattern similar to that of wt. In agreement with this result, this position is not especially well conserved in linear STIs allowing the presence of charged (both positive and negatively charged) and Pro residues. All the mutations explored in loop 2 had similar elution patterns to the wt sequence. This should be expected since this loop is solvent exposed and on the opposite side to loop 1.
The only mutation affecting trypsin binding in loop 3 was represented by mutant P16A. The rest of the mutants in this loop behaved similar to the wt sequence. This loop is partially exposed in the structure of several linear STIs with trypsin and it shows significant sequence heterogeneity among the different STIs. Position 16, however, is usually occupied in other STIs by hydrophobic residues (mainly Leu and Met), which could explain the observed behavior of mutant P16A. None of the mutations in loop 5, besides G25P, had an adverse effect on trypsin binding. It is interesting to remark that mutants Y26A and Y26W showed a similar elution profile to the wt sequence (Fig. 3). This position is very well conserved among different STIs being occupied mainly by either aromatic (Tyr or Phe) or in some cases IIe and His. Analysis of the structure of linear Cucurbita pepo typsin inhibitor-II (CPTI-II, which shares ≈75% sequence homology with MCoTI-I) complexed with bovine trypsin[27] shows that this position makes a direct contact with trypsin Tyr151 residue, which is highly conserved among different trypsin homologs. Intriguingly, mutants R22W and N24W seemed to slightly outcompete the rest of the library members including MCoTI-I wt in our binding competing experiments (Figs. 2B and 3). These residues are in close proximity to Tyr26 and they could help to further stabilize the aromatic interaction described before between the MCoTI-I mutants and trypsin.
The position corresponding to Val1 at the end of loop 6 was also explored by including mutants V1A and V1S in this study. This position favors the presence of hydrophobic residues (mainly Val, Met and IIe) among different STIs, although hydrophilic and charged residues are also found in some STIs. Visual inspection of the CPTI-II-trypsin complex,[27] for example, reveals that this position is in close proximity to trypsin Trp215. This aromatic residue is highly conserved among the different trypsin homologs indicating that this interaction may be important to the stabilization of the complex. Consistent with this finding, replacement of Val1 in MCoTI-I by Ser and Ala produced mutants that consistently showed a weaker affinity than MCoTI-I wt.
In summary, these data provide significant insights into the structural constraints of the MCoTI cyclotide framework and the functional elements for trypsin binding. To our knowledge, this is the first time that the biosynthesis of a genetically-encoded library of MCoTI-based cyclotides containing a comprehensive suite of amino acid mutants is reported. Craik and co-workers have also recently reported the chemical synthesis of a complete suite of Ala mutants for the cyclotide Kalata B1 (KB-1).[17] These mutants were fully characterized structurally and functionally. Their results indicated that only two of the mutations explored (KB-1 W20A and P21A, both located in loop 5, see Scheme 1) prevented folding.[17] The mutagenesis results obtained in our work show similar results highlighting the extreme robustness of the cyclotide scaffold to mutations. Only two of the 27 mutations studied in the cyclotide MCoTI-I, G25P and I20G, affected negatively the adoption a native cyclotide fold. Intriguingly, the rest of the mutations allowed the adoption of a native fold as indicated by ES-MS analysis and their ability to bind trypsin (or NMR in the case of K4A). These results should provide an excellent starting point for the effective design of MCoTI-based cyclotide libraries for rapid screening and selection of de novo cyclotide sequences with specific biological activities.
The libraries used in this work were produced either in vitro by GSH-induced cyclization/folding or by in vivo self-processing of the corresponding precursor proteins. In both cases the results were similar indicating that this approach is quite general for the production of complex libraries. Importantly, in-vivo biosynthesis of cyclotide-based libraries may have tremendous potential for drug discovery. This study shows that MCoTI-cyclotides may provide an ideal scaffold for the biosynthesis of large combinatorial libraries inside of living E. coli cells. Coupled to an appropriate in-vivo reporter system, this library may rapidly be screened using high throughput technologies such as fluorescence activated cell sorting.[29, 30]
Experimental Section
See the Supporting Information for experimental details.
Supplementary Material
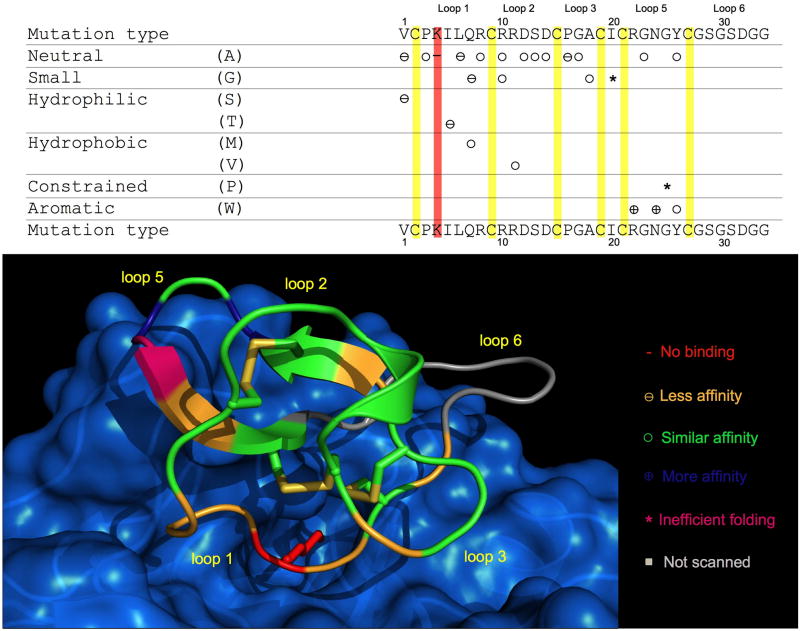
Figure 4.
Summary of the relative affinities for trypsin of the different MCoTI-I mutants studied in this work. A model of cyclotide MCoTI-I bound to trypsin is shown at the bottom indicating the position of the mutations. The side-chain of residue Lys4 is shown in red bound to specificity pocket of trypsin. The model was produced by homology modeling at the Swiss model workspace (http://swissmodel.expasy.org//SWISS-MODEL.html) [33] using the structure of CPTI-II-trypsin complex (PDB code: 2btc) [27] as template. Structure was generated using the PyMol software package.
Acknowledgments
This work was supported by funding from the School of Pharmacy at the University of Southern California and Lawrence Livermore National Laboratory.
References
- 1.Craik DJ, Simonsen S, Daly NL. Curr Opin Drug Discov Devel. 2002;5:251. [PubMed] [Google Scholar]
- 2.Craik DJ, Cemazar M, Wang CK, Daly NL. Biopolymers. 2006;84:250. doi: 10.1002/bip.20451. [DOI] [PubMed] [Google Scholar]
- 3.Clark RJ, Daly NL, Craik DJ. Biochem J. 2006;394:85. doi: 10.1042/BJ20051691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craik DJ, Cemazar M, Daly NL. Curr Opin Drug Discov Devel. 2006;9:251. [PubMed] [Google Scholar]
- 5.Colgrave ML, Craik DJ. Biochemistry. 2004;43:5965. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 6.Saether O, Craik DJ, Campbell ID, Sletten K, Juul J, Norman DG. Biochemistry. 1995;34:4147. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 7.Greenwood KP, Daly NL, Brown DL, Stow JL, Craik DJ. Int J Biochem Cell Biol. 2007;39:2252. doi: 10.1016/j.biocel.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Craik DJ, Clark RJ, Daly NL. Expert Opin Investig Drugs. 2007;16:595. doi: 10.1517/13543784.16.5.595. [DOI] [PubMed] [Google Scholar]
- 9.Kolmar H. Curr Opin Pharmacol. 2009 doi: 10.1016/j.coph.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Ireland DC, Colgrave ML, Daly NL, Craik DJ. Adv Exp Med Biol. 2009;611:477. doi: 10.1007/978-0-387-73657-0_205. [DOI] [PubMed] [Google Scholar]
- 11.Daly NL, Love S, Alewood PF, Craik DJ. Biochemistry. 1999;38:10606. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]
- 12.Avrutina O, Schmoldt HU, Kolmar H, Diederichsen U. Eur J Org Chem. 2004;204:4931. [Google Scholar]
- 13.Thongyoo P, Tate EW, Leatherbarrow RJ. Chem Commun (Camb) 2006:2848. doi: 10.1039/b607324g. [DOI] [PubMed] [Google Scholar]
- 14.Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, Tate EW. Org Biomol Chem. 2008;6:1462. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- 15.a) Kimura RH, Tran AT, Camarero JA. Angew Chem Int Ed Engl. 2006;45:973. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]; b) Kimura RH, Tran AT, Camarero JA. Angew Chem. 2006;118:987. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- 16.Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Chembiochem. 2007;8:1363. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- 17.Simonsen SM, Sando L, Rosengren KJ, Wang CK, Colgrave ML, Daly NL, Craik DJ. J Biol Chem. 2008;283:9805. doi: 10.1074/jbc.M709303200. [DOI] [PubMed] [Google Scholar]
- 18.Craik DJ, Daly NL, Mulvenna J, Plan MR, Trabi M. Curr Protein Pept Sci. 2004;5:297. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Trinh Hong T, Pham TT, Le Nguyen D. Biochemistry. 2000;39:5722. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 20.Heitz A, Hernandez JF, Gagnon J, Hong TT, Pham TT, Nguyen TM, Le-Nguyen D, Chiche L. Biochemistry. 2001;40:7973. doi: 10.1021/bi0106639. [DOI] [PubMed] [Google Scholar]
- 21.Felizmenio-Quimio ME, Daly NL, Craik DJ. J Biol Chem. 2001;276:22875. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
- 22.Avrutina O, Schmoldt HU, Gabrijelcic-Geiger D, Le Nguyen D, Sommerhoff CP, Diederichsen U, Kolmar H. Biol Chem. 2005;386:1301. doi: 10.1515/BC.2005.148. [DOI] [PubMed] [Google Scholar]
- 23.Heitz A, Avrutina O, Le-Nguyen D, Diederichsen U, Hernandez JF, Gracy J, Kolmar H, Chiche L. BMC Struct Biol. 2008;8:54. doi: 10.1186/1472-6807-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Camarero JA, Pavel J, Muir TW. Angew Chem Int Ed. 1997;37:347. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; b) Camarero JA, Pavel J, Muir TW. Angew Chem. 1998;110:361. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 25.Camarero JA, Muir TW. J Am Chem Soc. 1999;121:5597. [Google Scholar]
- 26.Austin J, Kimura RH, Woo YH, Camarero JA. Amino Acids. 2009 doi: 10.1007/s00726-009-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helland R, Berglund GI, Otlewski J, Apostoluk W, Andersen OA, Willassen NP, Smalas AO. Acta Crystallogr D Biol Crystallogr. 1999;55:139. doi: 10.1107/S090744499801052X. [DOI] [PubMed] [Google Scholar]
- 28.Kratzner R, Debreczeni JE, Pape T, Schneider TR, Wentzel A, Kolmar H, Sheldrick GM, Uson I. Acta Crystallogr D Biol Crystallogr. 2005;61:1255. doi: 10.1107/S0907444905021207. [DOI] [PubMed] [Google Scholar]
- 29.Kimura RH, Steenblock ER, Camarero JA. Anal Biochem. 2007;369:60. doi: 10.1016/j.ab.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Sancheti H, Camarero JA. Adv Drug Deliv Rev. 2009 doi: 10.1016/j.addr.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clamp M, Cuff J, Searle SM, Barton GJ. Bioinformatics. 2004;20:426. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 32.Kuipers BJ, Gruppen H. J Agric Food Chem. 2007;55:5445. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- 33.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.