Abstract
Background
Overexpression of the PGM2 gene encoding phosphoglucomutase (Pgm2p) has been shown to improve galactose utilization both under aerobic and under anaerobic conditions. Similarly, xylose utilization has been improved by overexpression of genes encoding xylulokinase (XK), enzymes from the non-oxidative pentose phosphate pathway (non-ox PPP) and deletion of the endogenous aldose reductase GRE3 gene in engineered Saccharomyces cerevisiae strains carrying either fungal or bacterial xylose pathways. In the present study, we investigated how the combination of these traits affect xylose and galactose utilization in the presence or absence of glucose in S. cerevisiae strains engineered with the xylose reductase (XR)-xylitol dehydrogenase (XDH) pathway.
Results
In the absence of PGM2 overexpression, the combined overexpression of XK, the non-ox PPP and deletion of the GRE3 gene significantly delayed aerobic growth on galactose, whereas no difference was observed between the control strain and the xylose-engineered strain when the PGM2 gene was overexpressed. Under anaerobic conditions, the overexpression of the PGM2 gene increased the ethanol yield and the xylose consumption rate in medium containing xylose as the only carbon source. The possibility of Pgm2p acting as a xylose isomerase (XI) could be excluded by measuring the XI activity in both strains. The additional copy of the PGM2 gene also resulted in a shorter fermentation time during the co-consumption of galactose and xylose. However, the effect was lost upon addition of glucose to the growth medium.
Conclusions
PGM2 overexpression was shown to benefit xylose and galactose fermentation, alone and in combination. In contrast, galactose fermentation was impaired in the engineered xylose-utilizing strain harbouring extra copies of the non-ox PPP genes and a deletion of the GRE3 gene, unless PGM2 was overexpressed. These cross-reactions are of particular relevance for the fermentation of mixed sugars from lignocellulosic feedstock.
Background
The use of renewable lignocellulosic biomass as raw material for the production of second-generation bioethanol is an alternative to sugar- and starch-containing feedstocks, which are also used for food and feed production. Lignocellulosic biomass can contain a significant fraction of pentose sugars such as xylose [1] that is not naturally fermented by the preferred microorganism for industrial bioethanol production, baker's yeast Saccharomyces cerevisiae [2]. Therefore recombinant xylose-fermenting S. cerevisiae strains have been generated to increase the overall ethanol yield from lignocellulosic biomass [3].
Xylose utilization by S. cerevisiae has been achieved either by expression of the P. stipitis XYL1 and XYL2 genes encoding the NAD(P)H-dependent xylose reductase (XR) and the NAD+-dependent xylitol dehydrogenase (XDH) [4-6] or by expression of genes encoding xylose isomerase (XI) [7,8]. When compared in an isogenic strain background, the XI strain displayed higher ethanol yield, whereas the XR-XDH strain had higher ethanol productivity [9]. Furthermore, the engineering of several additional steps has been shown to benefit xylose conversion to ethanol, including the overexpression of the xylulokinase (XK) gene [10,11], the overexpression of genes encoding the enzymes of the non-oxidative pentose phosphate pathway (non-ox PPP) [12,13] and the deletion of the aldose reductase gene encoded by GRE3 [14]. In the context of lignocellulose fermentation, these genetic modifications must be evaluated in the presence of other lignocellulose derived sugars (glucose, galactose, mannose and arabinose) and together with genetic modifications that favour the utilization of these sugars. In the case of galactose, the overexpression of the PGM2 gene encoding phosphoglucomutase has been shown to improve galactose utilization both under aerobic [15] and under anaerobic conditions [16].
In the current investigation, we therefore evaluated genetic modifications improving xylose and galactose utilization separately and in combination in S. cerevisiae strains expressing the XR-XDH pathway. The resulting strains were characterized in medium containing xylose, a mixture of either xylose and galactose or of xylose, galactose and glucose.
Results
Overexpression of the PGM2 gene in xylose-utilizing strains
To evaluate the influence of PGM2 overexpression on xylose utilization, the PGM2 gene was chromosomally integrated under the control of the truncated HXT7 promoter [17] in strain TMB 3320, already harbouring the P. stipitis XR-XDH pathway and genetic modifications that improve xylose consumption: high XK and high non-ox PPP activity as well as deletion of the GRE3 gene. The resulting strain was named PGM2-PPP-XYL (Table 1). The corresponding control strain lacking only the extra copy of PGM2 gene was also constructed and named Control-PPP-XYL (Table 1). Additionally, strains lacking the overexpression of non-ox PPP genes and the deletion of the GRE3 gene were constructed and named Control-XYL and PGM2-XYL (Table 1).
Table 1.
S. cerevisiae strains and plasmids used in this study
| S. cerevisiae strains and corresponding abbreviations | Strain background | Relevant genotype/phenotype | Reference |
|---|---|---|---|
| TMB 3320 | TMB 3043 | CEN.PK2-1C, Δgre3 his3::HIS3 PGK1p-XK1-PGK1t, PGK1p-TAL1-PGK1t, PGK1p-RKI1-PGK1t, PGK1p-TKL1-PGK1t, PGK1p-RPE1-PGK1t, TRP1, leu2::LEU2 ADH1p-XYL1-ADH1t PGKp-XYL2-PGKt ura3 | Garcia Sanchez et al., manuscript in preparation |
| CEN.PK 113-11C | MATa his3Δ1 ura 3-52 MAL2-8c SUC2 | [42] | |
| TMB 3137 "Control-PPP-XYL" | TMB 3320 | ura3::URA3 YIplac211 | This work |
| TMB 3138 "PGM2-PPP-XYL" | TMB 3320 | ura3::URA3 YIplac211 HXT-PGM2 | This work |
| TMB 3141 | CEN.PK 113-11C | his3:: HIS3 YIpXR/XDH/XK | This work |
| TMB 3139 "Control-XYL" | TMB 3141 | his3:: HIS3 YIpXR/XDH/XK ura3::URA3 YIplac211 | This work |
| TMB 3140 "PGM2-XYL" | TMB 3141 | his3:: HIS3 YIpXR/XDH/XK, ura3::URA3 YIplac211 HXT-PGM2 | This work |
| TMB 3128 "Control m" | CEN.PK 113-11C | his3::HIS3 YEplacHXT URA3 | [16] |
| TMB 3129 "PGM2 m " | CEN.PK 113-11C | his3::HIS3 YEplacHXT7'p-PGM2 URA3 | [16] |
| TMB 3135 " Control i" | CEN.PK 113-11C | his3::HIS3 ura3::URA3 | [16] |
| TMB 3136 "PGM2 i" | CEN.PK 113-11C | his3::HIS3 ura3::URA3 Ylplac211 HXT-PGM2 | [16] |
| Plasmids | |||
| YIplac211 | URA3 | [43] | |
| YIplac211 HXT-PGM2 | YIplac211, HXT7'p-PGM2-PGKt URA3 | This work | |
| YIpXR/XDH/XK | ADHp-XYL1-ADHt, PGKp-XYL2-PGKt, PGKp-XK-PGKt, HIS3, β-lactamase | (Eliasson, Christensson et al. 2000) | |
Aerobic growth on xylose and galactose
The four xylose-utilizing strains were compared with respect to aerobic growth on 50 g l-1 xylose to assess how overexpression of PGM2 affected xylose utilization. In agreement with their genetic makeup, strains overexpressing the non-ox PPP genes and carrying a deletion of the GRE3 gene (Control-PPP-XYL and PGM2-PPP-XYL) had a three- to four-fold higher growth rate on xylose than strains Control-XYL and PGM2-XYL (Table 2). On the other hand, PGM2 overexpression did not significantly affect the aerobic growth on xylose.
Table 2.
Maximum specific growth rate (μmax ± standard deviation) on 50 g l-1 galactose or 50 g l-1 xylose under aerobic conditions and using YNB medium.
| Carbon source | ||
|---|---|---|
| Strain | 50 g l-1 galactose | 50 g l-1 xylose |
| Control-PPP-XYL | 0.21 ± 0.01 | 0.038 ± 0.014 |
| PGM2-PPP-XYL | 0.32 ± 0.02 | 0.041 ± 0.008 |
| Control-XYL | 0.23 ± 0.01 | 0.012 ± 0.002 |
| PGM2-XYL | 0.27 ± 0.01 | 0.012 ± 0.003 |
| Control m | 0.22 ± 0.01 (*) | ND |
| PGM2 m | 0.30 ± 0.02 (*) | ND |
| Control i | 0.24 ± 0.02 (*) | ND |
| PGM2 i | 0.32 ± 0.01 (*) | ND |
(*) Taken from [16]. ND: Not determined. For both carbon sources, the inocula were prepared in defined medium with 20 g l-1 glucose.
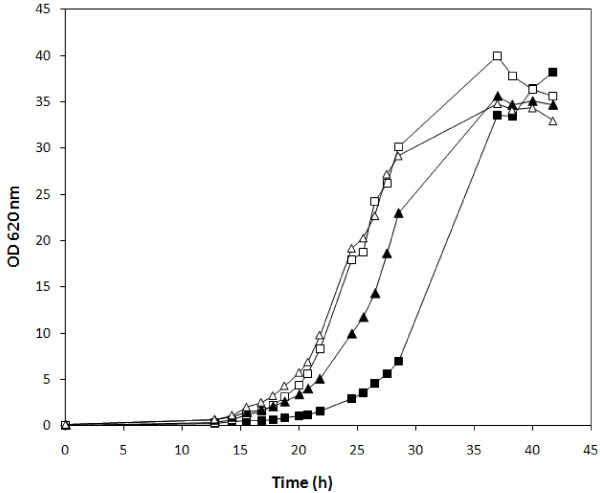
The four xylose-utilizing strains were also compared with respect to aerobic growth on 50 g l-1 galactose (Figure 1). We confirmed in both strain backgrounds that galactose utilization was increased by overexpression of the PGM2 gene (Figure 1; Table 2). Strains PGM2-XYL and PGM2-PPP-XYL differed slightly in their maximum specific growth rate (μmax) values: 0.27 ± 0.01 and 0.32 ± 0.02 h-1 (Table 2), respectively. These μmax values are in the same range as those of the two previously described S. cerevisiae strains overexpressing only PGM2, PGM2 m and PGM i [16] (Table 1 and 2). In contrast, the absence of PGM2 overexpression delayed growth on galactose for both xylose-utilizing strains, significantly more for strain Control-PPP-XYL than for strain Control-XYL (Figure 1), whereas their maximum specific growth rates were similar and in the same range as previously described for strains Control i and Control m, 0.21 and 0.23 h-1 respectively, [16] (Table 1 and 2). When PGM2 was overexpressed, the difference between the two strains disappeared and the lag phase was shortened (Figure 1).
Figure 1.
Aerobic growth in batch culture on YNB medium with 50 g l-1 galactose with inocula grown in YNB medium with 20 g l-1 glucose. S. cerevisiae strains: Control-PPP-XYL (filled square), PGM2-PPP-XYL (open square), Control-XYL (filled triangle), PGM2-XYL (open triangle).
Anaerobic fermentation of xylose
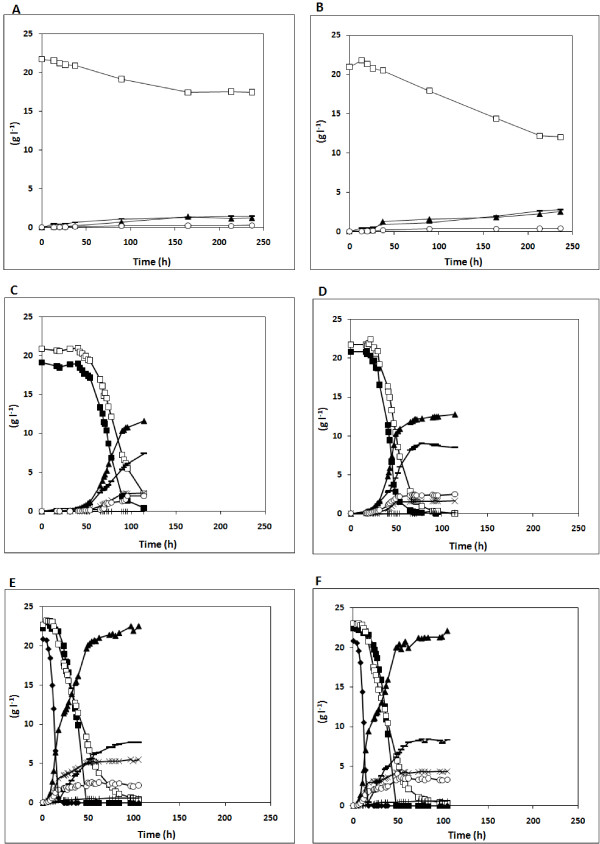
The effect of PGM2 overexpression on xylose utilization was also compared under anaerobic conditions with strains Control-PPP-XYL and PGM2-PPP-XYL (Figures 2A and 2B). In contrast to aerobic conditions, the μmax in 20 g l-1 xylose medium was 1.9-fold higher (p-value < 0.09) for the strain overexpressing the PGM2 gene: 0.023 ± 0.002 h-1 vs. 0.012 ± 0.004 h-1. This strain also consumed 112% more xylose than the control strain (Figures 2A and 2B). The increased xylose consumption resulted in a two-fold increase in biomass and a 20% increase in ethanol yield, whereas glycerol and xylitol yields decreased five- and two-fold, respectively (Table 3).
Figure 2.
Sugar consumption and product formation during anaerobic batch fermentation in defined medium. Strains: Control-PPP-XYL (A, C, E) and PGM2-PPP-XYL (B, D, F) with 20 g l-1 xylose (A, B), 20 g l-1 xylose and 20 g l-1 galactose (C, D) or 20 g l-1 xylose, 20 g l-1 galactose and 20 g l-1 glucose (E, F). Inocula were grown in defined medium with 20 g l-1 glucose. Symbols: galactose (filled square), xylose (open square), glucose (filled diamond), ethanol (filled triangle), biomass (DW) (open circle), acetate (plus symbol), glycerol (multiplication symbol), xylitol (minus symbol).
Table 3.
Parameters analyzed during anaerobic batch fermentation on different sugar mixtures.
| Yield | |||||
|---|---|---|---|---|---|
| Carbon source | Strain | a Biomass g/g substrate | a Glycerol g/g substrate | b Xylitol g/g xylose | a Ethanol g/g substrate |
| Xylose | Control-PPP-XYL | 0.040 ± 0.006 | 0.056 ± 0.017 | 0.29 ± 0.04 | 0.25 ± 0.06 |
| PGM2-PPP-XYL | 0.076 ± 0.002 | 0.011 ± 0.000 | 0.15 ± 0.07 | 0.30 ± 0.06 | |
| Galactose + Xylose | Control-PPP-XYL | 0.056 ± 0.007 | 0.061 ± 0.004 | 0.29 ± 0.05 | 0.31 ± 0.02 |
| PGM2-PPP-XYL | 0.064 ± 0.004 | 0.042 ± 0.001 | 0.37 ± 0.04 | 0.32 ± 0.00 | |
| Galactose + Xylose + Glucose | Control-PPP-XYL | 0.061 ± 0.002 | 0.138 ± 0.028 | 0.39 ± 0.04 | 0.32 ± 0.00 |
| PGM2-PPP-XYL | 0.070 ± 0.001 | 0.115 ± 0.003 | 0.40 ± 0.03 | 0.32 ± 0.01 | |
aBiomass, glycerol and ethanol yields were calculated by plotting the consumed sugar/s (g l-1) against the produced biomass (g biomass l-1), the produced glycerol (g glycerol l-1) or the produced ethanol (g ethanol l-1) respectively on the phase of constant and highest values. bXylitol yields were calculated by plotting the consumed xylose (g xylose l-1) against the produced xylitol (g xylitol l-1) on the phase of constant and highest values.
Anaerobic fermentation of mixed sugars
Anaerobic fermentation of a mixture of xylose and galactose was further investigated for strains Control-PPP-XYL and PGM2-PPP-XYL (Figures 2C and 2D; Table 3). In the presence of galactose, the xylose consumption rate increased for both strains compared to when xylose was the sole carbon source (Figures 2A and 2B). Moreover, xylose and galactose were co-consumed, with galactose being the preferred sugar. When the PGM2 gene was overexpressed, the fermentation of both sugars proceeded faster (Figure 2D). In addition, the biomass yield increased slightly, whereas the glycerol yield decreased 1.5-fold and the xylitol yield increased 1.3-fold (Table 3).
The two strains were then investigated in anaerobic fermentation of a mixture of xylose, galactose and glucose (Figures 2E and 2F; Table 3). As expected, the repressing sugar glucose was consumed first and xylose and galactose consumption started only when the glucose concentration was below 5 g l-1. In contrast to the fermentation of the xylose and galactose mixture (Figures 2C and 2D), xylose started being consumed before galactose while galactose was more readily depleted.
In the presence of glucose, the fermentation time was almost identical for the two strains, regardless of the presence of an additional copy of the PGM2 gene. Also glycerol, xylitol and ethanol yields were similar for the two strains, whereas the biomass yield increased 1.15-fold in the PGM2-overexpressing strain. Glucose fermentation increased the glycerol yield two- to three-fold.
XI activity
To rule out that the increased growth on xylose arose from a XI activity of Pgm2p, the XI activity was determined in galactose-grown cells of strains Control m and PGM2 m. The PGM activity had previously been shown to be five to six times higher in the strain carrying an extra copy of PGM2 integrated on the genome as compared to the control strain [16]. We were not able to detect any XI activity in cell extracts of either strain Control m or strain PGM2 m (data not shown).
Discussion
The present study investigated the cross-reactions that may occur in S. cerevisiae when two independent genetic engineering strategies for the improvement of the utilization of two different carbon sources, namely, galactose and xylose, were combined. The work demonstrated that PGM2 overexpression improved not only galactose but also anaerobic xylose utilization in recombinant xylose-utilizing S. cerevisiae strains. The beneficial effect was also observed in mixtures of xylose and galactose, but it disappeared in the presence of glucose. In parallel, the improved aerobic growth on galactose as a result of PGM2 overexpression, which had previously been observed in different strains [15,16], was maintained in xylose-utilizing strains. In contrast, aerobic growth on galactose was significantly delayed in the engineered xylose-utilizing strain harbouring extra copies of the non-ox PPP genes and a deletion of the GRE3 gene, unless PGM2 was overexpressed.
The anaerobic growth rate on xylose increased up to 1.9 times when PGM2 was overexpressed. In addition, the total amount of consumed xylose increased 2.1-fold (Figure 2) and the ethanol yield was 20% higher (Table 3). PGM2 upregulation has been observed in several genome-wide transcription analyses of strains developed for improved xylose utilization. The PGM2 level was 4.8- and 3.9-fold higher on xylose and xylose-glucose media under aerobic conditions and 1.6-fold higher on glucose-xylose medium under anaerobic conditions in an anaerobically growing evolved strain as compared to the non-growing parental strain [18]. In another genome-wide transcription analysis that compared several improved xylose-growing S. cerevisiae strains, all structural genes from the galactose pathway were derepressed, resulting, among others, in PGM2 upregulation [19]. Still, there is no immediate connection between the enzyme PGM2 and xylose metabolism. We assayed cell extracts for XI activity and confirmed that PGM2 is an intramolecular transferase isomerase [20,21] rather than an intramolecular oxidoreductase isomerase such as XI [22,23].
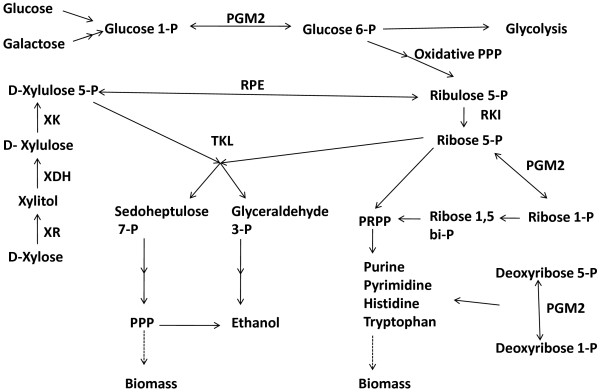
Another connection between PGM2 activity and xylose metabolism could be that PGM2 catalyzes phosphotransferase reaction(s), which enhance anaerobic growth by increasing the pool of intermediary metabolites required for biosynthetic reactions. Phosphomannomutase activity has been demonstrated for S. cerevisiae PGM1 and PGM2 [24]. Furthermore, the human homolog to S. cerevisiae PGM2 has been shown to be more active as phosphopentomutase than as phosphoglucomutase [20]. It could notably use ribose 1-P, ribose 5-P, deoxyribose 1-P and deoxyribose 5-P as substrates [20], suggesting that S. cerevisiae PGM2 may be able to convert ribose 5-P to ribose 1-P and vice versa (Figure 3). Xylose is catabolized via ribose 5-P, an intermediate of the non-ox PPP, which via ribose 1-P is a precursor of histidine and tryptophan biosynthesis as well as of purine, pyrimidine and ultimately nucleic acid biosynthesis (Figure 3). PGM2 overexpression may thus increase the xylose flux and pull xylose catabolism by converting ribose 5-P to ribose 1-P and further to biomass formation (Figure 3). In fact, increased biomass production was obtained, regardless of carbon source, when the PGM2 gene was overexpressed (Table 3). Despite a large standard deviation, the increased biomass formation was accompanied by a reduced glycerol formation (Table 3), which may reflect a reduced requirement for synthesis of biomass intermediates [25]. Finally, PGM2 overexpression reduced xylitol formation in anaerobic xylose fermentation, which serves as an additional indication of an increased xylose flux [9].
Figure 3.
Metabolic reactions involving the PGM2 (adapted from [41]). Abbreviations: PPP, Pentose phosphate pathway; PRPP, phosphoribosyl pyrophosphate; RPE, ribulose 5-phosphate 3-epimerase; RKI, ribose 5-phosphate ketol-isomerase; TKL, transketolase; -P, phosphate.
Co-fermentation of xylose and galactose increased the rate of xylose utilization compared with pure xylose fermentation. Furthermore, both sugars were depleted faster when PGM2 was overexpressed while the ethanol yield remained unchanged. Enhanced xylose uptake in the presence of galactose has previously been reported [26]. Galactose induces the galactose catabolizing enzymes of Leloir pathway [27], including the galactose transporter Gal2p [28]. In addition to transporting galactose, Gal2p has been shown to transport xylose [29], and its expression in S. cerevisiae is induced 1000-fold when the yeast is exposed to galactose [27], which was found also to benefit xylose utilization. PGM2 overexpression increases the intracellular pool of glucose 6-P [30], which has been shown to activate glycolysis [31-33]. However, when glucose was added to the medium, the benefit of PGM2 overexpression for xylose and galactose co-fermentation was abolished, in agreement with the fact that the glucose 6-P pool is higher in glucose than in galactose grown cells [34]. Thus the xylose and galactose fermentation time was reduced both by glucose and by PGM2 overexpression, but the effect was not additive.
Conclusions
Genetic modifications that were known to improve individually xylose and galactose utilization were combined and evaluated on various sugar combinations. PGM2 overexpression was shown to benefit xylose and galactose fermentation, alone or in combination. In contrast, galactose fermentation was impaired in the engineered xylose-utilizing strain harbouring extra copies of the non-oxidative PPP genes and a deletion of the GRE3 gene, unless PGM2 was overexpressed. Knowledge on these cross-reactions is of particular relevance in the context of bioethanol production from mixtures of sugars derived from lignocellulosic feedstock.
Methods
Strains
Escherichia coli DH5α and JM101 (Life Technologies, Rockville, MD, USA) were used for cloning. Plasmids and S. cerevisiae strains (Table 1) were stored at -80°C in 15% glycerol. Freshly streaked plates from frozen stocks were used to inoculate precultures.
Molecular biology techniques
Standard molecular biology techniques were used for cloning [35]. Restriction enzymes, T4-DNA ligase and Shrimp Alkaline Phosphatase (SAP) used for cloning were obtained from Fermentas (Vilnius, Lithuania). Yeast genomic DNA was purified with Easy-DNA Kit (Invitrogen, Groningen, The Netherlands). E. coli DH5α competent cells were prepared as described before [36] and transformed using the calcium chloride method [37]. Yeast was transformed using the lithium acetate method [38]. Plasmids were extracted from bacteria either with GeneJET TM (Fermentas, Vilnius, Lithuania) or with Qiagen Mini Plasmid Purification kit (Qiagen GmbH, Hilden, Germany). The QIAquick kit was used for gel extraction of DNA fragments from agarose gel as well as for purification of amplified DNA products. Taq polymerase (Fermentas, Vilnius, Lithuania) was used for analytical PCR, while Pfu-polymerase (Fermentas, Vilnius, Lithuania) and Pwo-polymerase (Roche Diagnostics GmbH, Mannheim, Germany) were used for high-fidelity polymerase chain reactions. The Abi-Prism Big-Dye cycle sequencing kit (Applied Biosystems, Weiterstadt, Germany) was used for DNA sequencing that was performed by BM Labbet AB (Furulund, Sweden).
Strain construction
Yeast strains carrying an extra copy of the PGM2 gene under a strong constitutive promoter and the corresponding control were constructed in two strain backgrounds. First, the xylose-utilizing strain TMB 3320 was transformed with EcoRV-linearized plasmids YIplac211 and YIplac211 HXT-PGM2 in the URA3 locus, to generate strain Control-PPP-XYL, TMB 3137, and strain PGM2-PPP-XYL, TMB 3138, respectively (Table 1).
In parallel, strain CEN.PK 113-11C was transformed with plasmid YIpXR/XDH/XK (Table 1) cleaved with PstI in the HIS3 locus. Yeast transformants that were selected in defined medium lacking histidine and supplemented with uracil were able to grow in defined medium with 50 g l-1 xylose. The resulting strain TMB3141 was further transformed with plasmids YIplac211 and YIplac211 HXT-PGM2 (Table 1) that were cleaved in the URA3 locus with EcoRV. The resulting strains were named Control-XYL, TMB 3139, and PGM2-XYL, TMB 3140, respectively (Table 1). Genomic DNA was extracted from strains Control-PPP-XYL, PGM2-PPP-XYL, Control-XYL and PGM2-XYL to verify integration at the correct loci using PCR.
Aerobic cultivation
E. coli was grown and selected on Luria-Bertani (LB) [35] with 100 mg/L ampicillin. For yeast growth, Yeast Nitrogen Base medium (YNB; Difco Laboratories/Becton Dickinson, Sparks, MD, USA) at a concentration of 6.7 g l-1 was supplemented either with 50 g l-1 galactose, 50 g l-1 xylose, or 20 g l-1 glucose. YNB liquid medium was buffered with potassium hydrogen phthalate (10.21 g l-1 phthalate, 2.1 g l-1 KOH, pH 5.5) [39]. The concentration of YNB was doubled when the sugar concentration was higher than 20 g l-1 to avoid nutrient limitation. Plates were supplemented with 20 g l-1 glucose and 20 g l-1 agar.
Precultures were grown in YNB with 20 g l-1 glucose until mid- to late exponential phase overnight in 50-ml tubes with 5 ml of growth medium. The precultures were used to inoculate aerobic batch cultures at OD620 nm 0.1-0.2 in cotton-stoppered baffled 500-ml flasks with 50 ml of growth medium and the relevant carbon source. The medium was supplemented with amino acids when using auxotrophic strains. Precultures and aerobic batch cultivation experiments were performed at 30°C and 180-200 rpm agitation (Gallenkamp INR-200, Leicester, UK) at least in duplicate.
Enzymatic activity
XI activity was measured in crude extracts of cells grown in YNB medium with 20 g l-1 galactose using the sorbitol dehydrogenase assay [8]. Cells were harvested in exponential phase, centrifuged at 5000 rpm for 5 min, washed with cold sterile water twice and permeabilized with Y-PER (Pierce, Rockford, IL, USA). The protein concentration was determined with Coomassie Protein Assay Reagent (Pierce, Rockford, IL, USA) using bovine serum albumin as standard. Reagents used to determine enzyme activity were purchased from Sigma-Aldrich (St. Louis, MO, USA). For every strain and condition, enzyme activity measurements of at least two independent biological replicates were performed.
Anaerobic fermentation
For anaerobic fermentation and corresponding precultivation, defined mineral medium was used [40]. The medium was supplemented with 0.4 g l-1 Tween 80 and 0.01 g l-1 ergosterol. The carbon source was either 20 g l-1 xylose, a mixture of 20 g l-1 xylose and 20 g l-1 galactose, or a mixture of 20 g l-1 xylose, 20 g l-1 galactose, and 20 g l-1 glucose. When the sugar concentration exceeded 20 g l-1, the concentration of Tween 80 and ergosterol was doubled.
Aerobic precultivation was performed at 30°C (Gallenkamp INR-200, Leicester, UK) and 180-200 rpm. The preculture medium contained 20 g l-1 glucose in phthalate buffer as described for the aerobic cultivation. A first preculture was grown until late exponential phase in 5 ml culture in 50-ml tubes and was used to inoculate a second aerobic preculture of 100 ml in 1000-ml cotton-stoppered baffled shake flasks. The second preculture was grown until late exponential phase, washed twice with sterile water after centrifugation at 5000 rpm for 10 min and used to inoculate anaerobic batch cultures at OD620 nm 0.1-0.2. Anaerobic batch fermentation was performed in either 3 l Biostat Bio Reactors (B. Braun Biotech International, Melsungen, Germany) or Applikon Bio Reactors (Applikon, Schiedam, The Netherlands) with a working volume of 1.5 l at 30°C and 200 rpm and pH controlled at 5.5 with 3 M KOH. Anaerobic conditions were obtained by sparging nitrogen gas containing less than 5 ppm O2 (AGA Gas, Sundbyberg, Sweden) at a flow rate of 0.2 l min-1 controlled by a gas mass flow meter (Bronkhorst, HI-TECH, Ruurlo, The Netherlands). Outlet carbon dioxide and oxygen were monitored by a Carbon Dioxide and Oxygen Monitor type 1308 (Brüel & Kjaer, Copenhagen, Denmark). Anaerobic fermentation experiments were performed at least in duplicate.
Analysis of metabolites
Galactose, xylose, glycerol, acetic acid and ethanol were separated by high-pressure liquid chromatography (HPLC) (Waters, Milford, MA, USA, or Beckman Instruments, Fullerton, CA, USA) with an Aminex HPX-87H ion exchange column (Bio-Rad, Hercules, CA, USA) at 45°C. The mobile phase was 5 mM H2SO4 at a flow rate of 0.6 ml min-1.
When xylose and galactose were present in the same sample xylose, galactose, glucose and xylitol were additionally separated and quantified by one or two in series connected HPX-87P (Bio-Rad, Hercules, CA, USA) (Waters, Milford, MA, USA) ion exchange column(s) at 85°C with water as the mobile phase at a flow rate of 0.5 ml min-1. A refractive index detector (RID-6A; Shimadzu, Kyoto, Japan) was used for quantification.
Cell dry weight was determined at least in duplicate at different time points during the fermentation experiment by filtering 5 ml of culture through a preweighed hydrophilic polyethersulfone 0.45 μM filter (Pall Life Sciences, Ann Arbor, MI, USA). Filters were then dried in a microwave and cooled prior to weight determination.
Abbreviations
PGM2: phosphoglucomutase isoform 2; PPP: Pentose phosphate pathway; non-ox: non-oxidative; XR: xylose reductase; XDH: xylitol dehydrogenase; XK: xylulokinase.
Competing interests
BHH is co-owner and chairman of the board of C5 Lignotechnologies in Lund AB.
RGS and MFGG declare that they have no competing interests.
Authors' contributions
RGS participated in the design of the study, performed all the experimental work and drafted the manuscript. BHH and MFGG participated in the design of the study and edited the manuscript. All authors read and approved the final manuscript.
Contributor Information
Rosa Garcia Sanchez, Email: rosa.garcia_sanchez@tmb.lth.se.
Bärbel Hahn-Hägerdal, Email: barbel.hahn-hagerdal@tmb.lth.se.
Marie F Gorwa-Grauslund, Email: marie-francoise.gorwa@tmb.lth.se.
Acknowledgements
The Swedish Energy Agency and the EU project NILE (New Improvements for Ligno-cellulosic Ethanol) are acknowledged for financial support.
References
- Hayn MSW, Klinger R, Steinmüller H, Sinner M, Esterbauer H. In: Bioconversion of Forest and Agricultural Plant Residues CAB International. Saddler JN, editor. Wallingford, UK; 1993. Basic research and pilot studies on the enzymatic conversion of lignocellulosics; pp. 33–72. [Google Scholar]
- Rudolf A, Karhumaa K, Hahn Hägerdal B. Yeast Biotechnology: Diversity and Applications. Vol. 3. Springer Netherlands; 2009. Ethanol production from traditional and emerging raw materials; pp. 489–513. full_text. [Google Scholar]
- Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- Kötter P, Amore R, Hollenberg CP, Ciriacy M. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet. 1990;18:493–500. doi: 10.1007/BF00327019. [DOI] [PubMed] [Google Scholar]
- Tantirungkij M, Nakashima N, Seki T, Yoshida T. Construction of xylose-assimilating Saccharomyces cerevisiae. J Fermentation Bioeng 1993. pp. 83–88. [DOI]
- Walfridsson M, Anderlund M, Bao X, Hahn-Hägerdal B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl Microbiol Biotechnol. 1997;48:218–224. doi: 10.1007/s002530051041. [DOI] [PubMed] [Google Scholar]
- Walfridsson M, Bao X, Anderlund M, Lilius G, Bülow L, Hahn-Hägerdal B. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Env Microbiol. 1996;62(12):4184–4190. doi: 10.1128/aem.62.12.4648-4651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MSM, de Laat WTAM, den Ridder JJ, Op den Camp HJM, van Dijken JP, Pronk JT. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 2003;4:69–78. doi: 10.1016/S1567-1356(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Garcia Sanchez R, Hahn-Hägerdal B, Gorwa-Grauslund MF. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Factor. 2007;6:5. doi: 10.1186/1475-2859-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho NWY, Chen ZD, Brainard AP. Genetically engineered Sacccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Env Microbiol. 1998;64:1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson A, Christensson C, Wahlbom CF, Hahn-Hägerdal B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Env Microbiol. 2000;66:3381–3386. doi: 10.1128/AEM.66.8.3381-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast. 2005;22:359–368. doi: 10.1002/yea.1216. [DOI] [PubMed] [Google Scholar]
- Johansson B, Hahn-Hägerdal B. The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res. 2002;2:277–282. doi: 10.1111/j.1567-1364.2002.tb00095.x. [DOI] [PubMed] [Google Scholar]
- Träff KL, Otero Cordero RR, van Zyl WH, Hahn-Hägerdal B. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Env Microbiol. 2001;67:5668–5674. doi: 10.1128/AEM.67.12.5668-5674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro C, Knudsen S, Regenberg B, Olsson L, Nielsen J. Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Env Microbiol. 2005;71:6465–6472. doi: 10.1128/AEM.71.11.6465-6472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Sanchez R, Hahn-Hägerdal B, Gorwa-Grauslund MF. PGM2 overexpression improves anaerobic galactose fermentation in Saccharomyces cerevisiae. Microb Cell Fact. 2010;9:40. doi: 10.1186/1475-2859-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf J, Zimmermann FK, Müller S. Simultaneous genomic overexpression of seven glycolytic enzymes in the yeast Saccharomyces cerevisiae. Enzyme Microb Technol. 2000;26:688–698. doi: 10.1016/S0141-0229(00)00160-5. [DOI] [PubMed] [Google Scholar]
- Sonderegger M, Jeppsson M, Hahn-Hägerdal B, Sauer U. Molecular basis for anaerobic growth of Saccharomyces cerevisiae on xylose, investigated by global gene expression and metabolic flux analysis. Appl Environ Microbiol. 2004;70:2307–2317. doi: 10.1128/AEM.70.4.2307-2317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson O, Jeppsson M, Sonderegger M, Skorupa Parachin N, Sauer U, Hahn-Hägerdal B, Gorwa-Grauslund MF. Identification of common traits in improved xylose-growing Saccharomyces cerevisiae for inverse metabolic engineering. Yeast. 2008;25:835–847. doi: 10.1002/yea.1638. [DOI] [PubMed] [Google Scholar]
- Maliekal P, Sokolova T, Vertommen D, Veiga-Da-Cunha M, Van Schaftingen E. Molecular identification of mammalian phosphopentomutase and glucose-1,6-bisphosphate synthase, two members of the alpha-D-phosphohexomutase family. J Biol Chem. 2007;282:31844–31851. doi: 10.1074/jbc.M706818200. [DOI] [PubMed] [Google Scholar]
- Joshi JG. Phosphoglucomutase from yeast. Meth Enzymol. 1982;89:599–615. doi: 10.1016/s0076-6879(82)89103-9. full_text. [DOI] [PubMed] [Google Scholar]
- Chen WP. Glucose isomerase (a Review) Process Biochem. 1980;15:36–41. [Google Scholar]
- Chen WP. Glucose isomerase (a Review) Process Biochem. 1980;15:30–35. [Google Scholar]
- Boles E, Liebetrau W, Hofmann M, Zimmermann FK. A family of hexosephosphate mutases in Saccharomyces cerevisiae. Eur J Biochem. 1994;220:83–96. doi: 10.1111/j.1432-1033.1994.tb18601.x. [DOI] [PubMed] [Google Scholar]
- Albers E, Larsson C, Lidén G, Niklasson C, Gustafsson L. Influence of the nitrogen source on Saccharomyces cerevisiae anaerobic growth and product formation. Appl Env Microbiol. 1996;62:3187–3195. doi: 10.1128/aem.62.9.3187-3195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinander NQ, Hahn-Hägerdal B. Influence of cosubstrate concentration on xylose conversion by recombinant, XYL1-expressing Saccharomyces cerevisiae: a comparison of different sugars and ethanol as cosubstrates. Appl Env Microbiol. 1997;63:1959–1964. doi: 10.1128/aem.63.5.1959-1964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo VP. Galactose transport in Saccharomyces cerevisiae I. nonmetabolized sugars as substrates and inducers of galactose transport system. J Bacteriol. 1968;95:1727–1731. doi: 10.1128/jb.95.5.1727-1731.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher T, Becker J, Gardonyi M, Hahn-Hägerdal B, Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002;148:2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
- de Jongh WA, Bro C, Ostergaard S, Regenberg B, Olsson L, Nielsen J. The roles of galactitol, galactose-1-phosphate, and phosphoglucomutase in galactose-induced toxicity in Saccharomyces cerevisiae. Biotechnol Bioeng. 2008;101:317–326. doi: 10.1002/bit.21890. [DOI] [PubMed] [Google Scholar]
- Gonçalves PM, Griffioen G, Bebelman JP, Planta RJ. Signalling pathways leading to transcriptional regulation of genes involved in the activation of glycolysis in yeast. Mol Microbiol. 1997;25:483–493. doi: 10.1046/j.1365-2958.1997.4811847.x. [DOI] [PubMed] [Google Scholar]
- Müller S, Boles E, May M, Zimmermann FK. Different internal metabolites trigger the induction of glycolytic gene-expression in Saccharomyces cerevisiae. J Bacteriol. 1995;177:4517–4519. doi: 10.1128/jb.177.15.4517-4519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E, Heinisch J, Zimmermann FK. Different signals control the activation of glycolysis in the yeast Saccharomyces cerevisiae. Yeast. 1993;9:761–770. doi: 10.1002/yea.320090710. [DOI] [PubMed] [Google Scholar]
- van den Brink J, Akeroyd M, van der Hoeven R, Pronk JT, de Winde JH, Daran-Lapujade P. Energetic limits to metabolic flexibility: responses of Saccharomyces cerevisiae to glucose-galactose transitions. Microbiology. 2009;155:1340–1350. doi: 10.1099/mic.0.025775-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritch ETM. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Inoue HN, Okayama HH. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-P. [DOI] [PubMed] [Google Scholar]
- Dagert M, Ehrlich SD. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979;6:23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Hahn-Hägerdal B, Karhumaa K, Larsson CU, Gorwa-Grauslund MF, Gorgens J, van Zyl WH. Role of cultivation media in the development of yeast strains for large scale industrial use. Microb Cell Fact. 2005;4:31. doi: 10.1186/1475-2859-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hägerdal B, Gorwa-Grauslund MF. The expression of a Pichia stipitis xylose reductase mutant with higher K(M) for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:665–673. doi: 10.1002/bit.20737. [DOI] [PubMed] [Google Scholar]
- BRENDA The Comprehensive Enzyme Information System. http://www.brenda-enzymes.org/
- Entian K, Kötter P. In: Yeast gene analysis. Brown JPA, Tuite MF, editor. Vol. 26. San Diego, CA: Academic Press; 1998. Yeast mutant and plasmid collections; pp. 431–449. full_text. [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]