Abstract
CRKL (CRK-Like) is an adapter protein predominantly phosphorylated in cells that express the tyrosine kinase p210BCR-ABL, the fusion product of a (9;22) chromosomal translocation causative for chronic myeloid leukemia (CML). It has been unclear, however, whether CRKL plays a functional role in p210BCR-ABL transformation. Here we show that CRKL is required for p210BCR-ABL to support IL-3-independent growth of myeloid progenitor cells and long-term outgrowth of B-lymphoid cells from fetal liver-derived hematopoietic progenitor cells. Furthermore, a synthetic phosphotyrosyl peptide that binds to the CRKL SH2 domain with high affinity blocks association of endogenous CRKL with the p210BCR-ABL complex and reduces c-MYC levels in K562 human leukemic cells as well as mouse hematopoietic cells transformed by p210BCR-ABL or the imatinib-resistant mutant T315I. These results indicate that the function of CRKL as an adapter protein is essential for p210BCR-ABL-induced transformation.
Keywords: CRKL, BCR-ABL, c-MYC, Leukemia, STAT5
Introduction
Patients with chronic myeloid leukemia (CML) harbor a Philadelphia chromosome (Ph) due to a translocation between chromosomes 9 and 22 (1). The resulting BCR-ABL fusion gene gives rise to a p210BCR-ABL oncoprotein (2, 3). Forced expression of the p210BCR-ABL protein renders hematopoietic cells growth factor-independent (4, 5) and induces proliferation of myeloid progenitors and bone marrow-derived B-lymphoid cells (6–9). Transplantation of p210BCR-ABL-transducedmurine bone marrow cells into lethally irradiated syngeneic mice induces a CML-like myeloproliferative disorder, demonstrating the leukemogenic potential of p210BCR-ABL in vivo (10–13). The tyrosine kinase activity of p210BCR-ABL is essential for its oncogenic potential both in vitro and in vivo (13, 14), hence the ABL inhibitor imatinib has been a successful treatment for CML (15).
CRKL (CRK-Like), a member of the CRK family of adapter proteins, consists of an N-terminal SH2 domain followed by two SH3 domains, SH3n and SH3c (16). Although related to CRK, CRKL is a distinct gene located on chromosome 22q11.21 (16). Overexpression of CRKL enhances p190BCR-ABL-induced transformation and leukemogenesis in fibroblasts and in a transgenic mouse model (17, 18). CRKL links tyrosine kinase substrates to downstream effectors containing SH3 binding motifs. CRKL is constitutively phosphorylated by BCR-ABL in neutrophils of CML patients and the degree of CRKL phosphorylation is a marker of BCR-ABL kinase activity (19, 20).
Direct binding of CRKL to p210BCR-ABL is mediated by association of the CRKL SH3n domain with a proline-rich motif in ABL (21). However, a mutant BCR-ABL lacking the SH3 binding motif still transforms myeloid cells (21), suggesting that CRKL remains associated with the p210BCR-ABL complex by interactions with other proteins. Cell-penetrating peptides designed to bind with high affinity to the CRKL SH3n domain block the proliferation of primary CML blast cells (22). These SH3 binding peptides also bind to the CRK gene products with high affinity (23), identifying a commonly-encountered difficulty in studies designed to provide definitive evidence for a role that CRKL may play in p210BCR-ABL induced transformation.
We and others generated mouse mutants with targeted disruptions at the Crkl locus (24, 25). Crkl-deficiency in 129/Sv and C57BL/6 backgrounds results in embryonic lethality that mimics 22q11 deletion syndrome also known as DiGeorge syndrome (24). The phenotype appears to be sensitive to genetic background and disappears in a mixed genetic background involving 129/Sv and Black Swiss (25). Using this Crkl “insensitive” background, it was shown that Crkl-deficiency did not inhibit leukemogenesis caused by p190BCR-ABL in a mouse model for acute lymphoblastic leukemia (ALL) (25). Disappearance of the embryonic phenotype, however, indicates that this genetic background provides an alternative mechanism that compensates for loss of Crkl.
Biological differences between p210BCR-ABL and p190BCR-ABL may also explain the results of the study mentioned above. p190BCR-ABL causes Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL), which is relatively refractory to imatinib therapy (26). A recent study demonstrates that a significant subset of leukemia cells from p190BCR-ABL transgenic mice is insensitive to imatinib and quickly takes over the culture particularly when stromal cells are present (27). Since imatinib is still effective at blocking the kinase activity of p190BCR-ABL in these resistant cells, this phenomenon suggests a kinase-independent mechanism in p190BCR-ABL-induced leukemia. p190BCR-ABL can sustain survival and proliferation of leukemia cells in the presence of imatinib (thus without substantial ABL tyrosine kinase activity) can explain the ability of p190BCR-ABL to induce leukemia in the transgenic model without Crkl. To directly address the requirement for CRKL for p210BCR-ABL transformation, we have therefore utilized hematopoietic progenitor cells from the fetal liver of Crkl-deficient mouse embryos in a Crkl sensitive genetic background. Our results are further supported by studies with a synthetic peptide that blocks association of CRKL with the p210BCR-ABL protein complex in mouse hematopoetic progenitor cells and in human K562 leukemia cells. The current study demonstrates an essential role for CRKL in p210BCR-ABL induced transformation of hematopoietic progenitor cells.
Materials and Methods
Mice
The Institutional Animal Use and Care Committees (IAUCC) of the University of Chicago and the Oregon Health & Science University approved animal studies. Crkl+/− mice were described previously (24). All mice and embryos were examined in a mixed 129S4/SvJaeSor X C57BL/6J background. Embryos were derived by timed matings between Crkl+/− parents.
Cell Lines
Certified Baf3 cells and K562 cells were obtained from the American Type Culture Collection (ATCC), Manassas, VA and grown in the recommended culture media. The Baf3 transfectant (expressing the p210BCR-ABL mutant T315I mutation) has been previously described (28). Transformed p210BCR-ABLB-lymphoid cells derived from Wt and Crkl-deficient fetal liver cells were generated as described below. None of the cell lines used in this study were cultured for longer than 6-months from initial purchase or characterization. No further authentication of cell line characteristics was done.
Retrovirus preparation
Bosc23 cells (28) were maintained in Dulbecco’s Modified Eagles Medium (DMEM)supplemented with 10% fetal bovine serum (FBS), 1 U/mL penicillin, and1 μg/mL streptomycin. Hematopoietic progenitor cells were infected with p210BCR-ABL retrovirus or control GFP retrovirus generated by transfecting Bosc23 cells with MSCV-p210-IRES-GFP (13), or empty MSCV-IRES GFP vector and the viral supernatant harvested at 48 hours post-transfection.
Myeloid and lymphoid outgrowth assays
Due to the lethal phenotype of Crkl−/ − embryos at late gestation (24) the fetal liver was harvested as a source of hematopoietic progenitor cells. For colony forming assays, Wild type (Wt), Crkl+/− or Crkl−/ − cells harvested from fetal livers of day 13.5 murine embryos were incubated overnight in pre-stimulation media (Iscoves Modified Dulbecco Medium (IMDM), 15% FBS, 5% WEHI-conditioned media, 1 μg/mL ciprofloxacin, murine IL-3 (6 ng/mL), IL-6 (10 ng/mL), and SCF (50 ng/mL). Cells were transduced with p210BCR-ABL expressing retrovirus in the presence of 2 μg/ml polybrene (Sigma-Aldrich, St. Louis, MO) by two rounds of spinoculation (29), and 8 x 104 cells per 35mm dish were plated in triplicate in Methocult M3234 medium (StemCell Technologies, Vancouver, Canada) with or without IL-3 (100 pg/mL). Colonies were scored at day 12.
Whitlock-Witte cultures were performed as described (8). Briefly, cells harvested from the fetal liver of E13.5 Wt, Crkl+/− or Crkl−/ − mouse embryos were infected with p210BCR-ABL or control retrovirus in lymphoid media (RPMI plus 10% FBS, 1 U/mL penicillin, 1 μg/mL streptomycin, 1% L-glutamine, and 0.05 mM 2-mercaptoethanol) containing 8 μg/mL polybrene by two rounds of spinoculation. Approximately 5 x 106 cells were plated per 60 mm dish in triplicate. Cultures were fed every 3 days by adding 2 mL lymphoid media. Viable non-adherent cells were counted at day 14 post-transduction and viability was determined by trypan blue dye exclusion.
Analysis of cell surface markers
Immunophenotyping of p210BCR-ABL-transformed lymphoid cells expanded from Whitlock-Witte cultures as well as fresh fetal liver cells from E13.5 Wt, Crkl+/− and Crkl−/ − embryos was performed by fluorescence-activated cell sorting (FACS) using a FACSCalibur instrument (BD Biosciences, San Jose, CA). Whitlock-Witte culture-derived lymphoid cells transformed with p210BCR-ABL were stained with PE-conjugated-B220, -Sca1, or -c-Kit. For analysis of hematopoietic progenitor populations in fetal liver, lineage-committed (Lin+) hematopoietic cells were identified with the biotinylated mouse lineage antibody cocktail (BD) and streptavidin-conjugated PE-Cy7 (Caltag Laboratories Inc, Burlingame, CA). Lineage negative (Lin−) cells were then stained with the following conjugated antibodies; PE-c-Kit, APC-Thy1.1, and FITC-Sca1. The percentage of stem cells in each fetal liver cell population was calculated by determining the number of Lin negative, c-KIT-, Sca1-, and Thy1.1-positive cells (Lin−, c-Kit+, Sca1+, Thy1.1+). To evaluate the composition of B cell progenitors in the fetal liverof Wt, Crkl+/−, and Crkl−/ − mice, cells were stained with B220-APC, CD43-PE, IgM-APC and IgD-APC. Based on immunophenotype, cells were characterized as pro-B (B220-low, CD43+, IgM−, IgD−), Pre-B (B220-low, CD43−, IgM−, IgD−), immature (B220-moderate, CD43−, IgM+, IgD−), or mature (B220-high, CD43−, IgM-low, IgD-high) B-cells. Monoclonal antibodies raised against B220, Sca-1, Thy1.1, c-Kit, and CD43 were purchased from BD; for IgM and IgD, from Southern Biotechnology Associates, Birmingham, AL.
Peptides, immunoprecipitation, and immunoblotting
The purity of synthetic peptides was greater than 95%. The amino acid sequences used were: pYELP peptide, GVSEpYELPEDPRWELPR; CRKL pY207 peptide, GIPEPAHApYAQPQTTTPLPA; and CRK pY221 peptide, GGPEPGPpYAQPSVNTPLPN. Control peptides included a tyrosine or phenylalanine residue in place of phosphotyrosine (pY). Co-immunoprecipitation, SDS-PAGE and immunoblotting were performed using standard protocols. In addition to fetal-liver derived mouse lymphoid cells, we used the human CML blast crisis cell line K562 and murine leukemia cell lines BaF3-p210BCR/ABL or BaF3- p210BCR/ABL with T315I mutation. Cell lysates were prepared in lysis buffer (15% glycerol, 1% NP40, 50 mM Tris pH 7.4, 0.2 M NaCl, 2.5 mM MgCl2) including protease inhibitor cocktail (Roche, Indianapolis, IN), 1 mM sodium orthovanadate and 10 mM NaF. For peptide inhibition experiments, phosphopeptide or control peptide was added to pre-cleared cell lysates and incubated for 1 hour at room temperature. Stable association of CRKL or CRK with p210BCR-ABL was assessed by co-immunoprecipitation with mouse anti-ABL monoclonal antibody (clone 8E9, Santa Cruz Biotechnology, Santa Cruz, CA) and anti-mouse IgG-conjugated Sepharose beads (Sigma), followed by SDS-PAGE and immunoblot analysis with rabbit anti-CRKL (C20) or CRK antibody (Santa Cruz Biotechnology). Prior control experiments performed with isotype-matched, irrelevant antibody confirm the specificity of bands detected in the high stringency anti-ABL immunoprecipitates.
To assess the biological effects of the phosphopeptide on BCR-ABL function,and to compare its activity to the ABL kinase inhibitor imatinib, PEP-1 (a 21 amino acid peptide carrier) was used to deliver phosphorylated or control peptides into the cell (30). The phosphorylated or control peptide was preincubated with PEP-1 for 1 hour at a molar ratio of 1:20. The peptide mix was then added to cell culture and incubated for the indicated amount of time before washout and lysis. Imatinib, kindly provided by E. Buchdunger (Novartis, Basel, Switzerland), was prepared freshly as a 10 mM stock solution in sterile PBS.
Immunoblots were performed according to a standard method, and probed using specific antibodies. To probe phosphorylation at Y649 in STAT5A and Y699 in STAT5B, we used mouse monoclonal anti-phospho STAT5 (clone ST5P-4A9, Invitrogen, Carlsbad, CA). Total STAT5 (STAT5A and STAT5B) was determined by rabbit anti-STAT5 (Santa Cruz Biotechnology). The BCL-X and MYC gene products were probed with rabbit anti-BCL-XL (Santa Cruz Biotechnology) and rabbit polyclonal anti-Myc (Upstate, Waltham, MA).
Cell cycle analysis
Cell cycle analysis was carried out by FACS.Cells were collected by centrifugation,washed with PBS, and permeabilized in 70% ethanol at −20°C for 30 minutes prior to DNA staining.Approximately 1 x 106 permeabilized cells were incubated with 50 μg/ml propidium iodide (Roche) or 3 μM 4’,6-diamidino-2-phenylindole (DAPI, Invitrogen), and 0.1 mg/ml RNase A in staining buffer (100 mM Tris, pH 7.4, 150 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, and 0.1% NP 40) for 15–30 minutes. Stained cells were analyzed in a FACSCalibur or BD LSR II flow cytometer (BD) using Modfit LT or FacsDIVA software.
Proliferation assays
p210BCR-ABL-transformed Wt and Crkl−/ − hematopoietic cells (5 x 104 cells/well) that were expanded from Whitlock-Witte cultures were plated into 24-well flat-bottomed plates in RPMI medium containing 5% FBS and 50 μM 2-mercaptoethanol. Cells were counted daily for 3 days. Results represent counts obtained from triplicate wells performed with cells derived from three independent Whitlock-Witte cultures.
Results
CRKL is required for p210BCR-ABL-induced IL-3-independent myeloid outgrowth
BCR-ABL expression in hematopoietic progenitor cells results in growth factor-independent myeloid colonies in methylcellulose (6). To assess the requirement of CRKL in this process, fetal liver cells harvested from day 13.5 Wt, Crkl+/− and Crkl−/ − embryos were infected with retrovirus that co-transduces p210BCR-ABL and GFP. Transduction efficiency of Wt, Crkl+/− and Crkl−/ − cells was similar as comparable numbers of cells were GFP-positive by FACS analysis following transduction (data not shown). Cells were seeded into methylcellulose in the presence or absence of IL-3. Growth factor-dependent and -independent colonies were counted on day 12. We found that p210BCR-ABL expression in Wt cells leads to the formation of cytokine-independent myeloid colonies of CFU-GM, CFU-M and CFU-GEMM lineages. In contrast, there was a 3-fold reduction in the number of growth factor-independent colonies formed by Crkl−/ − cells (Fig. 1A). The number of Crkl+/− colonies was reduced by 2-fold and there were significantly fewer cells formed by Crkl−/ − cells than Crkl+/− cells, suggesting a dosage-sensitive effect of CRKL on p210BCR-ABL-induced transformation (Fig. 1A). In the presence of IL-3, Wt, Crkl+/− and Crkl−/ −cells gave similar numbers of myeloid colonies (Fig. 1B). There was no difference in the relative proportion of colonies of CFU-GEMM, CFU-GM or CFU-M lineages formed by Wt, Crkl+/− or Crkl−/ −cells (Fig. 1C). Importantly, all colonies were GFP-positive by fluorescence microscopy and contained the provirus by PCR screening (data not shown). These data suggest that 1) Crkl deficiency does not compromise the generation of hematopoietic colonies in response to physiological stimuli and 2) decreased myeloid colony formation in Crkl−/ − cultures is not due to decreased transduction efficiency of early myeloid progenitors in the absence of CRKL.
Figure 1.
CRKL is required for p210BCR-ABL-induced growth factor independence in myeloid cells. Equal numbers of fetal liver cells from E13.5 Wt, Crkl+/− and Crkl−/ − littermate embryos were infected with p210BCR-ABL retrovirus and cultured (A) alone or (B) supplemented with 100-pg/mL mouse IL-3. Myeloid colonies were counted 12 days post-infection. Averages with SD from six independent assays in triplicate are shown: *p = 0.01 and 0.007 for Crkl+/− and Crkl−/ − fetal liver cells compared to Wt, respectively; **p = 0.0005 between Crkl−/ − cells and Crkl+/− cells. (C) Differences in relative proportion of colonies of CFU-GM, CFU-M or CFU-GEMM lineages formed by Wt, Crkl+/− or Crkl−/ − cells.
CRKL is required for p210BCR-ABL-induced lymphoid outgrowth
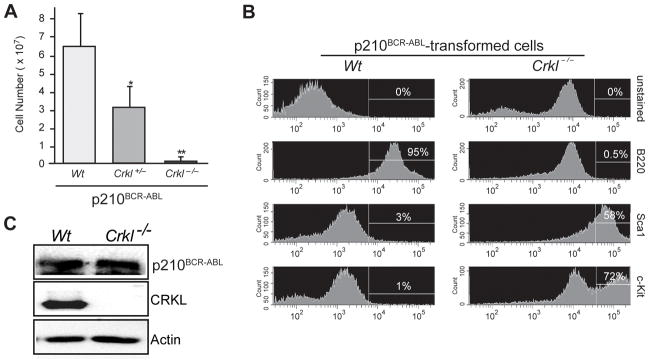
To further explore the requirement of CRKL for transformation by p210BCR-ABL, we assessed outgrowth of Crkl−/ − lymphoid cells induced by p210BCR-ABL. Hematopoietic progenitor cells harvested from the fetal livers of 13.5 day-old Wt, Crkl+/− and Crkl−/ − embryos were transduced with p210BCR-ABL retrovirus and cultured under conditions favoring outgrowth of transformed lymphoid cells (8). A similar percentage of Wt, Crkl+/− and Crkl−/ −cells were GFP-positive following retroviral transduction with p210BCR-ABL (data not shown), but fewer cells were observed in Crkl+/− (p = 0.05)and Crkl−/ − (p = 0.0005) cultures at day 14 post-transduction compared to Wt cultures (Fig. 2A). We also observed fewer cells in Crkl−/ − relative to Crkl+/− cultures, suggesting a dosage-sensitive effect of CRKL on p210BCR-ABL-induced lymphoid transformation. Consistent with previous reports (31), all cells emerging from Wt fetal liver expressed the B220 antigen; however, Crkl−/ − cells were largely B220 negative and expressed markers of more primitive progenitors, such as c-Kit and Sca-1 (Fig. 2B). There was no difference in p210BCR-ABL levels showed between these cells (Fig. 2C) indicating that poor outgrowth of cells from Crkl−/ − fetal liver was not due to poor expression of p210BCR-ABL.
Figure 2.
The requirement for CRKL in generating p210BCR-ABL B-lymphoid cells. (A) Fetal liver cells infected with p210BCR-ABL retrovirus were cultured under conditions favoring outgrowth of transformed B-lymphocytes. Following incubation at 37°C for 14 days, viable cells were counted. Shown are averages with SD from three independent assays in triplicate with Wt, Crkl+/− or Crkl−/ − matched littermates: for Crkl+/−and Crkl−/ − compared to Wt, p = 0.05* and p = 0.0005**, respectively. (B) The p210BCR-ABL-transformed lymphoid cells from Wt cultures were B220 positive whereas Crkl−/ − cells were largely B220 negative. Crkl−/ −cells expressed markers of more primitive progenitors, such as c-Kit and Sca1. (C) Immunoblotting of BCR-ABL and CRKL in Wt and Crkl-deficient p210BCR-ABL-transformed lymphoid cells with anti-ABL and anti-CRKL antibodies, with ACTIN as a loading control.
These results suggest that p210BCR-ABL fails to induce B220+ lymphoid cells in the absence of CRKL, but could also be attributable to skewed progenitor populations in Crkl+/− and Crkl−/ − cells compared to Wt cells. We therefore investigated by FACS whether the composition of hematopoietic progenitor cells differed between fetal liver cells derived from Wt, Crkl+/− and Crkl−/ − embryos. The percentage of stem cells (Lin-, c-Kit+, Sca-1+, Thy1.1+) was the same in Wt, Crkl+/− and Crkl−/ − fetal liver cells (Fig. 3). Moreover, we found no difference in the percentage of pro-B (B220-low, CD43+, IgM-, IgD-), pre-B (B220-low, CD43-, IgM-, IgD-), immature B (B220-moderate, CD43-, IgM+, IgD-) or mature B (B220-high, CD43-, IgM-low, IgD-high) cells in the fetal liver of Wt, Crkl+/−, or Crkl−/ −embryos (Fig. 3). We conclude that reduction in B cell outgrowth from Crkl−/ − and Crkl+/− fetal liver in Whitlock-Witte cultures was not due to a deficiency in B cell progenitors. This finding is consistent with previous findings that there was no difference in the ability of hematopoietic progenitor cells derived from Crkl−/ − fetal livers to reconstitute the bone marrow of irradiated Rag2−/ − recipients compared to Wt with similar numbers of circulating B220+ cells (32).
Figure 3.
Crkl deficiency does not affect distribution of stem cell and B cell progenitor populations in fetal liver cells isolated from E13.5 Wt, Crkl+/− and Crkl−/ − mouse embryos. FACS profiling of stem cells (Thy1.1+ and Sca1+), pre-B cells (B220 low), pro-B (IgM-low, CD43+), immature (CD43−, IgM+), and mature (CD43−, IgM-low) B cell progenitors revealed no significant differences in the distribution of cells from Wt, Crkl+/−or Crkl−/ − embryos.
Phosphotyrosine-mediated interactions play a major role in association of CRKL with the p210BCR-ABL complex
Biological functions of adapter proteins such as CRKL rely on their ability to link partners through protein interaction domains (16). We previously demonstrated that CRKL associates with fibroblast growth factor receptors, FGFR1 and FGFR2, through its SH2 domain (33). Based on a CRKL SH2 domain-binding motif identified in FGFR1, we synthesized a phosphotyrosyl peptide (pYELP) exhibiting high affinity for the CRKL SH2 domain but ~30-fold weaker binding to the SH2 domain of CRK (34). Although the peptide sequence was based on an FGFR1 autophosphorylation site, pYELP peptide efficiently disrupted association of endogenous CRKL with the p210BCR-ABL protein complex when added to cell lysates from Wt cells (Fig 4A). Conversely, this peptide had no inhibitory effect on association of CRK with the p210 protein complex (Fig. 4A). In the same experiment, control peptides without phosphotyrosine (YELP and FELP) did not affect association of CRKL with the p210BCR-ABL complex. Further, We observed pYELP peptide also inhibited their association in vivo in a dose-dependent manner when the peptide is introduced into the cell along with a cell-membrane penetrating peptide carrier, PEP-1 (30) (Fig. 4A). Of note, appropriate control experiments were performed with an isotype-matched, irrelevant antibody and that these experiments confirmed the specificity of bands detected in the high stringency anti-ABL immunoprecipitates (Data not shown). In total, these results indicate that pYELP peptide can block the adapter function of CRKL in vitro and in vivo.
Figure 4.
pYELP peptide inhibits association of BCR-ABL and CRKL leading to G0/G1 cell cycle arrest and cell death. (A) A phosphotyrosyl peptide, pYELP, dissociates CRKL from the p210BCR-ABL complex in vitro. The 17 amino acid peptide pYELP or control peptides without phosphotyrosine (YELP and FELP) was added to cell lysates prepared from B-lymphoid cells derived from p210BCR-ABL transduced Wt fetal liver. After 1-hour incubation at room temperature, stable association of CRKL or CRK with p210BCR-ABL was assessed by co-immunoprecipitation with anti-ABL antibody followed by immunoblot analysis with anti-CRKL or CRK antibody. (B) pYELP peptide inhibits association of CRKL with p210BCR-ABL in vivo. B-lymphoid cells derived from Wt fetal liver transduced with p210BCR-ABL retrovirus cultured in Whitlock-Witte media were incubated with pYELP or similar phosphopeptide corresponding to CRKL pY207 or CRK pY221 along with PEP-1 carrier peptide for 2 hours before cell lysates were prepared. Association of CRKL and p210BCR-ABL was evaluated by co-immunoprecipitation.
CRKL is constitutively phosphorylated at Y207 in leukemia cells expressing p210BCR-ABL (35). Y207 and Y221 in CRKL and CRK, respectively, are thought to play a role in regulating adapter functions through intramolecular associations (34, 36). Interestingly, however, phosphotyrosyl peptides based on the internal SH2 domain-binding motif in CRKL and CRK (pY207 or pY221 peptide, respectively) failed to dissociate these adapters from the p210BCR-ABL complex at concentrations up to 10 μM (Fig. 4B and data not shown), providing evidence that CRKL functions as an adapter despite phosphorylation at Y207 in leukemia cells.
We utilized the ability of pYELP peptide to dissociate CRKL from the p210BCR-ABL complex in Wt B-lymphoid cells transformed by p210BCR-ABL. Treatment with either pYELP peptide or imatinib led to minor decreases in the number of cells in G0/G1, S, and G2, whereas the population in subG0/G1 was markedly increased (p< 0.01) in mouse B-lymphoid cells transformed by p210BCR-ABL in Whitlock-Witte culture (Fig. 5A). Treatment of K562 cells with either pYELP peptide or imatinib led to a significant increase in G0/G1 population (p < 0.01) and an increase in the sub G0/G1 population (P < 0.01) accompanied by decreases in the S and G2 populations (p< 0.01, Fig. 5B). These results indicate that blockage of CRKL-mediated pathways or ABL catalytic activity had consistently similar effects in both cell types, manifested as increased apoptosis in p210BCR-ABL-transformed B-lymphoid cells or as G1 arrest in K562 cells.
Figure 5.
pYELP peptide mediates dissociation of BCR-ABL and CRKL leading to G0/G1 cell cycle arrest with reduced cell survival. (A) p210BCR-ABL − transformed mouse B-lymphoid cells isolated by Whitlock-Witte culture were treated with either pYELP, control peptide (CTR), or imatinib and cell cycle analysis was performed by DAPI staining using FACS 12 hours after the treatment. (B) K562 cells were treated with either pYELP or imatinib and cell cycle analysis was performed as described above. The percentage of cells in various stages of the cell cycle is shown to the right of cell cycle profiles. Results are displayed as the mean ± SD of results obtained from four independent experiments. The asterisk indicates a significant difference from the control cells at p < 0.01.
CRKL is required for p210BCR-ABL-induced high BCL-XL and phospho-STAT5 levels
p210BCR-ABL constitutively activates the transcription factor STAT5 (encoded by the Stat5A and Stat5B genes), which is required for transforming activity (37, 38). It has been reported that p210BCR-ABL promotes expression of the prosurvival factor BCL-XL through STAT5 (39). Given the adapter role of CRKL in BCR-ABL-mediated activation of STAT5 signaling cascades, we investigated whether the phenotypic defect observed in p210BCR-ABL-transformed Crkl−/ − hematopoietic cells was a result of failure of BCR-ABL to activate STAT5 in the absence of CRKL. We observed a striking decrease in p-STAT5 in CRKL-deficient p210BCR-ABL-transformed cells relative to their Wt counterparts as well as a corresponding reduction in BCL-XL (Fig. 6A). Consistent with this, p210BCR-ABL transformed Crkl−/ − cells proliferated significantly more slowly than transformed Wt cells (Fig 6B). Next we determined whether the effect of the blocking peptide pYELP on the phosphorylation of STAT5 (at Y694 or Y699 in STAT5A or STAT5B, respectively) in both mouse and human cells. Upon treatment of p210BCR-ABL transformed lymphoid cells with pYELP peptide, phosphorylation of STAT5 was reduced in a dose-dependent manner, whereas control peptides had little effect (Fig. 6C, left panel). Similar effects of pYELP peptide were also observed in the human CML cell line K562 (Fig. 6C, right panel). In both cases, reduced pSTAT5 levels were again associated with a similar decrease in BCL-XL levels (Fig. 6C). Taken together, these results indicate an important role for CRKL in the cell survival network that p210BCR-ABL aberrantly activates.
Figure 6.
CRKL is required for p210BCR-ABL-induced STAT5-dependent downstream molecular cascades. A) Cell lysates from p210BCR-ABL – transformed Crkl-deficient and Wt lymphoid cells were immunoblotted with antibodies recognizing phospho-STAT5 and BCL-XL, with ACTIN as a loading control. (B) Growth rates of BCR-ABL transformed Wt and Crkl−/ − cells. Cells were plated in triplicate, and viable cell counts were determined every 24 h by trypan blue exclusion. (C) pYELP inhibits p210-induced upregulation of BCL-XL and tyrosine phosphorylation of STAT5. p210BCR-ABL-transformed mouse B-lymphoid cells isolated by Whitlock-Witte culture and K562 cells were treated with pYELP or control peptides (YELP, FELP). Cell lysates were prepared 12 hours after peptide treatment. Immunoblots were probed with anti-BCL-XL, anti-phospho-STAT5 (Y694), or anti-STAT5 antibodies.
CRKL is required for p210BCR-ABL-induced high c-MYC levels
The proto-oncogene c-MYC is required for BCR-ABL transformation (40), and plays an essential role in G1 to S transition in cell cycle (41). We therefore determined whether c-MYC levels were dependent on CRKL. We found that Crkl−/ −lymphoid cells expressing BCR-ABL had a striking reduction in c-MYC protein levels compared to wild-type cells (Fig. 7A). This difference in Crkl-deficient lymphoid cells may again be attributable to their primitive differentiation status (Fig. 2) rather than CRKL-dependent properties. In order to address this issue, we used pYELP peptide to dissociate CRKL from the p210BCR-ABL protein complex in Wt B-lymphoid cells transformed by p210BCR-ABL. As expected, treatment with the ABL inhibitor imatinib resulted in reduced c-MYC protein levels in B-lymphoid cells derived from Wt fetal liver by p210BCR-ABL (Fig. 7B). When pYELP peptide was introduced into transformed B-lymphoid cells using the carrier peptide PEP-1, pYELP efficiently inhibited c-MYC protein levels (Fig. 7B). In contrast, control peptides FELP or YELP without tyrosine phosphorylation had little effect. pYELP peptide also had a similar effect on c-MYC levels in the CML blast crisis K562 cells (Fig. 7C).
Figure 7.
CRKL is required for p210BCR-ABL-induced high c-MYC levels. (A) c-MYC protein levels were determined in p210BCR-ABL-transformed Wt or Crkl−/ − lymphoid cells derived from fetal liver. The blot was re-probed with anti-actin antibody for loading controls. (B) c-MYC levels were decreased by pYELP treatment in p210BCR-ABL-transformed B-lymphoid cells from Wt fetal liver. Peptide was added to B-lymphoid cells at different concentrations and resulting cell lysates were probed with c-MYC and c-ABL antibodies. (C) Blocking CRKL adapter function with pYELP peptide decreases c-MYC levels in murine p210BCR-ABL-transformed B-lymphoid cells from Whitlock-Witte cultures and in human K562 CML cells. Note that pYELP reduces c-MYC levels in both cell types. (D) pYELP treatment decreased c-MYC levels in Ba/F3 cells transformed with WT p210BCR-ABL and T315I mutant p210BCR-ABL. Note that although T315I is resistant to imatinib, the cells still respond to the treatment with pYELP peptide. Cells were treated with either pYELP peptide mixed with the carrier peptide PEP-1 or imatinib for 2 hours. Cell lysates were probed with antibodies against c-MYC or c-ABL.
Recent studies revealed a rise of imatinib-resistant CML cells in some patients treated with imatinib (42). Therefore we tested the effect of pYELP peptide on BaF3 cells expressing p210BCR-ABL Wt or the p210BCR-ABL T315I mutant, which is insensitive to imatinib. We found that imatinib failed to reduce c-MYC levels in cells that express the p210BCR-ABL T315I mutant, while treatment with pYELP peptide was able to reduce c-MYC levels in these cells (Fig. 7D). These results provide evidence that signaling pathways mediated by CRKL play an essential role in the induction or maintenance of elevated levels of the oncoprotein c-MYC in p210BCR-ABL transformed cells.
Discussion
Since the discovery that CRKL is the major substrate of BCR-ABL in CML patient neutrophils (19, 20, 43), the role of this signaling protein in p210BCR-ABL leukemogenesis has been the focus of much attention. To address the role of CRKL in transformation by p210BCR-ABL, we analyzed the ability of p210BCR-ABL to transform hematopoietic progenitor cells derived from mice harboring a null mutation in the Crkl locus. In the current study we used a Crkl-sensitive genetic background in which Crkl-deficiency results in a lethal phenotype during late embryogenesis (24). On the contrary, while another Crkl-knockout strain was used to assess the role of CRKL in p190BCR-ABL-induced leukemogenesis, the genetic background used was insensitive to CRKL as evidenced by disappearance of an overt phenotype in Crkl−/ − homozygous mice (25).
We found that p210BCR-ABL transduced Crkl−/ − progenitors showed a marked decrease in growth factor-independent myeloid colony formation as compared to its Wt counterparts. Likewise, we found that compared to Wt, transformation of Crkl−/ − progenitor cells by p210BCR-ABL resulted in greatly reduced numbers of lymphoid cells under Whitlock-Witte conditions. BCR-ABL provides both proliferative and survival signals (44, 45), and a reduction in myeloid colony formation as well as failed outgrowth of B-lymphoid populations in Whitlock-Witte cultures indicates a reduced ability of p210BCR-ABL to promote survival and/or proliferation in Crkl-deficient cells.
Our results suggest that p210BCR-ABL-transformed mouse B-lymphoid cells are particularly sensitive to CRKL-dependent pathways for survival (Fig. 5). Consistent with our results using a CRKL SH2 blocking peptide, p210BCR-ABL-transformed Crkl−/ − fetal liver cells outgrown from Whitlock-Witte cultures showed an abnormal cell cycle profile as well as reduced BCL-XL levels compared to p210BCR-ABL-transformed Wt counterparts. These CRKL-deficient lymphoid cultures, however, presented cell markers different from B-lymphoid cells (Fig. 2), thus making it difficult to directly interpret these results in light of a role that CRKL may play in p210BCR-ABL-induced transformation. Anecdotal evidence for compromised survival included the inability of p210BCR-ABL-transduced Crkl−/ − cells to recover from a regular freeze-thaw cycle widely used to store cell/tissue culture in liquid nitrogen or ultra low-temperature freezer, whereas Wt fetal liver counterparts transformed by p210BCR-ABL were easily re-established from low temperature storage. This was however an obstacle to further investigation of Crkl−/ − cells after introduction of p210BCR-ABL by additional experimental manipulations such as transfection.
It has been reported that p210BCR-ABL induces expression of the anti-apoptotic gene BCL-XL (39). One explanation for our observations above is that loss of CRKL as a transcriptional co-activator of STAT5 may reduce STAT5-dependent transcription of BCL-XL, thus rendering susceptibility to apoptosis. Tyrosine phosphorylation at Y694 or Y699 in STAT5A and 5B, respectively, is essential for STAT5 dimerization required for its transcriptional activity (46). Thus, reduction of tyrosine phosphorylation of STAT5 at these sites in p210BCR-ABL-transformed cells upon treatment with pYELP peptide likely contributes to increased apoptotic cell death.
CRKL is associated with STAT5 in p210BCR-ABL expressing cells (47), and GST fusion proteins carrying either the SH2 or the first of the two SH3 domains of CRKL were able to pull-down STAT5 in lysates of cells expressing p210BCR-ABL (40, 48). As the tyrosine phosphorylation site of STAT5 is essential for dimerization required for its transcriptional activity, binding of CRKL SH2 domain to this site would be counterproductive to the function of STAT5. While it is possible that the SH3 domain may mediate physical interaction in the cell, CRKL-STAT5 complexes are not found in cells that do not express p210BCR-ABL. Thus, interactions between CRKL and STAT5 may be stabilized in a tertiary protein complex in which CRKL SH2 domain may also plays a role. It remains to be resolved how the CRKL-STAT5 complex is maintained in p210BCR-ABL expressing cells.
Previous studies have indicated that c-MYC plays an essential role in p210BCR-ABL-mediated transformation (40, 48). The c-MYC oncoprotein is overexpressed in many human neoplasms and has been found to regulate cell cycle (41). It is noteworthy that MYC is a putative STAT5 target gene (49, 50). However, p210BCR-ABL can transform STAT5A/B compound homozygous bone marrow cells (51), thus suggesting that c-MYC may be induced by STAT5-dependent and -independent pathways downstream of p210BCR-ABL. Our results indicate that disruption of CRKL SH2 mediated interactions results in a reduction of c-MYC protein levels in p210BCR-ABL expressing cells. Therefore, CRKL is likely involved in multiple pathways. Future studies are warranted to elucidate more precise pathways that the adapter protein CRKL mediates in p210BCR-ABL induced leukemogenesis. As CRKL SH2 blocking peptide can reduce c-MYC levels in the imatinib-resistant p210BCR-ABL mutant T315I, analysis of CRKL-mediated pathways may provide useful therapeutic targets for treatment of CML patients.
Acknowledgments
We thank R. Duggan and D. Leclerc (University of Chicago Flow Cytometry Facility) for technical assistance; P. Nash, and D.L. Guris for critical reading of the manuscript.
This work was supported in part by grants from the NCI, a Specialized Center of Research Award from the Leukemia and Lymphoma Society, a Clinical Scientist Award from the Burroughs Wellcome Fund, and the T. J. Martell Foundation to B.J.D; grants from the Leukemia Research Foundation, the HHMI Research Resources Program, and the NIDCR/NIH (R01 DE015883) to A.I.
References
- 1.Kurzrock R, Gutterman JU, Talpaz M. The molecular genetics of Philadelphia chromosome-positive leukemias. N Engl J Med. 1988;319(15):990–8. doi: 10.1056/NEJM198810133191506. [DOI] [PubMed] [Google Scholar]
- 2.Bartram CR, de Klein A, Hagemeijer A, et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306(5940):277–80. doi: 10.1038/306277a0. [DOI] [PubMed] [Google Scholar]
- 3.Heisterkamp N, Stephenson JR, Groffen J, et al. Localization of the c-ab1 oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature. 1983;306(5940):239–42. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 4.Daley GQ, Baltimore D. Transformation of an interleukin 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210bcr/abl protein. Proc Natl Acad Sci U S A. 1988;85(23):9312–6. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hariharan IK, Adams JM, Cory S. bcr-abl oncogene renders myeloid cell line factor independent: potential autocrine mechanism in chronic myeloid leukemia. Oncogene Res. 1988;3(4):387–99. [PubMed] [Google Scholar]
- 6.Gishizky ML, Witte ON. Initiation of deregulated growth of multipotent progenitor cells by bcr-abl in vitro. Science. 1992;256(5058):836–9. doi: 10.1126/science.1375394. [DOI] [PubMed] [Google Scholar]
- 7.Gishizky ML, Witte ON. BCR/ABL enhances growth of multipotent progenitor cells but does not block their differentiation potential in vitro. Curr Top Microbiol Immunol. 1992;182:65–72. doi: 10.1007/978-3-642-77633-5_8. [DOI] [PubMed] [Google Scholar]
- 8.McLaughlin J, Chianese E, Witte ON. In vitro transformation of immature hematopoietic cells by the P210 BCR/ABL oncogene product of the Philadelphia chromosome. Proc Natl Acad Sci U S A. 1987;84(18):6558–62. doi: 10.1073/pnas.84.18.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young JC, Witte ON. Selective transformation of primitive lymphoid cells by the BCR/ABL oncogene expressed in long-term lymphoid or myeloid cultures. Mol Cell Biol. 1988;8(10):4079–87. doi: 10.1128/mcb.8.10.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247(4944):824–30. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 11.Elefanty AG, Hariharan IK, Cory S. bcr-abl, the hallmark of chronic myeloid leukaemia in man, induces multiple haemopoietic neoplasms in mice. Embo J. 1990;9(4):1069–78. doi: 10.1002/j.1460-2075.1990.tb08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelliher MA, McLaughlin J, Witte ON, Rosenberg N. Induction of a chronic myelogenous leukemia-like syndrome in mice with v-abl and BCR/ABL. Proc Natl Acad Sci U S A. 1990;87(17):6649–53. doi: 10.1073/pnas.87.17.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Ren R. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 1998;92(10):3829–40. [PubMed] [Google Scholar]
- 14.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247(4946):1079–82. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 15.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 16.Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20(44):6348–71. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- 17.Hemmeryckx B, van Wijk A, Reichert A, et al. Crkl enhances leukemogenesis in BCR/ABL P190 transgenic mice. Cancer Res. 2001;61(4):1398–405. [PubMed] [Google Scholar]
- 18.Senechal K, Heaney C, Druker B, Sawyers CL. Structural requirements for function of the Crkl adapter protein in fibroblasts and hematopoietic cells. Mol Cell Biol. 1998;18(9):5082–90. doi: 10.1128/mcb.18.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols GL, Raines MA, Vera JC, Lacomis L, Tempst P, Golde DW. Identification of CRKL as the constitutively phosphorylated 39-kD tyrosine phosphoprotein in chronic myelogenous leukemia cells. Blood. 1994;84(9):2912–8. [PubMed] [Google Scholar]
- 20.ten Hoeve J, Arlinghaus RB, Guo JQ, Heisterkamp N, Groffen J. Tyrosine phosphorylation of CRKL in Philadelphia+ leukemia. Blood. 1994;84(6):1731–6. [PubMed] [Google Scholar]
- 21.Heaney C, Kolibaba K, Bhat A, et al. Direct binding of CRKL to BCR-ABL is not required for BCR-ABL transformation. Blood. 1997;89(1):297–306. [PubMed] [Google Scholar]
- 22.Kardinal C, Konkol B, Schulz A, et al. Cell-penetrating SH3 domain blocker peptides inhibit proliferation of primary blast cells from CML patients. Faseb J. 2000;14(11):1529–38. doi: 10.1096/fj.14.11.1529. [DOI] [PubMed] [Google Scholar]
- 23.Posern G, Zheng J, Knudsen BS, et al. Development of highly selective SH3 binding peptides for Crk and CRKL which disrupt Crk-complexes with DOCK180, SoS and C3G. Oncogene. 1998;16(15):1903–12. doi: 10.1038/sj.onc.1201714. [DOI] [PubMed] [Google Scholar]
- 24.Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27(3):293–8. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- 25.Hemmeryckx B, Reichert A, Watanabe M, et al. BCR/ABL P190 transgenic mice develop leukemia in the absence of Crkl. Oncogene. 2002;21(20):3225–31. doi: 10.1038/sj.onc.1205452. [DOI] [PubMed] [Google Scholar]
- 26.Ottmann OG, Druker BJ, Sawyers CL, et al. A phase 2 study of imatinib in patients with relapsed or refractory Philadelphia chromosome-positive acute lymphoid leukemias. Blood. 2002;100(6):1965–71. doi: 10.1182/blood-2001-12-0181. [DOI] [PubMed] [Google Scholar]
- 27.Mishra S, Zhang B, Cunnick JM, Heisterkamp N, Groffen J. Resistance to imatinib of bcr/abl p190 lymphoblastic leukemia cells. Cancer Res. 2006;66(10):5387–93. doi: 10.1158/0008-5472.CAN-05-3058. [DOI] [PubMed] [Google Scholar]
- 28.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90(18):8392–6. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotani H, Newton PB, 3rd, Zhang S, et al. Improved methods of retroviral vector transduction and production for gene therapy. Hum Gene Ther. 1994;5(1):19–28. doi: 10.1089/hum.1994.5.1-19. [DOI] [PubMed] [Google Scholar]
- 30.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol. 2001;19(12):1173–6. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 31.Scherle PA, Dorshkind K, Witte ON. Clonal lymphoid progenitor cell lines expressing the BCR/ABL oncogene retain full differentiative function. Proc Natl Acad Sci U S A. 1990;87(5):1908–12. doi: 10.1073/pnas.87.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson AC, Marks RE, Fields PE, Imamoto A, Gajewski TF. T cell development and function in CrkL-deficient mice. Eur J Immunol. 2003;33(10):2687–95. doi: 10.1002/eji.200324294. [DOI] [PubMed] [Google Scholar]
- 33.Moon AM, Guris DL, Seo JH, et al. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev Cell. 2006;10(1):71–80. doi: 10.1016/j.devcel.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo JH, Suenaga A, Hatakeyama M, Taiji M, Imamoto A. Structural and functional basis of a role for CRKL in a fibroblast growth factor 8-induced feed-forward loop. Mol Cell Biol. 2009;29(11):3076–87. doi: 10.1128/MCB.01686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Jong R, ten Hoeve J, Heisterkamp N, Groffen J. Tyrosine 207 in CRKL is the BCR/ABL phosphorylation site. Oncogene. 1997;14(5):507–13. doi: 10.1038/sj.onc.1200885. [DOI] [PubMed] [Google Scholar]
- 36.Kobashigawa Y, Sakai M, Naito M, et al. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat Struct Mol Biol. 2007;14(6):503–10. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- 37.Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers CL. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene. 1996;13(2):247–54. [PubMed] [Google Scholar]
- 38.Sillaber C, Gesbert F, Frank DA, Sattler M, Griffin JD. STAT5 activation contributes to growth and viability in Bcr/Abl-transformed cells. Blood. 2000;95(6):2118–25. [PubMed] [Google Scholar]
- 39.Gesbert F, Griffin JD. Bcr/Abl activates transcription of the Bcl-X gene through STAT5. Blood. 2000;96(6):2269–76. [PubMed] [Google Scholar]
- 40.Sawyers CL, Callahan W, Witte ON. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70(6):901–10. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 41.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2(10):764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 42.Quintas-Cardama A, Kantarjian H, Cortes J. Flying under the radar: the new wave of BCR-ABL inhibitors. Nat Rev Drug Discov. 2007;6(10):834–48. doi: 10.1038/nrd2324. [DOI] [PubMed] [Google Scholar]
- 43.Oda A, Miyakawa Y, Druker BJ, et al. Crkl is constitutively tyrosine phosphorylated in platelets from chronic myelogenous leukemia patients and inducibly phosphorylated in normal platelets stimulated by thrombopoietin. Blood. 1996;88(11):4304–13. [PubMed] [Google Scholar]
- 44.Hess P, Pihan G, Sawyers CL, Flavell RA, Davis RJ. Survival signaling mediated by c-Jun NH(2)-terminal kinase in transformed B lymphoblasts. Nat Genet. 2002;32(1):201–5. doi: 10.1038/ng946. [DOI] [PubMed] [Google Scholar]
- 45.Jena N, Deng M, Sicinska E, Sicinski P, Daley GQ. Critical role for cyclin D2 in BCR/ABL- induced proliferation of hematopoietic cells. Cancer Res. 2002;62(2):535–41. [PubMed] [Google Scholar]
- 46.Stoecklin E, Wissler M, Moriggl R, Groner B. Specific DNA binding of STAT5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol Cell Biol. 1997;17(11):6708–16. doi: 10.1128/mcb.17.11.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhodes J, York RD, Tara D, Tajinda K, Druker BJ. CrkL functions as a nuclear adaptor and transcriptional activator in Bcr-Abl-expressing cells. Exp Hematol. 2000;28(3):305–10. doi: 10.1016/s0301-472x(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 48.Afar DE, Goga A, McLaughlin J, Witte ON, Sawyers CL. Differential complementation of Bcr- Abl point mutants with c-MYC. Science. 1994;264(5157):424–6. doi: 10.1126/science.8153630. [DOI] [PubMed] [Google Scholar]
- 49.Huang M, Dorsey JF, Epling-Burnette PK, et al. Inhibition of Bcr-Abl kinase activity by PD180970 blocks constitutive activation of STAT5 and growth of CML cells. Oncogene. 2002;21(57):8804–16. doi: 10.1038/sj.onc.1206028. [DOI] [PubMed] [Google Scholar]
- 50.Lord JD, McIntosh BC, Greenberg PD, Nelson BH. The IL-2 receptor promotes lymphocyte proliferation and induction of the c-MYC, bcl-2, and bcl-x genes through the trans-activation domain of STAT5. J Immunol. 2000;164(5):2533–41. doi: 10.4049/jimmunol.164.5.2533. [DOI] [PubMed] [Google Scholar]
- 51.Sexl V, Piekorz R, Moriggl R, et al. STAT5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl- and bcr/abl-induced transformation are independent of STAT5. Blood. 2000;96(6):2277–83. [PubMed] [Google Scholar]