Abstract
Signals originating from the cancer-associated fibroblasts (CAFs) may positively regulate the proliferation and tumorigenicity in prostate cancer. In this study, we investigated whether CAFs may regulate the biology of prostate cancer stem cells (CSCs), using a conditional Pten deletion mouse model of prostate adenocarcinoma to isolate both CAF cultures and CSC-enriched cell fractions from the tumors. CSCs that were isolated, possessed self-renewal, spheroid-forming, and multipotential differentiation activities in tissue culture, segregating with a cell fraction exhibiting a signature expression phenotype, including Sca-1 (high), CD49f (high), CK5 (high), p63 (high), survivin (high), Runx2 (high), CD44 (low), CD133 (low), CK18 (low), and androgen receptor (low). CSC spheroid-forming efficiency was differentially influenced by the nature of fibroblasts in a co-culture system: compared to mouse urogenital sinus mesenchyme or normal prostate fibroblasts, CAFs enhanced spheroid formation with the spheroids displaying generally larger sizes and more complex histology. Graft experiments showed that CSCs admixed with CAFs produced prostatic glandular structures with more numerous lesions, high proliferative index, and tumor-like histopathologies, compared to those formed in the presence of normal prostate fibroblasts. Together, our findings underscore a significant role of CAFs in CSC biology.
Introduction
The important contribution of stroma to the genesis and progression of a variety of tumors has been described (1–4). Stromal cells are known to stimulate epithelial cell growth through their ability to produce extracellular matrix and by secretion of growth factors and cytokines, and to support angiogenesis (1, 2, 5). There is evidence that fibroblastic cells from prostate tumors, termed as “cancer-associated” fibroblasts (CAFs) can enhance the tumorigenic potential of the epithelial compartment (6, 7). Over the past five years, in the field of solid tumors, an enormous attention has been placed on the study of cancer stem cells (CSCs). CSCs are considered to be a minor population of tumor-initiating or tumorigenic cells within the tumor that can self-renew while simultaneously giving rise to tumor cells. Their stem cell-like properties may be responsible for solid tumor initiation, homeostasis, progression, metastasis and recurrence (8–11). Since bulk of the tumor cells have only limited proliferation ability and are non-tumorigenic, CSCs might be central to the mechanisms in the cancer. Recent studies in solid tumors have shown the findings of such cells in brain, breast, colon, lung, liver, pancreas, ovarian, head and neck, melanoma and prostate cancers (9, 12–19). Considering the known role of fibroblasts of the tumor microenvironment in cancer, it is now important to ask if these cells do serve critical functions in the biology of CSCs.
Supported by our ability to follow progression, regression and relapse of prostate cancer in living mice (20) along with the ability to obtain primary cultures of CAFs at specific stages of growth of the tumor (21), we are set to begin a critical analysis of the effects of CAFs on CSCs isolated from the same tumors. In this report, we first describe isolation and characterization of a small subpopulation of epithelial cells, enriched for putative CSCs from prostate adenocarcinomas of the conditional Pten deletion mouse model (20), and then demonstrate that CAFs isolated from these tumors can significantly support and potentiate the stemness and growth properties of the CSCs present in the isolated epithelial subpopulations.
Material and Methods
Animals
The conditional Pten deletion mouse model with simultaneous activation of the luciferase reporter (cPten−/−L), used in the current work was described previously (20). For tissue grafting, NOD.SCID mice purchased from NCI-Frederick, were used.
Cell sorting
Single cell suspensions from minced prostate tissues were obtained following the published protocol (20). For magnetic cell sorting (MACS), the cells were stained with biotinylated “Lin” antibodies (against CD45, CD31 and Ter119; BD Bioscience; 0.1μg/106 cells) for 10 minutes on ice. After washing with the cell staining buffer, Lin− cell fraction was separated from Lin+ cells using the DYNAV CELLection™ Biotin Binder Kit (invitrogen) following the manufacturer’s protocol. Lin− Sca-1+ cells were separated from other cells in the Lin− fraction using the same kit and biotinylated Sca-1 antibody (Biolegend). For fluorescence-activated cell sorting (FACS), cells were stained with biotinylated Lin antibodies followed by PE/Cy5-conjugated streptavidin (Biolegend; 0.2 μg/106 cells), PE/Cy5-conjugated Sca-1 antibody (Biolegend; 0.1μg/106 cells), and PE-conjugated CD49f antibody (Biolegend; 0.25 μg/106 cells). Stained cells were then examined using BD FACSAria™ Cell Sorting System with BD FACSDiva™ software.
Assays for spheroid formation
Culturing and passaging conditions were adapted and modified from published protocols (22–24). Briefly, sorted prostate cells were counted and suspended in 1:1 Matrigel (BD Bioscience)/PrEGM (Lonza) in a total volume of 250 μL. The mixture was placed in a well of a 24-well plate, solidified at 37°C, and then cultured in PrEGM. Stromal cells were seeded inside an insert, pore size: 8.0 μm (BD Bioscience) above the matrigel. This mixture was cultured at 37°C, and half of PrEGM was changed every 3 days. Spheroids were counted at 14 days after plating. For serial passages, spheroids formed in Matrigel were digested in 500 μl of dispase (BD Bioscience) at 37 °C for 30 mins followed by treatment with DMEM/F12 medium (Invitrogen) containing 10% fetal bovine serum (FBS), collagenase (Sigma; 1 mg/ml), hyaluronidase (Sigma; 1 mg/ml) and DNase I (Sigma; 1 μg/ml) for 30 mins and then in 0.05% Trypsin/EDTA for 10 min. After passing through a 40-μm filter, cells were counted and re-plated.
Stromal cells
UGSM was isolated following published procedures (22, 25). NPFs and CAFs were isolated as described before (21). The passage number of stromal cells used in this work was limited to 3–7 for UGSM and 3–10 for NPFs and CAFs.
Immunostainings
The Matrigel layer in the wells harboring spheroids was covered with OCT, sectioned (8 μm thickness) at −25 °C, and stained by either H&E or immunofluorescence (20). Primary antibodies against p63 (1:100; Santa Cruz), CK8 (1:100; TROMA-1 antibody; Developmental Studies Hybridoma Bank, University of Iowa), CK5 (1:1000; Covance), androgen receptor (AR; 1:100; Santa Cruz), Ki67 (1:400; Vector Laboratories), Nkx3.1 (1:400; Abcam), Cre (1:1000; Covance) or Vimentin (1:50; Cell Signaling) were used. Immunohistochemical analysis of parallel paraffin sections of 4% PFA-fixed tissues was done by a modified avidin-biotin complex (ABC) technique (20).
Renal grafting
Epithelial cells with or without stromal cells were mixed in 50 μL neutralized rat tail collagen type I (BD Bioscience) and placed in the middle part of a well in a 12-well plate. The grafts were cultured in Bfs medium (26) overnight at 37°C prior to transplanting under the renal capsules of 8 to 12 week-old NOD.SCID male mice.
PCR analyses
Total cellular RNA (100 ng), extracted by RNAqueous-Micro Kit (Ambion) was reverse-transcribed by random hexamers using qScript™ cDNA Synthesis Kit (Quanta), and the reverse transcription reaction (1μL) was then subjected to PCR amplification using FastStart Universal SYBR Green Master (Roche). PCR signals were recorded and analyzed in Stratagene MX3000P qPCR system with MxPro software (Stratagene; v4.01). For DNA quantitative PCR, genomic DNA was extracted by Pico Pure™ DNA Extraction Kit (Arcturus). DNA sample (100 ng) was mixed with the primer set and FastStart Universal SYBR Green Master (Roche). PCR reactions were performed and analyzed using the same qPCR system and software. Primer sets are listed in Supplementary Table S1.
Statistical analysis
All data were presented as means ± SE. Statistical calculations were done with Microsoft Excel analysis tools. Differences between individual groups were analyzed by paired t test or chi-square, as appropriate. P values of <0.05 were considered statistically significant.
Results
In vitro stem cell properties
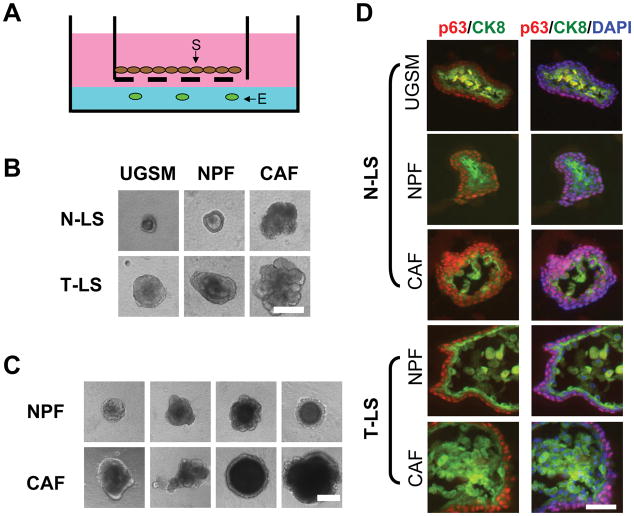
Lin−Sca-1+ (LS) cells isolated from either prostate tumors (T) or the prostatic proximal region of normal (N) prostates were tested for their growth in a modified Matrigel colony assay system (27, 28). As illustrated in Fig. 1A, epithelial cells (104) were mixed with 50% Matrigel in the well, and primary stromal cells (104), namely, UGSM, NPF or CAF, were seeded into the inserts placed above the Matrigel layer. Spheroids formed from T-LS cells, in general, appeared to be larger than the spheroids from N-LS cells (Fig 1B; Supplementary Fig. S1A). Frequently, T-LS spheroids also presented a dense structure in the intra-lumen region (Fig. 1B). While the influence of UGSM and NPFs on the spheroid morphology was not found to be significantly different from one another, CAFs seemed to exert a profound effect on the spheroid morphology as seen by their larger, denser and complex appearance (Fig. 1B, C and D). This effect of CAFs was seen on both N-LS and T-LS cells. N-LS spheroids grown with UGSM or NPFs had two distinct cell layers containing either p63+ cells or CK8+ cells (Fig. 1D, top). The sizes of N-LS spheroids co-cultured with CAFs were generally larger than those formed in the presence of UGSM or NPFs. Cells with co-expression of p63 and CK8 were found to be increased in T-LS spheroids compared to N-LS spheroids (Supplementary Fig. S1B). T-LS spheroids co-cultured with CAFs showed not only enlarged size and p63 and CK8 double positive cells in the inner cell layer, but also multiple layers of CK8+ cells in the intra-lumen region (Fig. 1D, bottom). Additionally, it was noted that the proportion of CK8+ cells was increased in the spheroids of either N-LS or T-LS origin by CAFs as compared to NPFs, and that in T-LS spheroids p63+ cell proportion was decreased in the presence of CAFs as compared to NPFs while in N-LS spheroids the effect was opposite (Supplementary Fig. S1B). Together, the results suggested that LS cells in the tumor model, like the normal prostate-derived LS cells might have the multi-potentiality to differentiate to other cell types, along with a unique attribute to form tumor-like glandular structures in vitro, especially when exposed to paracrine signaling molecules from CAFs.
Figure 1. Analyses of spheroids formed from putative normal and cancer stem cells.
A. Illustration of the placement of an insert (pore size: 8.0 μm) containing stromal cells on top of the Matrigel for the co-culture experiments (E: epithelial cells; S: stromal cells). B. Phase contrast images of representative spheroids formed from either N-LS or T-LS cells after 14 days of co-culture with primary stromal cells from different sources. Bar, 100 μm. C. Illustration of the different sizes of the spheroids formed from tumor LSC cells after co-culturing with either NPFs or CAFs. Bar, 100 μm. D. Comparative immunohistochemical analysis of spheroids formed from normal and tumor LS cells co-cultured with UGSM, NPFs or CAFs. Sections of spheroids were analyzed by co-immunofluorescence using antibodies against the basal cell marker p63 (red) and luminal cell marker CK8 (green). DAPI was used for labeling cell nuclei. Bar, 25 μm.
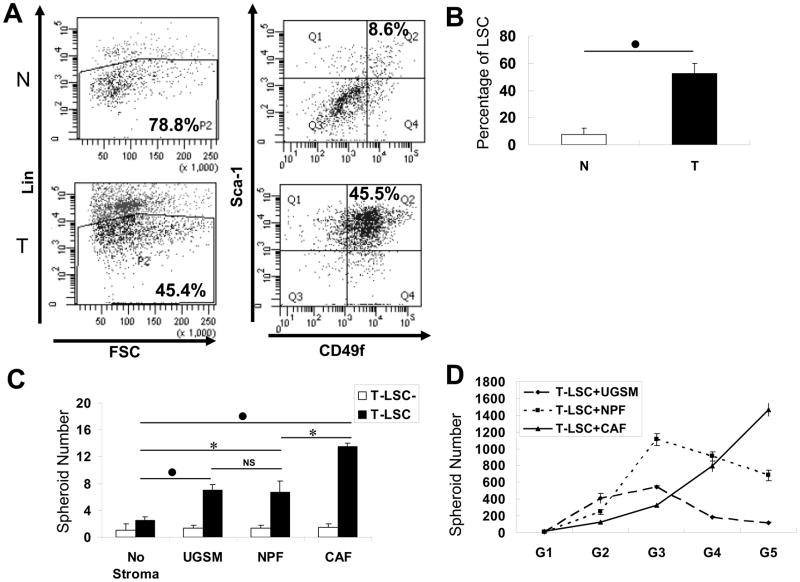
Along with the selection for Sca-1 marker, single cells from normal proximal prostatic tissues or tumors were also enriched based on another putative murine stem cell surface marker, CD49f (27). Cells separated by Lin (Fig. 2A, left) showed the percentage of Lin− cells in the whole input cell portion to be 78.8% in normal prostatic tissue that was collected from a mouse of 12 months of age, and 45.4% in the prostate tumor from an age-matched cPten−/−L mouse. The Lin− fraction was then sorted by the expression levels of Sca-1 and CD49f. The Sca-1 and CD49f double positive cells were named LSC cells and the total remainder as LSC− cells (Fig. 2A, right). The percentage of LSC cells in the Lin− cell population in this normal prostatic tissue was 8.6% (7.27±4.85%, mean from 6 animals) while that in the tumor counterpart was 45.5% (52.37±7.74%, mean from 7 animals), a seven fold increase (p<0.01) in the tumor tissue (Fig. 2B). When LSC or LSC− cells isolated from the same tumor were assayed, the spheroid-forming ability of T-LSC cells, not T-LSC− cells, could be significantly increased when co-cultured with stromal cells (Fig. 2C). The efficiency of spheroid-formation by T-LSC cells increased by about 3-fold in the presence of normal primary stromal cells, UGSM (p<0.01) or NPFs (p<0.05), but by 5.4-fold when T-LSC cells were grown with CAFs (p<0.01). There was no significant difference between UGSM and NPFs in this stimulatory effect. Similar results were obtained from the analyses of T-LSCs from two other tumors comparing the effect of NPFs and CAFs (p<0.05) as shown in Supplementary Fig. S1C. For the analysis of self-renewal ability, spheroids formed from T-LSC cells were dissociated and reseeded in fresh matrigel keeping the input epithelial and stromal cell numbers the same. This serial propagation was repeated another four times. The differences in the influence of UGSM, NPFs or CAFs on spheroid formation in these subsequent generations (G2 to G5) were variable. UGSM and NPFs were better than CAFs at G2 and G3, and CAFs were approximately equal to NPFs but better than UGSM at G4. Compared to UGSM or NPFs, CAFs appeared to be more efficient at G5 (Fig. 2D). At G4 and G5, the number of spheroids formed declined from that in G3 when grown with UGSM or NPFs. However, the T-LSC cells with CAFs displayed a continued trend in increase with serial passages. While stromal cells, in general, enhanced the spheroid-forming efficiency of T-LSC in the first generation, it remains to be determined if T-LSC alone may exhibit a different growth potential at subsequent generations.
Figure 2. Spheroid formation analysis with the LSC cell subpopulations.
A. Illustration of the FACS plots used for the isolation of LSC and LSC− cell subpopulations. Cells isolated from normal (N) and tumor (T) prostates were first segregated by Lin markers (left), and Lin− cells were further separated by the expression levels of Sca-1 and CD49f (right). Cells with the Lin−Sca-1+CD49f+ phenotype (Q2) were labeled as LSC; others (Q1, 3, and 4) as LSC−. B. Comparison of the proportion of LSC cell fraction in Lin− cell subpopulation between normal and tumor prostates. C. Analysis of the spheroid-forming abilities of T-LSC and T-LSC− cells in the 3D co-culture system with stromal cells of different sources. T-LSC (104) or T-LSC− (104) cells, isolated from the same tumor tissue were cultured with the same number of UGSM, NPFs, or CAFs for 14 days. D. Spheroids generated from T-LSC cells were passaged for 5 generations, each time in the presence of the same type of stromal cells as indicated. In panels B and C, statistical evaluation of the difference between two marked groups is indicated by *, for p<0.05, ●, for p<0.01, or NS, for not significant.
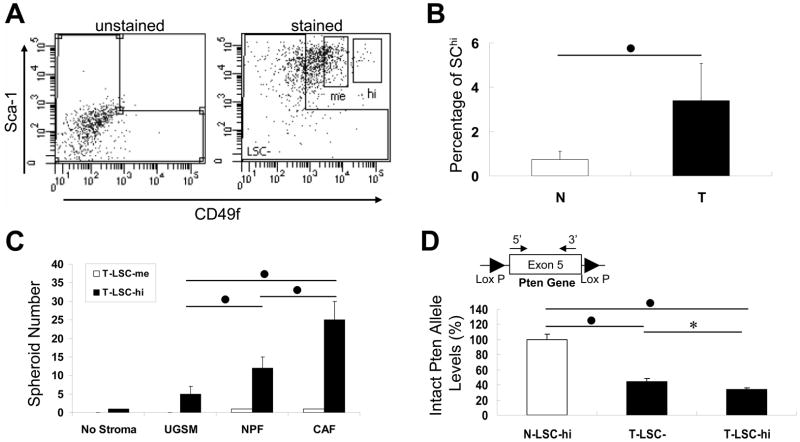
We isolated a subpopulation of LSC that expressed highest levels of CD49f, which we marked as LSChi and the remainder as LSCme denoting a medium level of CD49f expression (Fig. 3A, right). The FACS plot on the left of Fig. 3A showed the gating that was used to demarcate LSC− cells. In this tumor collected from a 12-month-old mouse, T-LSChi cells constituted 2.3% of the T-Lin− cell population. Analysis of results from 6 normal prostate and 11 prostate tumors indicated that the content of LSChi cells increased from 0.73±0.38% in normal to 3.39±1.68% in the tumor subpopulation (Fig. 3B). T-LSChi, not T-LSCme, cells were found capable of forming spheroids. The spheroid-forming efficiency of T-LSChi that was grown with UGSM cells significantly increased to 2.4-and 5.0-fold when UGSM was replaced by NPFs and CAFs, respectively (p<0.01) (Fig. 3C).
Figure 3. Analysis of the T-LSChi cell subpopulation.
A. FACS plot showing separation of T-LSChi or T-LSCme cells from the T-LSC cell subpopulation. Unstained cells were used for gating T-LSC− cells in the Lin− fraction. Labeled cells with the highest CD49f and Sca-1 levels were labeled as T-LSChi, and those with medium CD49f expression levels were T-LSCme. B. Comparison of the percentage of LSChi cell subpopulation in the Lin− cell subpopulation between normal (N) and tumor (T) prostates. Accumulated data from 6 normal and 11 tumored mice demonstrated that the LSChi cell subpopulation in tumors was 4- to 5-fold higher than that in the normal prostate. C. Analysis of the spheroid-forming ability with T-LSChi and T-LSCme cells co-cultured with different stromal cells. D. Detection of Pten gene deletion in T-LSChi cells. Genomic DNA extracted from N-LSChi, T-LSC− and T-LSChi cells were examined using primers specific to Pten exon5 DNA sequence by real-time PCR. Intact Pten allele level detected in N-LSChi cells was set to 100%. The location of primers inside Pten exon 5 is shown above the bar of T-LSChi cells. In panels B–D, statistical significance of the difference between a marked pair is indicated by *, for p<0.05, ●, for p<0.01.
Gland-forming potential of the cell subpopulations
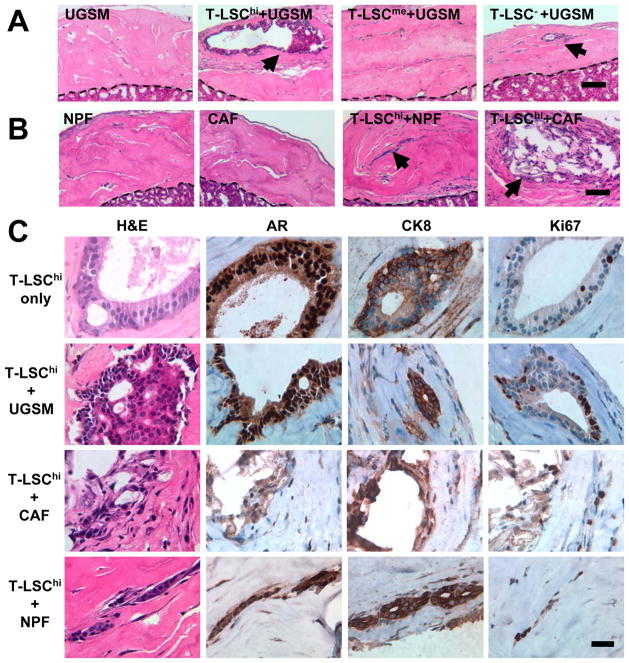
To determine the tissue regeneration characteristics, grafts were produced by mixing T-LSChi, T-LSCme or T-LSC− cells with UGSM cells for transplanting under renal capsules. The H&E staining of the representative tissue sections from grafts collected after ten weeks are shown in Fig. 4A. Grafts from T-LSChi cells displayed prostatic glandular structures containing multiple compact cell layers mimicking a tumor-like histology. Cells located in the structures showed enlarged nuclei (Fig. 4C; H&E). The incidence of glandular structure formation by T-LSChi cells was 100% (Table 1). However, no such structures were found in grafts formed from T-LSCme cells. Two out of six grafts formed from T-LSC− cells contained small glandular structures with multiple cell layers. IHC staining confirmed that the grafts formed from T-LSChi cells with UGSM were mostly composed of AR, CK8 and CRE positive cells (Fig. 4C; Supplementary Fig. S2C). Since UGSM cells were prepared from embryos lacking the Cre transgene, any contaminating epithelial cells therein should also be negative for CRE staining. Of a total of eight glandular structures from three different grafts examined in details, we detected CRE staining in all (100%). Practically all cells (98–100%) within the structures stained for CRE expression (data not shown). Thus, it was considered unlikely that the structures formed from T-LSChi were derived from contaminating epithelial cells present in the UGSM preparation. To determine whether T-LSChi cells alone could have tumorigenic potential, grafts containing only tumor T-LSChi cells (104), without UGSM cells, were prepared and transplanted into four animals. A single glandular structure was found in one of these grafts. This lesion in this graft resembled murine prostate intraepithelial neoplasia (PIN) with multiple layers of cells with enlarged nuclei (Fig. 4C; top row). The majority of the cells inside this glandular structure were AR and CK8 positive with a few also displaying proliferation (Ki67 positivity).
Figure 4. In vivo tissue regeneration analysis with tumor-derived cell subpopulations.
A. Hematoxylin and eosin (H&E) staining of sections of grafts generated from different tumor cell fractions with UGSM cells. The dashed line separates graft from the kidney tissues. Glandular structures are indicated by arrows. Bar, 100 μm. B. H&E staining of sections of grafts from NPFs alone, CAFs alone, and T-LSChi cells with either NPFs or CAFs. Arrows mark glandular structures. Bar, 100 μm. C. Comparison of the glandular structures at higher magnification. Sections were stained by H&E, and for AR, CK8 or Ki67. Bar, 25 μm.
Table 1.
Detection of prostatic glandular structures in grafts.
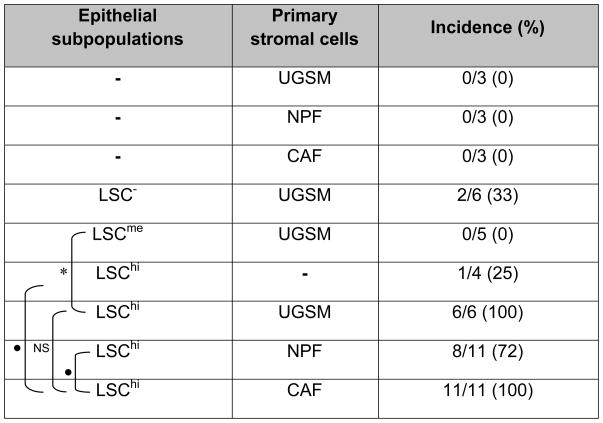 |
Grafts containing tumor epithelial subpopulation (104) and a type of primary stromal cells (104) were transplanted under kidney capsules. After 10 weeks, each animal was sacrificed, the kidney with the graft isolated, fixed, and then thin tissue sections were stained to determine the presence or absence of microscopically detectable glandular structures in the grafts. Statistical evaluation of the difference in incidence of detection of glandular structures between a marked pair of individual groups is indicated by
for p<0.05,
for p<0.01, or NS, for not significant.
The in vivo effects of NPFs and CAFs were also examined. Grafts formed from these cells alone did not contain any detectable glandular structures (Fig. 4B). Eight of eleven grafts formed from T-LSChi and NPFs were found to contain glandular structures (Table 2; Fig. 4B); their sizes were variable. All grafts (11 of 11) generated from T-LSChi and CAFs were found to form glandular structures and, like with NPF, also of varied sizes. However, as illustrated in Supplementary Fig. S2A, the number of these structures detected within a graft was higher with CAFs (4 to 10) than with NPFs (2 to 4). An estimation of the areas covered by the glandular structures in each graft showed that the cumulative values were larger in the presence of CAFs than NPFs (p <0.05). These results are shown in Supplementary Fig. S2B. IHC staining of sections of these grafts for AR, CK8 and Ki67 are shown in Fig. 4C, and for NKX3.1, CK5, Vimentin and CRE in Supplementary Fig. S2D. In general, the expression of AR, CK8, NKX3.1 and CRE was detected in the majority of the cells within the structures with a small number of cells staining for CK5. The NKX3.1 staining results implied that the CSC under study could differentiate into NKX3.1 expressing cells which might be the same as the luminal epithelial cells since all prostatic luminal cells express NKX3.1. Vimentin-positive fibroblastic cells were detected mostly outside the structures, although a few could also be detected inside in the case of T-LSChi + CAFs structures (Supplementary Fig. S2D). We also detected a large number of proliferative cells with Ki67 expression in these glandular structures, and the proliferation index in T-LSChi + CAFs was found to be approximately 3.5-fold higher than that in T-LSChi + NPFs (Supplementary Fig. S2C).
Analysis of selected gene expression and Pten deletion in tumor LSChi cells
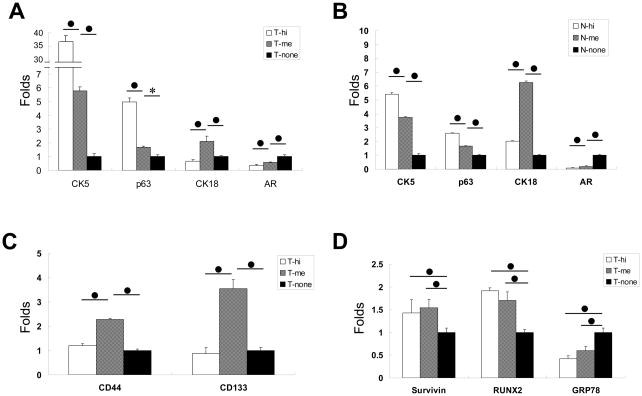
For further characterization of the tumor T-LSChi cells, RNA expression of specific candidate genes in T-LSChi, T-LSCme and T-LSC− cells was examined. We analyzed the expression of markers for the luminal cell (CK18), basal cell (CK5 and p63), and androgen receptor (AR) in the three subpopulations of cells. Real-time quantitative PCR values obtained from each PCR reaction were normalized to that of β-actin. The mean ratio of a given gene expression relative to β-actin in LSC− cells was set as 1 and the expression levels in LSChi or LSCme cells were calculated as fold changes relative to that in LSC− cells. The results showed that T-LSChi cells expressed higher CK5 and p63 but lower CK18, compared to T-LSCme cells, which in turn appeared to express higher levels of CK5, p63 and CK18 than T-LSC− cells. Of the three cell subpopulations, T-LSChi cells also expressed the lowest levels of AR (Fig. 5A). Similar RNA expression patterns were also observed in N-LSChi, N-LSCme and N-LSC− cells, indicating that T-LSChi shared traits with their normal counterparts (Fig. 5B). Recognizing that CD44 and CD133 were described earlier as putative surface markers for human normal and prostate cancer stem cells (19, 28), and CD133, also as an epithelial stem cell marker in murine prostate (29), we examined the expression of these genes in the three subpopulations. Contrary to our expectation, we detected highest RNA levels of CD44 and CD133 in the T-LSCme cells followed by T-LSChi and T-LSC− cells (Fig. 5C, left). We also analyzed expression of tumor-related genes Survivin, Runx2 and Grp78. We recently described that Survivin and RUNX2 are highly expressed in both mouse and human prostate cancers (30, 31), and that loss of GRP78 can inhibit the progression of prostate cancer in the Pten deletion mouse model of prostate cancer (32). A high level of expression of both Survivin and Runx2 genes was detected in T-LSChi and T-LSCme cells compared to T-LSC− cells. In contrast, Grp78 expression in the T-LSC− cells was higher than in the other two cell subpopulations (Fig. 5D), implying that GRP78 might be more relevant to terminally differentiated cancer cells in the model.
Figure 5. Analyses of gene expression in LSChi cells.
The RNA expression levels of different markers in LSChi (hi), LSCme (me) and LSC− (none)cells isolated from either normal (N) or tumor (T) prostate tissues were examined by real-time quantitative PCR. A. Levels of CK5, p63, CK18, and AR expressed in the tumor subpopulations. B. Similar analysis with the subpopulations from the normal prostate. C. Analysis of CD44 and CD133 expression in tumor cell subpopulations. D. Analysis of expression of Survivin, Runx2 and Grp78 in the tumor cell subpopulations. In each panel, statistical significance of the difference in expression level of a gene between a marked pair is indicated by *, for p<0.05, or ●, for p<0.01.
Next we attempted to determine the Pten allelic status in the T-LSChi cells. For this purpose, genomic DNA extracted from tumor T-LSChi and T-LSC− cells, and also from N-LSChi cells derived from a normal prostate of a littermate control animal (with floxed Pten alleles, floxed L transgene, but no Cre transgene) was subjected to real-time quantitative PCR. The pair of primers (PtenEX5-forward and PtenEX5-Reverse) used is located inside Pten exon 5 that is flanked by two LoxP sites. In the event Cre recombination occurred in all cells of the T-LSChi subpopulation, no PCR product would be expected. PCR values obtained from each PCR reaction were normalized to that of tubulin. The mean ratio of the DNA extracted from N-LSChi relative to tubulin was set to be 100% because these mice did not have Cre gene to induce Pten exon 5 deletion. The results showed that compared to N-LSChi cells, 43.9±4.2% Pten alleles in T-LSC− cells (p<0.01) contained exon 5 and this number reduced to 33.5±2.5% (p<0.05) in T-LSChi cells (Fig. 3D). Thus, it appeared that approximately two-thirds of Pten alleles lost their exon 5 in T-LSChi cells. It could not, however, be discerned if the majority of the cells had both alleles or one allele deleted and what might be the deletion status in the various cell types segregating as T-LSChi cells.
Discussion
It is becoming increasingly clear that normal tissue stem cells are localized in a defined microenvironment that provides specific factors for the maintenance of the properties of the stem cells as well as for the regulation of a balance between proliferation, differentiation and quiescence of these cells (33–36). In prostate cancer, there is strong evidence that signals originating from the cancer-associated fibroblasts (CAFs) could significantly enhance the tumorigenicity of cancer cells. As a central role for CSCs is being ascribed for tumor homeostasis and progression (37–39), we wished to inquire if CAFs may regulate the biology of prostate CSCs. This is a critical question for the hypothesis that terminally differentiated cancer cells may have only limited proliferation ability and CSCs, with asymmetric division to both self-renew and differentiate may indeed be responsible for the growth and progression of the tumor. In this report, we describe a mouse model of prostate adenocarcinoma from which CSC-enriched epithelial cells were derived to examine the effects of CAFs that were also generated from the tumors of the same model.
Interest in this study is three-fold. First, a modified method is described for the isolation of epithelial cell fractions retaining a small number of cells with properties of putative CSCs. A cell fraction from this tumor model is shown to possess self-renewal and spheroid-forming abilities along with multipotentiality for differentiation in vitro, and the ability to form tumor-like glandular structures in vivo under appropriate conditions. The selection for tumor cells (T-LSChi) with high levels of expression of both Sca-1 and CD49f surface markers appears to discriminate between these cells from those with high Sca-1 and medium CD49f levels (T-LSCme). While a cell fraction contained in the T-LSChi subpopulation displays spheroid-forming ability and the capability to generate prostate glandular structures, the T-LSCme subpopulation is practically devoid of these capabilities. The CSC-enriched T-LSChi is still mostly composed of non-CSC epithelial cells as evident from the efficiency of in vitro spheroid-forming ability, although it is likely that viability of all cells in the subfractions may not withstand the steps used for the isolation. The bulk of the cells in the T-LSChi compartment may represent transit-amplifying cells and terminally differentiated cells, and, thus, indicating that the markers used like Sca-1 and CD49f, are shared with non-CSC cells, but still, use of these markers in a quantitative manner, as shown here, does contribute to enrichment of prostate CSCs from the tumors of this mouse model.
Second, we have observed a significant difference in the pattern of relative expression of certain relevant genes in the T-LSChi and T-LSCme subpopulations. While expressions of basal cell markers CK5 and p63 are stronger in T-LSChi relative to T-LSCme, the T-LSCme fraction is found to express higher levels of CK18 and AR as compared to T-LSChi, although the level of AR is significantly reduced in both subgroups in comparison to the T-LSC− cells. The same general pattern is found in the respective subpopulations from the normal mouse prostate. Thus, it appears that CSC-enriched fraction from the prostate tumor of the Pten deletion model contain cells with the characteristics of the similarly enriched fraction from the normal mouse prostate. Two other putative epithelial stem cell markers were examined. The levels of each of CD44 and CD133 transcripts appear to be significantly higher in T-LSCme subpopulation of the tumors relative to either T-LSChi or T-LSC− groups of cells, implying that these two markers may not characterize the CSCs of the tumors in the Pten deletion mouse model. We also examined expression levels of three cancer-related genes: Survivin, Runx2 and Grp78. Survivin, a member of the inhibitor of apoptosis (IAP) protein family, is highly expressed in human cancer (40). In the conditional Pten deletion mice, we demonstrated a strong correlation between increased levels of Runx2 transcription factor with the growth of the tumor (31), an observation that is very similar to what we described for Survivin protein levels in the same model (30). Moreover, Runx2 appears to be a major regulator of Survivin gene transcription in prostate cancer cells (31). GRP78, a major ER chaperone is reported to be highly induced in a wide range of tumors including prostate cancer (41), and we described that loss of GRP78 in the prostatic epithelium can prevent prostate tumor formation in the Pten deletion model (32). Here, we find that while Grp78 expression is higher in the T-LSC− population compared to either T-LSChi or T-LSCme subpopulations, the pattern is opposite in the case of Survivin or Runx2. Survivin and Runx2 are expressed in both T-LSChi and T-LSCme groups of cells at levels even higher than the bulk of the cancer cells represented in the T-LSC− fraction. Based on these results, we project that high levels of expression of Survivin and Runx2 might be associated with both CSCs and transit-amplifying cells as it is with many cancer cells. However, this contention remains to be tested at the level of individual cells, a task that is difficult at this time in the absence of definitive markers for the cell types under study.
Third, for the first time, we demonstrate that the spheroid-forming efficiency of the CSC-enriched cells is differentially influenced by the fibroblasts in co-cultures. The modified spheroid-forming co-culture system we used has the promise to be a powerful method to facilitate the studies of paracrine signalings in interactions between stromal fibroblasts and CSCs. Because fibroblasts are located on the insert above the matrigel layer, there is no direct cell-cell contact between the two cell groups in this system. An important finding from such analysis is that as compared to UGSM or NPFs, CAFs enhance spheroid formation in the first generation by approximately two-fold. In vivo, the grafts grown from the T-LSChi cells are found to contain multiple glandular structures in each case, although grafts formed with CAFs appear to exhibit higher proliferative index as compared to those formed with NPFs. The observations with CAFs underscore a role of CAFs in CSC biology, and open up possibilities for better identifying the responsible molecular interactions.
In summary, our study describes a process refined to enrich the putative CSC population using the surface marker phenotype of Lin−Sca-1hiCD49fhi from the prostate adenocarcinomas of the Pten deletion model and demonstrates that such cells have the capacity to form tumor-like structures in spheroids in vitro and grafts in vivo. The CSCs appear to retain properties of normal tissue stem cells, such as, the potential to self-renew and to generate differentiated progenies. Most notably, we present evidence that CAFs could enhance both the stemness and growth potentials of the CSCs. It is likely that these new clues could be further developed to better understand the biology of CSCs in prostate cancer and potentially, in cancers, in general.
Supplementary Material
Acknowledgments
Grant Support: NIH RO1 CA113392 and NIH RO1 CA59705 (P. Roy-Burman), and in part, by supports to C-P. Liao and H. Adisetiyo from the California Institute for Regenerative Medicine training grant T1-004.
We thank Joseph Jeong for help with the procedure of tissue grafting under renal capsule, Amy Lee for sequences of Grp78 primer set, Cynthia Cunningham for help with FACS analyses and all members of the Roy-Burman Laboratory for assistance in various aspects of this work.
References
- 1.Hu M, Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008;18:27–34. doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–49. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 3.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 4.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–9. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 6.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–11. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunha GR, Hayward SW, Wang YZ, Ricke WA. Role of the stromal microenvironment in carcinogenesis of the prostate. Int J Cancer. 2003;107:1–10. doi: 10.1002/ijc.11335. [DOI] [PubMed] [Google Scholar]
- 8.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 9.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–7. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Tan BT, Park CY, Ailles LE, Weissman IL. The cancer stem cell hypothesis: a work in progress. Lab Invest. 2006;86:1203–7. doi: 10.1038/labinvest.3700488. [DOI] [PubMed] [Google Scholar]
- 12.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 13.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 14.Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–56. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 16.Fang D, Nguyen TK, Leishear K, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–37. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 17.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrandina G, Bonanno G, Pierelli L, et al. Expression of CD133-1 and CD133-2 in ovarian cancer. Int J Gynecol Cancer. 2008;18:506–14. doi: 10.1111/j.1525-1438.2007.01056.x. [DOI] [PubMed] [Google Scholar]
- 19.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 20.Liao CP, Zhong C, Saribekyan G, et al. Mouse models of prostate adenocarcinoma with the capacity to monitor spontaneous carcinogenesis by bioluminescence or fluorescence. Cancer Res. 2007;67:7525–33. doi: 10.1158/0008-5472.CAN-07-0668. [DOI] [PubMed] [Google Scholar]
- 21.Yang S, Pham LK, Liao CP, Frenkel B, Reddi AH, Roy-Burman P. A novel bone morphogenetic protein signaling in heterotypic cell interactions in prostate cancer. Cancer Res. 2008;68:198–205. doi: 10.1158/0008-5472.CAN-07-5074. [DOI] [PubMed] [Google Scholar]
- 22.Xin L, Lukacs RU, Lawson DA, Cheng D, Witte ON. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25:2760–9. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- 23.Lang SH, Stark M, Collins A, Paul AB, Stower MJ, Maitland NJ. Experimental prostate epithelial morphogenesis in response to stroma and three-dimensional matrigel culture. Cell Growth Differ. 2001;12:631–40. [PubMed] [Google Scholar]
- 24.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci U S A. 2005;102:6942–7. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci U S A. 2003;100 (Suppl 1):11896–903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65:8887–95. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]
- 27.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 28.Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539–45. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- 29.Leong KG, Wang BE, Johnson L, Gao WQ. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–8. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- 30.Yang S, Lim M, Pham LK, et al. Bone morphogenetic protein 7 protects prostate cancer cells from stress-induced apoptosis via both Smad and c-Jun NH2-terminal kinase pathways. Cancer Res. 2006;66:4285–90. doi: 10.1158/0008-5472.CAN-05-4456. [DOI] [PubMed] [Google Scholar]
- 31.Lim M, Zhong C, Yang S, Bell AM, Cohen MB, Roy-Burman P. Runx2 regulates survivin expression in prostate cancer cells. Lab Invest. 2010;90:222–33. doi: 10.1038/labinvest.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y, Wey S, Wang M, et al. Pten null prostate tumorigenesis and AKT activation are blocked by targeted knockout of ER chaperone GRP78/BiP in prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:19444–9. doi: 10.1073/pnas.0807691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–44. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston LA. Competitive interactions between cells: death, growth, and geography. Science. 2009;324:1679–82. doi: 10.1126/science.1163862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takao T, Tsujimura A. Prostate stem cells: the niche and cell markers. Int J Urol. 2008;15:289–94. doi: 10.1111/j.1442-2042.2008.02047.x. [DOI] [PubMed] [Google Scholar]
- 36.Nikitin AY, Matoso A, Roy-Burman P. Prostate stem cells and cancer. Histol Histopathol. 2007;22:1043–9. doi: 10.14670/HH-22.1043. [DOI] [PubMed] [Google Scholar]
- 37.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–2. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 38.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–3. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang SH, Frame FM, Collins AT. Prostate cancer stem cells. J Pathol. 2009;217:299–306. doi: 10.1002/path.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 41.Daneshmand S, Quek ML, Lin E, et al. Glucose-regulated protein GRP78 is up-regulated in prostate cancer and correlates with recurrence and survival. Hum Pathol. 2007;38:1547–52. doi: 10.1016/j.humpath.2007.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.