Abstract
Factors involved in the cellular response to double strand break (DSB) DNA damage have been identified as potential therapeutic targets that would greatly sensitize cancer cells to radiotherapy and genotoxic chemotherapy. They could disable the repair machinery and/or reinstate normal cell cycle checkpoint leading to growth arrest, senescence and apoptosis. It is now clear that a major aspect of the DNA damage response occurs through specific interactions with chromatin structure and its modulation. It implicates highly dynamic post-translational modifications of histones that are critical for DNA damage recognition/signaling, repair of the lesion and release of cell cycle arrest. Therefore, drugs that target the enzymes responsible for these modifications or the protein modules reading them have very high therapeutic potential. This review presents the current state of knowledge on the different chromatin modifications and their roles in each step of eukaryotic DSB DNA damage response.
1/ Introduction
In order to preserve its genomic integrity, the eukaryotic cell needs to be protected against agents that cause DNA damage. Indeed, cellular DNA is continuously exposed to exogenous (such as chemicals, UV-radiation, ionizing radiation) as well as endogenous (reactive oxygen species, alkylating agents such as S-adenosylmethionine etc…) insults known to induce various DNA lesions (1). To counteract these injuries, the cell has developed highly conserved DNA-damage responses (DDRs) that activate different repair pathways specifically adapted to the type of damage. These include (i) base-excision repair, (ii) nucleotide-excision repair, (iii) mismatch repair, and (iv) double-strand break repair (DSB) which is the most deleterious form of DNA damage since it can lead to loss of genetic material (2). DSB are mainly repaired by homologous recombination (HR) and non-homologous end-joining (NHEJ). HR uses the undamaged homologous chromosome or sister chromatid as a template to copy the missing information at the break. In contrast, NHEJ consists in the direct ligation of the two broken ends, which can produce short deletions.
In eukaryotic cells, the DNA-damage repair occurs in the context of chromatin. The chromatin is a DNA-protein structure that exists as a repetition of the basic unit called the nucleosome. A nucleosome is formed by an octamer of histones, containing two copies of each H2A, H2B, H3 and H4, wrapped with 146bp of DNA. The chromatin is a dynamic structure that regulates DNA accessibility during essential nuclear events such as replication, transcription, recombination and DNA-damage repair. Modulation of chromatin compaction can be regulated by different processes: introduction of histone variants into the nucleosome which confers different bio-physical features; post-translational histone modifications mainly occurring on histone tails protruding from the nucleosome; ATP-dependant chromatin remodeling complexes that have the ability to disrupt, evict or slide the entire nucleosome on the chromatin fiber; and histone chaperones which assist in nucleosome assembly/disassembly (3).
After DNA damage induction, the chromatin needs to be in an “open” state in order to allow the repair factors to access to the DNA molecule. This DDR process requires multiple steps including the initial signaling of the break, the access to the DNA for efficient repair, and the restoration of the chromatin to its initial state. In this review, we describe these steps of DDR involved in the DSB repair. Drugs that target the chromatin modifiers or readers implicated DNA damage response have very high therapeutic potential (see (4-6) and accompanying Focus reviews (7-10)).
2/ Recognition and Signaling of DNA-damage: Key role of γ-H2AX
When DNA damage occurs in the cell, the priority is to detect it and to signal it for repair. Even though these processes have been intensively studied, it is still not clear which factor arrives first at the break to recognize it and induce the DDR. Among all chromatin modifications linked to DSB damage response (Table 1), it is clear that phosphorylation of the H2A variant H2AX occurs within few minutes following the break, and is probably the first histone modification appearing in its vicinity (reviewed in (8, 11)). This phosphorylation occurs in a unique conserved SQE motif in the C-terminal tail (at serine 129 (S129) of yeast H2A or S139 of the H2AX human variant, so-called γ-H2AX (12, 13)). The kinases responsible for this modification have been identified as phosphatidylinositol 3-kinase-related kinases (PIKKs): the ataxia-telangiectasia mutated (ATM), ATM- and Rad3-related (ATR), and the DNA-dependant protein kinase (DNA-PK). ATM and DNA-PK principally function after ionizing radiation (IR) whereas ATR responds to replication stress and ultra-violet (UV) irradiation (14-16). In human cells, γ-H2AX spreads over more than 1Mb on each side of the break (50kb in yeast) (13, 17-20), thus amplifying the repair signal, which make it easily detectable by immunofluorescence, and commonly used as a bio-marker of DNA-damage nuclear foci (for review, see (21)). Moreover, mice deficient for γ-H2AX are radiosensitive and show chromosomal aberrations, strengthening the critical role of γ-H2AX in DDR (22).
Table1.
Histone modifications influencing DNA-damage response.
| Step of DDR | Histone residue (human) | Type of modification | Enzyme |
|---|---|---|---|
| 1/ Signaling | H2AX S139 | Phosphorylation | ATM/ATR, DNA-PK |
| H2A/H2AX | Ubiquitination | RNF8/RNF168 | |
| H4 K20 | Methylation | Set8/Suv4-20 | |
| H3 K79 | Methylation | Dot1 | |
| H2AX Y142 | Dephosphorylation | EYA1 | |
| H4 K91 | Mono-ubiquitination | BBAP | |
| H2AZ K126/133 | Sumoylation | ? | |
| 2/ Opening | H4/H2A(X) | Acetylation | Tip60/yNuA4 |
| H3 K9 | Acetylation | Gcn5, CBP/p300 | |
| 3/ Restoring | H2AX S139 | Dephosphorylation | yPph3/hPP4, PP2A, PP6, Wip1 |
| H3/H4 K | Deactetylation | Sin3/Rpd3, Sir2, Hst1/3/4 | |
| H4 S1 | Phosphorylation | Casein Kinase 2 (CK2) | |
| H2B S14 | Phosphorylation | Ste20 | |
| H3 K56 | Acetylation | yRtt109, CBP/p300, Gcn5 | |
| H3 K14, K23 | Acetylation | Gcn5 | |
| H4 K5, K12 | Acetylation | Hat1 | |
| H4 K91 | Acetylation | Hat1 | |
| H2A K119 | Mono-ubiquitination | Ring1b/Ring2 | |
It is still poorly understood which DSB sensor induces H2AX kinase recruitment. Different models have been proposed to explain ATM relocalization and activation at the break. First, it has been suggested that conformational changes of the DNA activate ATM at the DSB. Another theory suggests that ATM activation is dependent on initial DNA damage detection by the MRN repair complex (Mre11, Rad50, Nbs1) (Fig. 1) (23). In addition, it has been shown that inactivation of human histone acetyltransferases (HAT) of H3 and H4, such as hMOF or TIP60, suppresses ATM activation (24-26). Surprisingly, it has been found that γ-H2AX foci do not form with the same dynamic on different chromatin regions after DNA-damage, but more efficiently in euchromatin (27, 28). This phenomenon can be due to either less DSBs generated in heterochromatin, or because heterochromatin features inhibit the large spreading of repair marks near DSBs. In addition, Iacovoni et al. demonstrated that γ-H2AX spreads in a bidirectional but not necessarily symmetrical manner, being influenced by the transcription state of the gene present on the DNA surrounding the DSB (29). Another group demonstrated different dynamics and factors regulating γ-H2AX domains proximal or distal to the break (30). Finally, Durocher and Robert groups have also used γ-H2AX to map in high-resolution the genome-wide fragile sites (31).
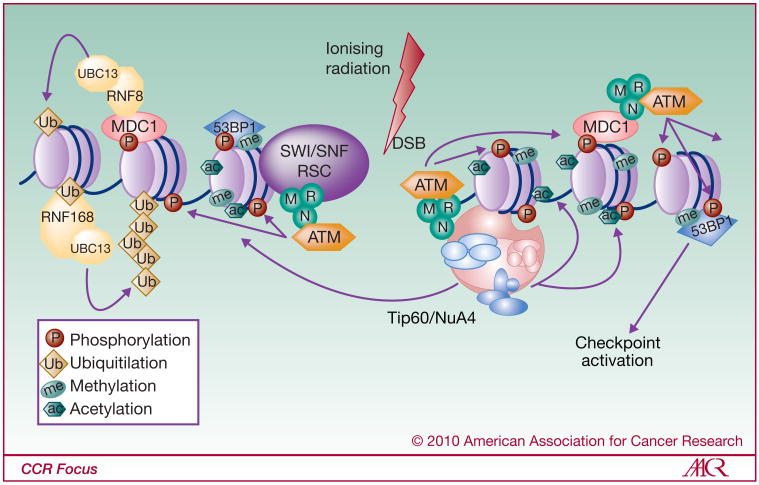
Figure 1.
Model of histone modifications and chromatin remodeling during DNA DSB repair,
Step 1: Recognition and signaling of a DSB. g-H2AX plays a key role in DNA-damage signaling, acting as a platform of assembly for the repair factors as well as for checkpoint proteins. Immediately following the apparition of a DSB, the MRN complex binds DNA-ends and participates in ATM kinase recruitment. ATM then rapidly phosphorylates H2AX histone variant at the site of the break. Phospho-H2AX, also called γ-H2AX allows the binding, retention and accumulation at the break of the complexes involved in the DDR. The simultaneous presence of the RSC remodeling complex at the break may facilitate the access of the recruited repair factors. Indeed, the mediator protein MDC1 is recruited to the DSB and binds γ-H2AX, where it promotes further ATM and MRN accumulation. As a consequence, γ-H2AX bi-directionally spreads out from the DSB (approximately 2Mb), thus increasing the accumulation of repair factors. MDC1 also recruits RNF8/UBC13 ubiquitin-ligase which ubiquitinates H2A and H2AX, which in turn, is recognized by RNF168-UBC13 H2AX-ubiquitin-ligase complex, resulting in the amplification of γ-H2AX poly-ubiquitination near the DSB. In parallel, γ-H2AX also permits TIP60 HAT recruitment at the break, followed by the acetylation of H2A and H4 histones and destabilization of the nucleosomes. In addition, phosphorylation of H2AX could induce conformational changes in the nucleosome, resulting in the exposition of H4K20me and H3K79me, recognized by the checkpoint protein 53BP1.
Following H2AX phosphorylation, DDR and repair factors accumulate at the break. Indeed, repair factors and checkpoint proteins (MRN, MDC1, BRCA1, 53BP1, UBC13/RNF8, RNF168) and chromatin-remodeling complexes (INO80, SWR1, TIP60-p400) will form foci that colocalize with γ-H2AX (Fig. 1) (22, 32-34). The phosphorylation of H2AX itself has been shown to not affect chromatin organization, but rather has a role in the localization of repair factors at the break (35). Although the presence of γ-H2AX is not required for the initial recruitment of signaling and repair factors (33), it is essential for their accumulation and retention at the break, and amplification of the signal (22, 32, 36). For example, MDC1 directly binds to γ-H2AX via its BRCT domain, and plays a critical role in the accumulation of Nbs1 (subunit of MRN), 53BP1 and ATM (Fig. 1) (37-39). As a consequence, Savic et al. have recently proposed a MDC1- and ATM-dependant γ-H2AX self-reinforcing mechanism that promotes a continued local H2AX phosphorylation (30). The very large domains of γ-H2AX surrounding DSBs are also thought to be binding platforms for cohesins that allow chromosome stability and keep DNA ends in close proximity for the repair process (19).
γ-H2AX is the best-characterized DNA-damage-induced modification, but it has been more recently demonstrated that ubiquitination also occurs rapidly at the break in response to DNA damage. In fact, γ-H2AX mediates the recruitment of the UBC13/RNF8 ubiquitin ligase complex, in a MDC1-dependant manner (40-42), resulting in the poly-ubiquitination of γ-H2AX and H2A at the DSB and this is coordinated with other ubiquitin and sumo ligases (Table1) (34, 43). It has also been shown that RNF8-dependant H2A-ubiquitynation is implicated in the recruitment of 53BP1 and BRCA1-Abraxas-RAP80 complex via direct binding of RAP80 with poly-UbH2AX (Fig. 2) (44-46). In addition, specific mono-ubiquitination of H2A has been reported at damage sites and can participate in local chromatin remodeling (47).
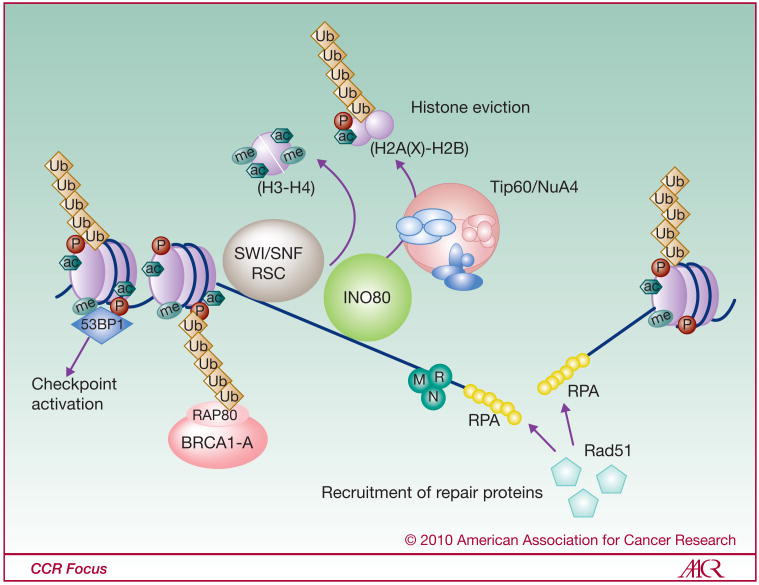
Figure 2.
Model of histone modifications and chromatin remodeling during DNA DSB repair,
Step 2: Opening of chromatin to repair the break. Once the DSB has been recognized and signaled, it is time to repair the break. Histones need to be removed from chromatin in the vicinity of the break to allow access to the DNA to the repair factors. Chromatin-remodelers are then recruited to the DSB. TIP60 complex recruited at the DSB comprises HAT activity as well as histone exchange ability. Following acetylation-dependant nucleosome destabilization, TIP60 complex can remove H2A(X)-H2B histone dimers from chromatin at the break. INO80 is also recruited at the break where it helps removing histones close to the DSB. The SWI/SNF/RSC/BRG-1 remodeling complex is also present at the break where it can associate with γ-H2AX and promote histone eviction or exchange. Such histone eviction allows association of the ssDNA-binding protein RPA with resected DNA and subsequent recruitment of repair factors such as Rad51. Moreover, BRCA1-A repair complex accumulates at the break through direct interaction of its RAP80 subunit with poly-UbH2A(X).
Methylation of histones H3K79 and H4K20 has been shown to be important in the DSB repair pathway. Even if these marks are not induced by DDR but are constitutively present on chromatin, there are evidences that H4K20me and H3K79me help in the recruitment of repair factors at the DSB. In fission yeast, H4K20me allows the recruitment of Crb2 (fission yeast homologue of 53BP1) through its Tudor domain (48), and Crb2 can also bind γ-H2AX through its BRCT domain (49). Mammalian 53BP1 can directly bind H4K20me2 (50) and this binding may work in conjunction with its BRCT domain-dependant binding to γ-H2AX (Fig1 and 2). Furthermore, H4K91 mono-ubiquitination by hBBAP plays a role in association with H4K20me in 53BP1 recruitment during DDR (51). In budding yeast, methylation of H3K79, catalyzed by yDot1 and promoted by H2BK123Ub, has been implicated in DNA repair. Both H3K79me and H2AS129ph are required for the recruitment of yRad9 (h53BP1) to chromatin, through direct recognition of histone marks by its Tudor and BRCT domains, allowing Rad53/Chk2-dependant checkpoint activation (52-55). It has been suggested that yRad9 is then phosphorylated by ATM, oligomerizes and forms a platform for DDR proteins resulting in checkpoint activation (53). The specific role of these constitutive methyl-marks during DNA repair could be explained by the fact that DNA damage may induce chromatin conformation changes, leading to the exposition of K20me and K79me, which then act like docking sites for the recruitment of signal inducers such as the checkpoint protein 53BP1; but some data cast doubts on such a model (52).
More recently, Xiao et al. identified that WSTF tyrosine (Y) kinase constitutively phosphorylates H2AX on Y142, and this phosphorylation is critical for the DDR (56). They show that H2AX Y142ph decreases when γ-H2AX is induced. They propose that dephosphorylation of H2AX Y142 could enhance MDC1 and ATM recruitment to extend and maintain γ-H2AX after DSB formation. At the same time, EYA1 was shown to be the phosphatase targeting H2AX Y142ph, influencing apoptotic or repair complexes recruitment to γ-H2AX in response to DNA damage (57).
3/ DNA-repair factors access to DNA: Chromatin needs to be remodeled at the break
To achieve accurate DNA damage repair, the chromatin needs to be opened in order to facilitate the access for the repair factors at the site of the DNA lesion. Histone-modifiers and ATP-dependant chromatin remodelers are recruited at the break to modulate the chromatin architecture. The destabilization of the nucleosome is thought to require acetylation of histones through the action of histone acetyltransferases (HATs) such as hTip60/yNuA4 (reviewed in (58)). Human Tip60 and yeast NuA4 HAT, as well as the INO80 and SWR1 Swi2-familly ATP-dependant remodelers are recruited to the DSB (Fig. 2), and they can directly interact with γ-H2AX through their common yArp4 subunit (mammalian BAF53) (17, 59-61). hTip60/yNuA4 is one the first modifiers appearing at the break where it acetylates H4 and H2A, and promotes the relaxation of the chromatin at the DSB (17, 62, 63). The mammalian TIP60 complex comprises both Tip60 HAT homologous of yEsa1 in NuA4, and p400/Domino homologous of ySWR1 (64). This overlap of HAT and remodeling activities show that they probably act together on chromatin. Indeed, a study in Drosophila showed that TIP60 can acetylate DNA-damage induced phosphoH2Av (a γ-H2AX-like histone variant), mediating the exchange with an unmodified H2Av at the DSB (65). However, whether this model applies to other species is currently unclear even though two studies showed persistence of γ-H2AX in mammalian cells depleted of Tip60 activity (66, 67). SWR1 is known to remodel chromatin through its ability to incorporate the H2A variant H2AZ at promoters and subtelomeric regions (68-70). Evidence from yeast models suggest that SWR1 may play a role in H2AZ deposition into chromatin surrounding DSB in absence of INO80 (71). In addition, Altaf et al. have recently reported that H2A and H4 acetylation by yNuA4 directly stimulates SWR1-dependant incorporation of H2AZ-containing H2A-H2B dimers into the nucleosome (72). However, the model of SWR1-dependant H2AZ incorporation at DSBs has been debated by Van Hattikum et al. whose studies have shown that there is no accumulation of H2AZ at the DSB during repair (60). Nevertheless, Kalocsay et al. have recently shown that H2AZ is transiently deposited close to the break, and its sumoylation in combination to Rad51 DNA-binding participates in the relocalization of a persistent DSB to the nuclear periphery (73). Thus, further studies need to investigate the function of SWR1 during DSB repair and whether Tip60/NuA4-dependant acetylation of H2A and H4 may be required for γ-H2AX exchange at the break. One report suggests that Tip60-dependent acetylation and removal of H2AX functions through stimulation of the histone ubiquitination by UBC13 (67).
yINO80 complex is rapidly recruited close to the DSB by the direct interaction of its Arp4 and/or Nhp10 subunits with γ-H2AX and influenced by NuA4-dependent acetylation (17, 59, 61). INO80 has been reported to mediate removal of core histones, containing or not H2AZ and H2AX, from region surrounding the DSB (Fig. 2). This nucleosome remodeling then allows resection of DNA at the DSB (60, 74). Furthermore, studies have also shown that yINO80 is required for maintaining high level of γ-H2AX during DNA-repair (71).
Another chromatin remodeler, RSC (member of the SWI/SNF family) is present at the break before INO80 and SWR1. In opposition to INO80 and SWR1, its presence at the break is not γ-H2AX-dependant, and it was shown to interact with MRN (Fig. 1) (75, 76). RSC has been shown to be required for yeast Tel1/Mec1 kinases (homologues of mammalian ATM/ATR) and yRad9 recruitment at the break (77), suggesting that RSC is an early sensor of the DSB (66). However, it is not clear whether RSC or MRN appears first at the break, their recruitment being dependant on each other (76, 78). Furthermore, it has been proposed that in mammalian cells, chromatin decondensation in the vicinity of DSB is dependant on ATP-dependant chromatin remodelers, but not on phosphorylation of H2AX, suggesting a very early function in DSB repair (79). In fact, mammalian SWI/SNF remodeler is critical for efficient induction of γ-H2AX (80), since inhibition of its catalytic core subunits, BRG1 and Brm, compromises phosphorylation of H2AX and γ-H2AX foci formation. More recently, Lee et al. have shown that SWI/SNF binds to γ-H2AX-containing nucleosomes via an interaction between its bromodomain-containing BRG1 subunit with GCN5-dependant acetylated H3, and this binding is important for DSB repair (81).
Altogether, data from the literature indicate that a specific combinations of chromatin marks and ATP-dependent chromatin remodeling allows binding of DNA repair factors and healing of the DNA lesion.
4/ Signaling the end of repair: restoration of chromatin to its initial state
Modulation of chromatin architecture mediated by chromatin modifiers and remodelers is an essential process for DNA damage repair. First, it allows repair factors to access to the damaged DNA (as discussed above), but it is also an important mechanism for switching off the DNA-damage signal. Thus, after repair has been completed, the cell clears the marks associated to DNA-damage signal and restores chromatin organization to its initial state. These processes are essential to recover from the checkpoint arrest and re-enter the cell cycle.
In order to signal to the cell that the repair process is achieved, γ-H2AX is eliminated from chromatin surrounding the repaired DSB, by either eviction or dephosphorylation (Fig. 3). It has been speculated that the presence of SWR1 at the break may allow γ-H2AX removal from chromatin surrounding DSB (82). Some data suggest that yINO80 and ySWR1 function antagonistically at the DSB, yINO80 maintaining the high level of γ-H2AX and ySWR1 replacing it with H2AZ variant (71). Another simple mechanism to get rid of γ-H2AX would be its dephosphorylation (Fig. 3). The HTP-C complex, containing the Pph3 phosphatase catalytic subunit, has been identified in yeast to be the γ-H2AX phosphatase, while the human phosphatase function is attributed to both PP2A and PP4C (83-86). These reports suggest that yPph3 and hPP4C (human Pph3 ortholog) have a function in the checkpoint termination. Interestingly, yPph3 is though to dephosphorylate γ-H2AX after its removal from chromatin, whereas hPP4C-dependant dephosphorylation appears to take place in the chromatin. It still remains to be determined if hPP2A dephosphorylates γ-H2AX directly on chromatin or displaced γ-H2AX. Very recently, other phosphatases have been characterized, hPP6 and hWip1 (87, 88). hPP6 has been shown to contribute to γ-H2AX dephosphorylation and subsequent checkpoint release (88). hWip1, whose expression is induced after DNA-damage, can bind to chromatin, co-localizes with γ-H2AX foci and regulates γ-H2AX dephosphorylation during recovery (87). It is interesting to notice that yeast γ-H2AX spread out over 50kb surrounding the DSB, but is less abundant as close as 1 or 2kb from the DSB, where INO80 and MRN are most present (17, 20). It is then logical to speculate that γ-H2AX is evicted from chromatin proximal to DSB, whereas it is dephosphorylated in the chromatin farther away. This model is supported by two different dynamics of γ-H2AX accumulation/removal depending on the position relative to the break (30).
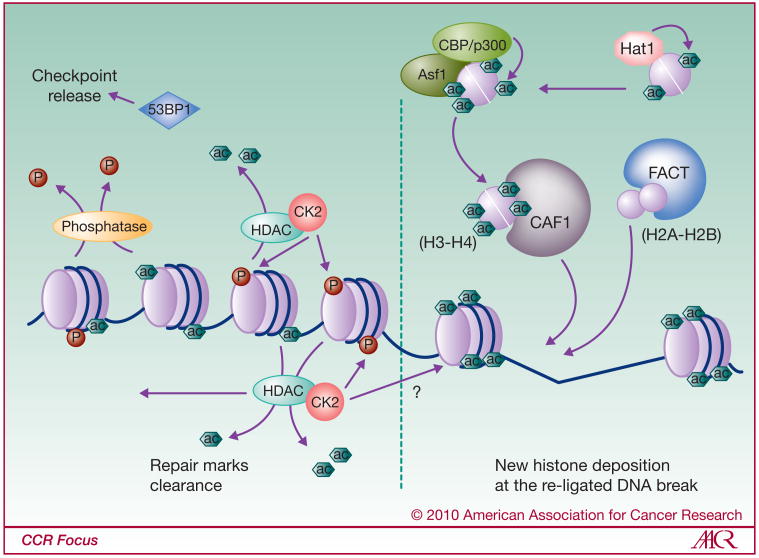
Figure 3.
Model of histone modifications and chromatin remodeling during DNA DSB repair,
Step 3: Chromatin restoration after DNA-break repair. When repair of the DSB is completed, the chromatin needs to be restored and the repair specific histone marks need to be removed in order to release repair factors and cell cycle checkpoints. Thus, γ-H2AX has to disappear from the repaired site. Phosphatases such as PP2A and PP4C dephosphorylate γ-H2AX and allow release of checkpoint factors like 53BP1. In order to restore chromatin, new histones are deposited onto the DNA. Histone chaperones such as FACT and CAF1 have been implicated in this process. Moreover, H3-H4 histones deposited by CAF1 are first acetylated by Hat1, and then by CBP/p300/Rtt109-Asf1, as marks of new synthesized histones. This incorporation of new histones is though to occur at the site of the repaired DNA. More distal to the site, repair marks are removed from nucleosomal histones in the chromatin context. Acetyl-marks associated with chromatin “opening” are eliminated by HDACs. ySin3/Rpd3 HDAC associates with Casein Kinase 2 (CK2) that is responsible for subsequent phosphorylation of H4S1, reinforcing nucleosome stability by blocking re-acetylation.
As we described above, the relaxation of chromatin for efficient repair involves HAT activities at the DSB, where they induce an increase of acetylation. Several histone deacetylases (HDAC) have thus been implicated in the DDR, but mainly once repair has been completed (Fig. 3) (89, 90). In opposition to HAT inducing chromatin “opening”, HDAC may have a role in chromatin restoration. In yeast, Sin3/Rpd3, Sir2 and Hst1 HDAC have been shown to facilitate DNA repair (89-91). In addition, mammalian Hdac3 has been linked to DNA damage repair, but its exact role is still unknown (92).
In yeast, Sin3/Rpd3 HDAC complex interacts with the casein kinase CK2 responsible for H4S1 phosphorylation, and this modification has been shown to increase at the break at the end of repair (Fig. 3) (91, 93). Interestingly, the phosphorylation of H4S1 inhibits acetylation of the adjacent lysine residues by NuA4, suggesting that H4S1ph appears at the break after repair completion to prevent new acetylation and to stabilize the nucleosome.
In mammalian cells, histone H2B has also been reported to be phosphorylated on S14 following DNA damage (94). H2BS14ph appears at late time points and accumulates in repair foci in a γ-H2AX-dependant manner. Phosphorylation of H2BS14 by hMst1 kinase has a role in apoptotic-dependant chromatin compaction, so we can propose that increased level of H2BS14ph at the site of repaired DNA may contribute to chromatin stabilization after restoration.
It is logical to predict that histone chaperones such as CAF1 (Chromatin Assembly Factor 1), Asf1 or FACT, are involved and required for chromatin remodeling. Indeed, they are recruited to the site of DNA damage, where they mediate nucleosome disassembly and reassembly (reviewed in (95)). To date, there is no evidence that chaperones play an active role in chromatin disassembly during DNA repair, whereas they are clearly implicated in nucleosome reassembly. In yeast and human, the CAF1 chaperone is recruited to UV-damaged sites and DSB, and with the help of Asf1, deposits H3-H4 onto the DNA (Fig. 3) (96-99). CAF1 is also required for hRing1b-dependant H2AK119 mono-ubiquitination, a mark involved in chromatin restoration after UV-induced damages (100). Asf1 association with yeast Rtt109 and human CBP/p300 or Gcn5 HAT is essential for H3K56 acetylation and, in yeast, acetylation of H3K56 is required for effective DDR (Fig. 3) (101-108). It has been suggested that Asf1 and CAF1 function would be mainly required for checkpoint recovery and chromatin restoration after repair, and that H3K56ac would signal the completion on chromatin reassembly (109, 110), followed by its deacetylation by Hst2/Hst3 sirtuins (111, 112).
In addition to H3K56ac, H3K14/K23 acetylation and acetyltransferase yHat1 are also linked to chromatin restoration. In fact, epistasis analysis has determined that yHat1 influences DSB-repair chromatin reassembly through an interaction with Asf1 but not CAF1 (Fig. 3) (113). Furthermore, Hat1 has been reported to acetylate free H4 on K5 and K12, and H4K5/K12ac would also play a role during DNA damage recovery (114, 115). Finally, while histone H4 ubiquitination has been linked to DDR, it is also known that acetylation of the same residue is important for chromatin assembly after repair (116). Further studies will be required to investigate the exact function of these marks during chromatin reassembly following DSB repair.
5/Conclusion-Future directions
Maintenance of genomic stability in eukaryotic cells required a tight regulation of histone modifications that accompany DDR. Although DNA-damage repair kinetics have been extensively studied, the precise order of histone modifiers, repair factors and remodelers recruitment remains imprecise. It appears more evident that a number of recruited factors regulate each other's accumulation and activation, rendering the study of the specific function of each factor more difficult. Here, we have mentioned the role of histone modifications during DNA DSB repair full process. It is clear that the timing and the cross-talk between histone marks are critical in the process of chromatin dynamic.
Moreover, beside histone marks appearing at the DSB (to signal, recruit repair factors and promote chromatin remodeling), other histone modifications induced by DNA-damage have been identified but not discussed in this review. Recent studies have shown an effect of DNA-damage induced histone modifications on transcription regulation. For example, ATM and histone ubiquitination have been linked to transcription silencing near DSBs (117). In addition, loss of H3T11 phosphorylation by hChk1 has been shown to repress transcription of cyclinB and cdk1 after DNA-damage induction, through loss of Gcn5-dependant promoter acetylation (118). hEco1, which is an acetyltransferase important for sister cohesion during S phase and DDR, has also been shown to repress transcription by interaction with the hLSD1 histone demethylase (119, 120).
It will be interesting then to continue to investigate the cooperation of the factors involved in the repair of DNA DSBs, but also the direct or indirect effect of DNA damage on non-repair processes such as specific transcription regulation.
Acknowledgments
We apologize to our colleagues for work that could not be cited due to space limitation. Work in our laboratories is supported by operating grants from the Canadian Institutes of Health Research (CIHR, MOP-14308/64289) and National Institute of Health (NIH, RO1 GM60443). SJK is a Leukemia and Lymphoma Society Scholar and JC holds a Canada Research Chair.
References
- 1.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–85. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 2.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–16. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Balch C, Montgomery JS, Paik HI, et al. New anti-cancer strategies: epigenetic therapies and biomarkers. Front Biosci. 2005;10:1897–931. doi: 10.2741/1668. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature reviews. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plummer R. Perspective on the pipeline of drugs being developed with modulation of DNA damage as a target. Clin Cancer Res. 2010;16 doi: 10.1158/1078-0432.CCR-10-0984. [DOI] [PubMed] [Google Scholar]
- 8.Redon CE, Nakamura AJ, Zhang Y, et al. Histone γH2AX and Poly(ADP ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res. 2010;16 doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annunziata CM, O'Shaughnessy J. Poly(ADP-Ribose) Polymerase as a novel therapeutic target in cancer. Clin Cancer Res. 2010;16 doi: 10.1158/1078-0432.CCR-10-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan N, Bristow RG. “Contextual” synthetic lethality/loss of heterozygosity: tumor hypoxia and modification of DNA repair. Clin Cancer Res. 2010;16 doi: 10.1158/1078-0432.CCR-10-0527. [DOI] [PubMed] [Google Scholar]
- 11.Thiriet C, Hayes JJ. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol Cell. 2005;18:617–22. doi: 10.1016/j.molcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–4. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 13.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 14.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–62. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 15.Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer research. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 16.An J, Huang YC, Xu QZ, et al. DNA-PKcs plays a dominant role in the regulation of H2AX phosphorylation in response to DNA damage and cell cycle progression. BMC Mol Biol. 2010;11:18. doi: 10.1186/1471-2199-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downs JA, Allard S, Jobin-Robitaille O, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–90. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–16. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unal E, Arbel-Eden A, Sattler U, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 20.Shroff R, Arbel-Eden A, Pilch D, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–11. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–86. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 22.Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–8. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 24.Gupta A, Sharma GG, Young CS, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–7. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma GG, So S, Gupta A, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–95. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–18. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iacovoni JS, Caron P, Lassadi I, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. Embo J. 2010;29:1446–57. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savic V, Yin B, Maas NL, et al. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szilard RK, Jacques PE, Laramee L, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nature structural & molecular biology. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–95. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 33.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature cell biology. 2003;5:675–9. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 34.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends in cell biology. 2009;19:207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Fink M, Imholz D, Thoma F. Contribution of the serine 129 of histone H2A to chromatin structure. Mol Cell Biol. 2007;27:3589–600. doi: 10.1128/MCB.02077-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassing CH, Chua KF, Sekiguchi J, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukas C, Melander F, Stucki M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. Embo J. 2004;23:2674–83. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou Z, Minter-Dykhouse K, Franco S, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 39.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–26. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 40.Mailand N, Bekker-Jensen S, Faustrup H, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 41.Kolas NK, Chapman JR, Nakada S, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–40. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huen MS, Grant R, Manke I, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–14. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–9. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobhian B, Shao G, Lilli DR, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marteijn JA, Bekker-Jensen S, Mailand N, et al. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol. 2009;186:835–47. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104:20759–63. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergink S, Salomons FA, Hoogstraten D, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–52. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–14. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura TM, Moser BA, Du LL, Russell P. Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol Cell Biol. 2005;25:10721–30. doi: 10.1128/MCB.25.24.10721-10730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Botuyan MV, Lee J, Ward IM, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Q, Dutt S, Xu R, et al. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell. 2009;36:110–20. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Javaheri A, Wysocki R, Jobin-Robitaille O, Altaf M, Cote J, Kron SJ. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc Natl Acad Sci U S A. 2006;103:13771–6. doi: 10.1073/pnas.0511192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nnakwe CC, Altaf M, Cote J, Kron SJ. Dissection of Rad9 BRCT domain function in the mitotic checkpoint response to telomere uncapping. DNA repair. 2009;8:1452–61. doi: 10.1016/j.dnarep.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–43. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toh GW, O'Shaughnessy AM, Jimeno S, et al. Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA repair. 2006;5:693–703. doi: 10.1016/j.dnarep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Xiao A, Li H, Shechter D, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–6. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–9. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 59.Morrison AJ, Highland J, Krogan NJ, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–75. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 60.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–25. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–88. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 62.Bird AW, Yu DY, Pray-Grant MG, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–5. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 63.Murr R, Loizou JI, Yang YG, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature cell biology. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 64.Auger A, Galarneau L, Altaf M, et al. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol. 2008;28:2257–70. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kusch T, Florens L, Macdonald WH, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–7. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 66.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ikura T, Tashiro S, Kakino A, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–40. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krogan NJ, Baetz K, Keogh MC, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci U S A. 2004;101:13513–8. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–8. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 71.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–49. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altaf M, Auger A, Monnet-Saksouk J, et al. NuA4-dependent acetylation of nucleosomal histone H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem. 2010;285:15966–77. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–43. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 74.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–83. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–61. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25:3934–44. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC functions as an early double-strand-break sensor in the cell's response to DNA damage. Curr Biol. 2007;17:1432–7. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–13. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kruhlak MJ, Celeste A, Dellaire G, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–34. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park JH, Park EJ, Lee HS, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–97. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–45. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Altaf M, Saksouk N, Cote J. Histone modifications in response to DNA damage. Mutation research. 2007;618:81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 83.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–9. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 84.Keogh MC, Kim JA, Downey M, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 85.Nakada S, Chen GI, Gingras AC, Durocher D. PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 2008;9:1019–26. doi: 10.1038/embor.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chowdhury D, Xu X, Zhong X, et al. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Macurek L, Lindqvist A, Voets O, Kool J, Vos HR, Medema RH. Wip1 phosphatase is associated with chromatin and dephosphorylates gammaH2AX to promote checkpoint inhibition. Oncogene. 2010;29:2281–91. doi: 10.1038/onc.2009.501. [DOI] [PubMed] [Google Scholar]
- 88.Douglas P, Zhong J, Ye R, Moorhead GB, Xu X, Lees-Miller SP. Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol Cell Biol. 2010;30:1368–81. doi: 10.1128/MCB.00741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jazayeri A, McAinsh AD, Jackson SP. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci U S A. 2004;101:1644–9. doi: 10.1073/pnas.0304797101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–13. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol. 2005;25:8179–90. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhaskara S, Chyla BJ, Amann JM, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheung WL, Turner FB, Krishnamoorthy T, et al. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol. 2005;15:656–60. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 94.Fernandez-Capetillo O, Allis CD, Nussenzweig A. Phosphorylation of histone H2B at DNA double-strand breaks. The Journal of experimental medicine. 2004;199:1671–7. doi: 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–95. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–74. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nabatiyan A, Szuts D, Krude T. Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells. Mol Cell Biol. 2006;26:1839–49. doi: 10.1128/MCB.26.5.1839-1849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moggs JG, Grandi P, Quivy JP, et al. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–18. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–93. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 100.Zhu Q, Wani G, Arab HH, El-Mahdy MA, Ray A, Wani AA. Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA repair. 2009;8:262–73. doi: 10.1016/j.dnarep.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hyland EM, Cosgrove MS, Molina H, et al. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–70. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–89. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–7. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–8. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 105.Collins SR, Miller KM, Maas NL, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–10. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 106.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–5. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 107.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–52. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsubota T, Berndsen CE, Erkmann JA, et al. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–12. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc Natl Acad Sci U S A. 2009;106:1151–6. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen CC, Carson JJ, Feser J, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–43. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23:109–19. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 112.Celic I, Masumoto H, Griffith WP, et al. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–9. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 113.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol. 2002;22:8353–65. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barman HK, Takami Y, Ono T, et al. Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem Biophys Res Commun. 2006;345:1547–57. doi: 10.1016/j.bbrc.2006.05.079. [DOI] [PubMed] [Google Scholar]
- 115.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 116.Ye J, Ai X, Eugeni EE, et al. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol Cell. 2005;18:123–30. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 141:970–81. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shimada M, Niida H, Zineldeen DH, et al. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–32. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 119.Choi HK, Kim BJ, Seo JH, Kang JS, Cho H, Kim ST. Cohesion establishment factor, Eco1 represses transcription via association with histone demethylase, LSD1. Biochem Biophys Res Commun. 2010;394:1063–8. doi: 10.1016/j.bbrc.2010.03.125. [DOI] [PubMed] [Google Scholar]
- 120.Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell. 2009;34:311–21. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]