Abstract
Purpose
Tissue biomarker discovery is potentially limited by conventional tumor measurement techniques, which have an uncertain ability to accurately distinguish sensitive and resistant tumors. Semi-automated volumetric measurement of CT imaging has the potential to more accurately capture tumor growth dynamics, allowing for more exact separation of sensitive and resistant tumors and a more accurate comparison of tissue characteristics.
Experimental Design
48 patients with early stage non-small cell lung cancer (NSCLC) and clinical characteristics of sensitivity to gefitinib were studied. High resolution computed tomography was performed at baseline and after 3 weeks of gefitinib. Tumors were then resected and molecularly profiled. Unidimensional and volumetric measurements were performed using a semi-automated algorithm. Measurement changes were evaluated for their ability to differentiate tumors with and without sensitizing mutations.
Results
44% of tumors had EGFR sensitizing mutations. ROC curve analysis demonstrated that volumetric measurement had a higher area-under-the-curve than unidimensional measurement for identifying tumors harboring sensitizing mutations (p = 0.009). Tumor volume decrease of >24.9% was the imaging criteria best able to classify tumors with and without sensitizing mutations (sensitivity 90%, specificity 89%).
Conclusions
Volumetric tumor measurement was better than unidimensional tumor measurement at distinguishing tumors based on presence or absence of a sensitizing mutation. Use of volume-based response assessment for development of tissue biomarkers could reduce contamination between sensitive and resistant tumor populations, improving our ability to identify meaningful predictors of sensitivity.
Introduction
Tumor imaging for response evaluation has a fundamental role in oncology care and drug development. A reduction in tumor size, or “response”, has conventionally suggested a tumor is biologically vulnerable to a treatment; yet it remains unclear what magnitude of size reduction is biologically meaningful. Published criteria for the classification of response are used widely in the reporting of clinical trial results,(1, 2) but studies of individual patient outcomes have failed to identify a clear survival benefit associated with a “partial response”.(3, 4) Importantly, criteria such as RECIST (Response Evaluation Criteria In Solid Tumors (1)) were developed for consistency with historical definitions of response and for minimizing the impact of measurement variability,(5, 6) but were not developed as biologically-based criteria for tumor response
The lack of a biologic basis to conventional response assessment becomes particularly important for tissue biomarker development, especially because the efficacy of newer targeted therapies is often limited to a subset of biologically vulnerable cancers. The development of tissue biomarkers predicting sensitivity to therapy are essential, yet biomarker studies are potentially limited by their use of conventional response criteria, which divide patients into “responders” and “non-responders” based on simple unidimensional tumor measurements that fail to accurately capture tumor growth dynamics. By treating tumors with RECIST-response as “sensitive”, and others as “resistant”, investigators compare the tissue characteristics of two arbitrarily defined subgroups.
The clearest example of the deficiency of RECIST is in the treatment of NSCLC with EGFR tyrosine kinase inhibitors (TKIs) like gefitinib. In advanced NSCLC, EGFR sensitizing mutations in exon 19 and 21 have recently been validated as the strongest biomarker predicting prolonged progression free survival (PFS) on gefitinib:(7) patients harboring a sensitizing mutation have a median PFS of 10 months, compared to 1.5 months for those lacking a sensitizing mutation.(8) However the RECIST response rate for EGFR TKIs in patients with sensitizing mutations is only 70%,(8–10) meaning that nearly one in three patients is classified as a “non-responder” despite gaining important clinical benefit. Published waterfall plots have shown that the vast majority of EGFR mutant cancers develop tumor shrinkage on TKI, though criteria for a “partial response” may not be met.(10, 11) By grouping tumors harboring a sensitizing mutation as part of the “resistant” pool, this arbitrary response threshold could impair our ability to identify meaningful tissue differences, potentially hampering the development of tissue biomarkers for novel therapies.
We hypothesized that more accurate imaging techniques would be better able to distinguish resistant tumors from those harboring sensitizing mutations and could thus aid biomarker development. Toward this goal, we developed a semi-automated algorithm for CT-based tumor volume measurement, which we found to be feasible in lung nodules with low variability.(12, 13) Using gefitinib therapy as a model, and by measuring the entire tumor mass before and after treatment, we proposed to find a biologically-based threshold that could optimally dichotomize lung cancers into sensitive (EGFR mutant) and resistant (EGFR wild-type) groups. Such a finding would support further investigations into the application volumetric response as a tool for biomarker research in therapies where a predictive tissue biomarker has not yet been identified.
Materials and Methods
Imaging and tissue data were obtained prospectively as an exploratory analysis within a phase II trial of neoadjuvant gefitinib in patients with NSCLC,(14) the results of which are being published separately. All patients had stage I or II NSCLC and were deemed both operable and resectable. To enrich the population for EGFR-mutant cancers, patients were only included if (1) they had a smoking history of less than 15 pack years, or (2) their tumors had histologic features of bronchioloalveolar cancer, characteristics associated with EGFR TKI sensitivity.(15) Between July 2004 and March 2008, 50 patients were treated. Nine patients were eligible based on histology and 41 based on smoking history. Patients received gefitinib daily for 3 weeks before surgery, and discontinued it two days before their operation. CT imaging was performed before starting gefitinib and before surgery. This study was approved by our institutional review board.
Tumor genomic analysis
At time of resection, tumor tissue was snap-frozen in liquid nitrogen and stored in a −80 degree freezer. Representative areas of these specimens were pathologically reviewed to confirm the diagnosis and presence of tumor. Genomic DNA was analyzed for the most common EGFR sensitizing mutations (exons 19 and 21) using previously described polymerase chain reaction (PCR) based methods.(16–18) EGFR wild-type (wt) tumors were also tested for KRAS mutations, which are found in a non-overlapping subset of lung adenocarcinomas that have been found to be resistant to EGFR TKI therapy.(18–20) If no EGFR or KRAS mutations were found, then the remaining EGFR exons (18 through 24) were assessed by standard dideoxynucleotide sequencing. Pre-treatment tissue was analyzed by the above method for the first 18 patients on the study, but this requirement was removed after we observed 100% concordance between the pretreatment and resection results.(21) Selected specimens that were found to be EGFR/KRAS wt were submitted for more detailed mutational testing performed by mass spectrometry.
Tumor imaging and measurement
Baseline CT of each patient was performed within two weeks prior to gefitinib initiation. A follow-up CT scan was done using the same imaging acquisition technique 3 weeks later, before surgery. Non-contrast enhanced diagnostic chest CTs were performed with a LightSpeed 16 scanner (GE Medical Systems, Milwaukee, WI) during a breath-hold. High-resolution images with 1.25mm slice thickness were reconstructed per our volumetric imaging protocol.(13) Two patients were excluded because 1.25mm slice thickness reconstructions were not performed as required by protocol, leaving 48 of 50 patients qualified for our analysis.
Tumor contours were semi-automatically delineated using a three-dimensional segmentation algorithm.(12, 13) To ensure appropriateness of the segmentation results, the computer-generated tumor contours were inspected by two radiologists blinded to mutation status and scan dates (PG, LHS), and suboptimal computer segmentation results were corrected by one of the radiologists. The areas defined by these final tumor contours were then summed across all CT slices to calculate a total tumor volume measurement. Tumor greatest diameter was measured from a single transverse image plane as described by the RECIST guidelines.(1) Changes in tumor measurements were calculated by subtracting the baseline from the follow-up measurement, divided by the baseline measurement.
Statistical analysis
Tumors were divided into molecular subgroups defined by presence or absence of a sensitizing mutation. The distribution of tumor measurement changes was compared among subgroups using a Wilcoxon rank-sum test. Using estimated 95% limits of agreement for unidimensional (−7.3%, +6.2%) and volumetric (−12.1%, +13.4%) measurement from a prior study of measurement reproducibility,(13) an exact McNemar's test was used to compare the proportion of measurement changes that fell outside these ranges of variability.
Measurement methods were then evaluated to explore how well they could differentiate between tumors with and without a sensitizing mutation. Nonparametric estimates of receiver operating characteristic (ROC) curves and the area under these curves (AUC) were calculated for both measurement methods. For the ROC calculations, the relative percent change of the unidimensional and volumetric measurements were multiplied by −1, thus using the percent decrease in measurement as a positive value to estimate the ROC curves and AUCs. To test whether the AUCs were equal, methods suggested by DeLong et al. were used.(22)
Youdens' index (YI) was calculated to determine what threshold for volumetric and unidimensional measurement change optimally dichotomized tumors with and without sensitizing mutations.(23) YI is equal to: sensitivity + specificity − 1. By choosing its maximum value as a diagnostic threshold, we place equal importance on sensitivity and specificity. For context, we also calculated the sensitivity and specificity of a RECIST-based threshold; for unidimensional measurement, we used a 30% decrease in diameter,(1) while for volumetric measurement we used the mathematically equivalent volumetric decrease (65%) from which RECIST was developed.(1, 5)
All tests were considered significant at the p ≤ 0.05 level. Analyses were done in Stata 10.0 for Windows (copyright 2007, StataCorp LP, College Station TX).
Results
Tumor measurements grouped by mutation status
21 of 48 tumors (44%) harbored an EGFR mutation. 27 (56%) were EGFR wt with 5 of these harboring a KRAS mutation. A median of 24 days passed between baseline and follow-up scans (range 20–41 days), with a median of 22 days on gefitinib (range 20–28 days). Unidimensional and volumetric tumor measurements are summarized in Table 1 for all 48 tumors and grouped by presence or absence of sensitizing mutation. As predicted biologically, there was a significant difference between the mean measurement changes of these subgroups of tumors, demonstrated both with unidimensional measurement (−11.1% vs. −2.6%, p=0.002) and volumetric measurement (−45.9% vs. −4.4%, p<0.001). Across all tumors, the overall magnitude of change is greater for volumetric measurement, consistent with the mathematical relationship between change in diameter and change in volume.(5)
Table 1.
Change in tumor measurements, grouped by mutation status, for each measurement method.
| All tumors | EGFR mutant | EGFR wild-type* | P | |
|---|---|---|---|---|
| N | 48 | 21 | 27 | |
| Unidimensional percent change (%) | ||||
| Median | −5.3 | −11.1 | −1.8 | 0.002 |
| IQR | [−13.1, 0.7] | [−26.9, −4.4] | [−7.0, 1.3] | |
| Volumetric percent change (%) | ||||
| Median | −23.9 | −45.9 | −4.4 | < 0.001 |
| IQR | [−44.8, 0.0] | [−54.1, −29.1] | [−16.5, 7.0] | |
Includes all KRAS mutant tumors
IQR: interquartile range
p: p-value from a Wilcoxon rank-sum test comparing EGFR mutant tumors versus EGFR wild-type tumors
Individual tumor measurement changes were compared to the aforementioned 95% limits of agreement, determined from a prior study.(13) 75% of volumetric measurement changes surpassed the expected range of variability, while only 46% of unidimensional changes surpassed the expected range of variability (p = 0.003). This indicates that the majority of unidimensional measurement changes after 3 weeks of treatment were indistinguishable from the expected changes due to variability in CT acquisition and measurement.
Using tumor measurements to dichotomize tumors with and without sensitizing mutations
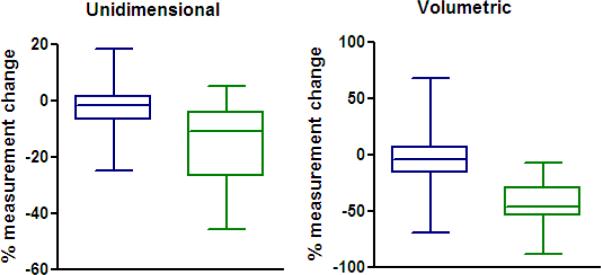
Figure 1 shows the distribution of tumor measurement changes for both measurement methods, grouping tumors by mutation status. With unidimesional measurement there is overlap between the interquartile ranges of the measurement changes from the two tumor subgroups, while there is better separation of these subgroups using volumetric measurement.
Figure 1.
Plots of unidimensional and volumetric measurement changes, grouped by absence (blue) or presence (green) of EGFR sensitizing mutation. There is a statistically significant difference between the molecular subtypes using both unidimensional (p=0.002) and volumetric (p<0.001) measurement, as is expected biologically. Note there is overlap between the interquartile ranges using unidimensional measurement, while there is better separation of groups using the volumetric measurement.
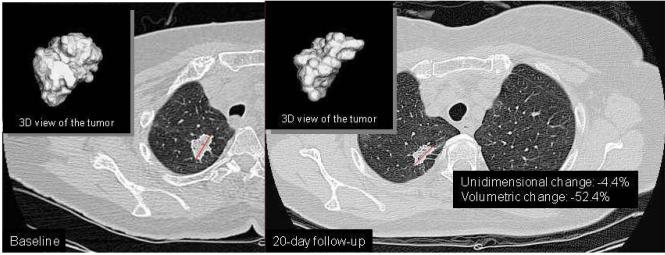
To determine which imaging method is a better diagnostic test for identifying tumors with sensitizing mutations, we calculated ROC curves (Figure 2). The ROC analysis evaluates each tumor measurement change as a potential threshold for a binary diagnostic test predicting EGFR mutation status. The ROC curve plots the sensitivity by 1-specificity for all such potential thresholds, and the AUC represents the strength of the test across multiple possible thresholds. We found that volumetric measurement has a significantly higher AUC than unidimensional measurement (p = 0.009). Figure 3 demonstrates this observation in one tumor harboring a sensitizing mutation, where volume measurement detected a change that was not detected by conventional unidimensional measurement.
Figure 2.
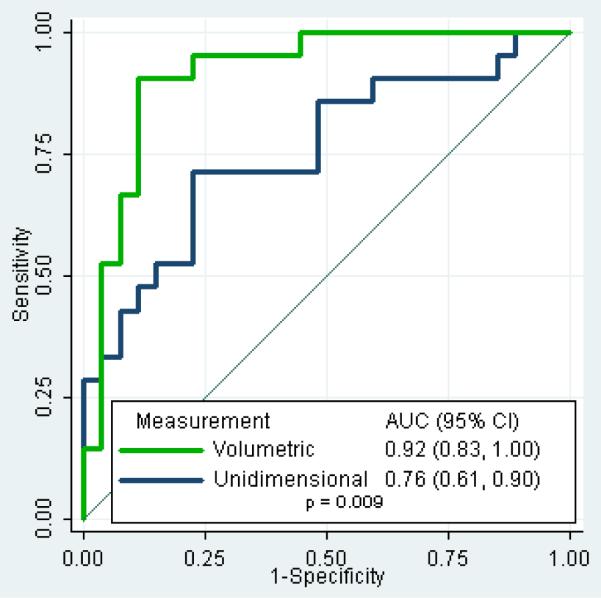
ROC curves for the ability of each measurement method to distinguish tumors based on presence or absence of a sensitizing mutation. For each possible percent measurement change, a specificity and sensitivity for predicting presence of a sensitizing mutation is calculated, and these are plotted on the X and Y axes. Volumetric measurement change is shown to have better diagnostic accuracy when the area under the curve (AUC) is compared (p=0.009).
Figure 3.
Detection of tumor change on CT, comparing volumetric and unidimensional measurement, for an EGFR mutant tumor at baseline (A) and at 20-day follow-up (B). Computer delineated tumor contours and diameter lines are superimposed on one image from each study date. Three-dimensional (3D) view of each segmented tumor is displayed at upper-left corner of the each panel. For this case, a significant change is detected using volume measurement (−52.4%), but not using unidimensional measurement (−4.4%).
Optimal thresholds for biomarker development
For each measurement method, we calculated which threshold had the highest combined sensitivity and specificity for dichotomizing tumors based on presence or absence of a sensitizing mutation (Table 2). For volumetric measurement, a threshold of 24.9% decrease (YI = 0.79) classified 90% of tumors with a mutation as “responders” and 89% of tumors without a mutation as “non-responders” (positive predictive value (PPV) = 86%, negative predictive value (NPV) = 92%). For unidimensional measurement, the threshold with the highest sensitivity and specificity was a 7.0% decrease (YI = 0.49), which classified 71% of tumors with a mutation as responders and 78% of tumors without a mutation as non-responders (PPV = 71%, NPV = 78%). Because a 7.0% decrease would be within the expected measurement variability of unidimensional measurement (−7.3%, +6.2%), we performed an ad hoc calculation to determine what threshold outside of that range had the highest YI. We determined that a 9.0% unidimensional decrease (YI = 0.38) would have a sensitivity of 52% and a specificity of 85% (PPV = 73%, NPV = 70%), and would be less influenced by measurement variability than a threshold of 7.0% decrease. These thresholds will require validation in an independent data set as a confirmation of their accuracy.
Table 2.
Sensitivity and specificity of different possible thresholds for dichotomizing tumors into EGFR mutant and EGFR wild-type groups.
| Threshold | Sens | Spec | |
|---|---|---|---|
| Unidimensional | |||
| Optimal* | 7.0% decrease | 71% | 78% |
| Alternate** | 9.0% decrease | 52% | 85% |
| RECIST | 30% decrease | 14% | 100% |
|
| |||
| Volumetric | |||
| Optimal* | 24.9% decrease | 90% | 89% |
| RECIST | 65% decrease | 14% | 96% |
Sens: Sensitivity = % of EGFR mutant tumors classified as responders
Spec: Specificity = % of EGFR wt tumors classified as non-responders
Threshold calculated to have highest summed sensitivity and specificity
Threshold outside of the 95% limits of agreement with the highest summed sensitivity and specificity
We then evaluated the effectiveness of RECIST-based thresholds at separating tumors based on presence or absence of a sensitizing mutation, using the conventional 30% decrease for unidimensional measurement and a mathematically equivalent 65% decrease for volumetric measurement.(5) Both thresholds had a high specificity (Table 2), but classified only 3 of 21 EGFR mutant tumors as responders (sensitivity of 14%). While these thresholds accurately classified EGFR wt tumors as non-responders, the high false-negative rate meant that only 59–60% of the non-responding tumors were actually EGFR wt (NPV of approximately 60%). Importantly, these results were calculated from applying RECIST-based thresholds after only 3 weeks of treatment, and therefore do not necessarily represent the accuracy of RECIST in biomarker analyses of full phase II trial results.
Discussion
The chance of a novel therapy achieving success in the clinical setting is vastly improved by the identification of a tissue biomarker predicting increased tumor sensitivity. For example, through the identification of EGFR sensitizing mutations in NSCLC, erlotinib and gefitinib have been transformed from one of many second-line therapies to the primary therapy for a significant subset of cancers. Alternatively, cetuximab is an EGFR targeted therapy that, despite positive phase III trial results,(24) has not received FDA approval for NSCLC and has garnered limited clinical enthusiasm in lung cancer therapy, in large part because candidate biomarkers (KRAS mutation, EGFR expression) have failed to show predictive ability.(24–27) While a recently published study in colorectal cancer has explored newer biomarkers of cetuximab response (amphiregulin, epiregulin),(28) the results were notable for marginal statistical significance. We hypothesize that such biomarker analyses could be strengthened by better use of imaging and biologically-based definitions of tumor response.
Our data indicate that biomarker studies like the one described above are limited by the modest ability of unidimensional measurement to distinguish tumors with and without sensitizing mutations. In our study, there was significant overlap between the unidimensional measurement changes of EGFR mutant and EGFR wt tumors; the optimal threshold for diameter measurement, a 7% decrease, had a disappointing sensitivity and specificity (71%, 78%). Volumetric measurement, however, was better able to dichotomize tumors with and without sensitizing mutations across multiple possible response thresholds, with a significantly better AUC by ROC curve analysis (p=0.009). Our data indicate that by using a 24.9% volume decrease to dichotomize tumors for biomarker analysis, sensitivity and specificity would be higher (90%, 89%), potentially improving the ability to identify meaningful tissue differences between the tumors being studied. These results will need to be validated in a larger, more generalizable patient population.
One possible value of volume measurement, aside from its ability to capture changes to the whole tumor mass, is that measurement variability appears to have a relatively limited impact on the measurement changes that are found. In a recently completed study, we demonstrated that for patients with NSCLC undergoing same-day repeat CT scans, the 95% limits of agreement for volumetric measurement were (−12.1% to +13.4%), while for unidimensional measurement they were (−7.3% to +6.2%).(13) In the present study, the tumor measurements were obtained using the same scanning technique, segmentation software, and measurement procedures as in this “rescan” study. Using unidimensional measurement, we found that the majority of measurement changes (54%) fell within the 95% limits of agreement, while with volumetric measurement a much smaller portion of measurement changes (25%) were within the 95% limits of agreement. Therefore measurement variability may have a greater impact on unidimensional measurement, potentially limiting its effectiveness in biomarker research.
Our data suggest that the use of RECIST-baseline thresholds for dichotomizing tumors according to presence of a sensitizing mutation is inaccurate in this setting. After 3 weeks of therapy, RECIST-based thresholds had a sensitivity of only 14%, though this is likely lower than would be found when applying these thresholds after a conventional 6 or 8 week treatment period. A more accurate approximation of the sensitivity of RECIST response in clinical trials would be derived from the 70% RECIST response rate for EGFR-mutant lung cancers treated with TKI. This is approximately equivalent to a “sensitivity” of 70% – while the remaining EGFR-mutant tumors are designated as non-responders, we believe the majority of these still gain benefit from this targeted therapy despite not meeting criteria for a partial response. Just as we found that a response threshold of less than a 30% decrease optimally dichotomizes tumors after 3 weeks of therapy, we suspect that a “minor response” would be better than a full RECIST response at dichotomizing tumors with and without sensitizing mutations. Others have found that minor response is predictive of better outcome to targeted therapy in metastatic colon cancer and in gastrointestinal stromal tumor;(29, 30) whether this is the case for TKI therapy in NSCLC is unclear, and requires further study.
As EGFR mutation status was our tissue biomarker of interest in this study, we used several precautions to ensure minimal error in molecular diagnostic testing. The use of surgical specimens ensured adequate tissue for analysis, and all specimens were reviewed pathologically to ensure an adequate population of cancer cells for tumor DNA extraction. Our PCR analysis, which is specific for exon 19 deletions/insertions and L858R mutations (making up >90% of all EGFR sensitizing mutations) is extremely sensitive and able to detect a mutation when present in as little as 5% of DNA.(16, 31) To identify rare mutations (<10% of total), direct DNA sequencing was performed, which is less sensitive but still identifies 80–90% of mutations. Together, these steps should therefore identify >98% of sensitizing mutations. Further mass spectrometry analysis was performed on four cases with significant tumor regression, identifying one additional rare EGFR sensitizing mutation. For the remaining 20 patients with no mutation identified, we believe the likelihood of there being an additional undetected EGFR mutation is less than 1–2%.
Conclusions
This analysis of tumor imaging changes after gefitinib therapy found that volumetric measurement can more accurately distinguish between tumors with and without sensitizing mutations than conventional unidimensional measurement. We recommend further study of tumor volume measurement as a tool for dichotomizing sensitive and resistant tumors in order to improve tissue biomarker development.
Statement of Translational Relevance.
The development of a tissue biomarker predicting sensitivity to a targeted therapy has become an essential step for the clinical success of a novel agent. However identification of predictive tissue biomarkers may be limited by the use of unidimensional tumor response assessment to subdivide tumors into sensitive and resistant populations because this measurement method does not fully characterize tumor growth dynamics. In this analysis we evaluate whether volumetric tumor measurement is better than unidimensional tumor measurement at distinguishing tumors with and without sensitizing mutations following treatment with a targeted therapy. As a model, we study the differential response of EGFR mutant and wild-type tumors to gefitinib therapy, because EGFR sensitizing mutations have recently been validated as highly predictive of improved progression free survival on gefitinib. Demonstrating that volumetric measurement allows better dichotomization of these molecular subtypes would suggest that this technology could have a role in improving tissue biomarker discovery for novel therapies.
Acknowledgements
We thank Carolina Montalvo, study coordinator, for her excellent effort in the data collection for this study over the past 5 years.
Financial Support - This work was supported in part by the grants R01CA05826, R21CA113653, and P01CA125143 from the National Cancer Institute, by the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center, and by a grant from the Carmel Hill Fund.
Footnotes
Role of the Sponsors – The funding organizations of this study had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Financial Disclosures – Dr. Pao reports serving as a consultant to AstraZeneca and MolecularMD. Dr. Kris reports serving as a consultant to AstraZeneca, Pfizer, and Boehringer Ingelheim.
Prior presentation – This material was presented in part as an oral presentation at the World Conference on Lung Cancer (IASLC) in San Francisco in August 2009.
Trial registration – Clinicaltrials.gov identifier: NCT00588445.
References
- 1.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. Journal of National Cancer Institute. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 2.Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47:207–14. doi: 10.1002/1097-0142(19810101)47:1<207::aid-cncr2820470134>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 3.Lara PN, Jr., Redman MW, Kelly K, et al. Disease control rate at 8 weeks predicts clinical benefit in advanced non-small-cell lung cancer: results from Southwest Oncology Group randomized trials. J Clin Oncol. 2008;26:463–7. doi: 10.1200/JCO.2007.13.0344. [DOI] [PubMed] [Google Scholar]
- 4.Birchard KR, Hoang JK, Herndon JE, Jr., Patz EF., Jr. Early changes in tumor size in patients treated for advanced stage nonsmall cell lung cancer do not correlate with survival. Cancer. 2009;115:581–6. doi: 10.1002/cncr.24060. [DOI] [PubMed] [Google Scholar]
- 5.James K, Eisenhauer E, Christian M, et al. Measuring response in solid tumors: unidimensional versus bidimensional measurement. J Natl Cancer Inst. 1999;91:523–8. doi: 10.1093/jnci/91.6.523. [DOI] [PubMed] [Google Scholar]
- 6.Moertel CG, Hanley JA. The effect of measuring error on the results of therapeutic trials in advanced cancer. Cancer. 1976;38:388–94. doi: 10.1002/1097-0142(197607)38:1<388::aid-cncr2820380156>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 7.Fukuoka M, Wu Y, Thongprasert S, et al. Biomarker analyses from a phase III, randomized, open-label, first-line study of gefitinib (G) versus carboplatin/paclitaxel (C/P) in clinically selected patients (pts) with advanced non-small cell lung cancer (NSCLC) in Asia (IPASS) J Clin Oncol. 2009;27 doi: 10.1200/JCO.2010.33.4235. Abstr 8006. [DOI] [PubMed] [Google Scholar]
- 8.Mok TS, Wu Y-L, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–67. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 10.Jackman DM, Miller VA, Cioffredi LA, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15:5267–73. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 12.Zhao B, Schwartz LH, Moskowitz CS, Ginsberg MS, Rizvi NA, Kris MG. Lung cancer: computerized quantification of tumor response--initial results. Radiology. 2006;241:892–8. doi: 10.1148/radiol.2413051887. [DOI] [PubMed] [Google Scholar]
- 13.Zhao B, James LP, Moskowitz CS, et al. Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology. 2009;252:263–72. doi: 10.1148/radiol.2522081593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kris MG, Pao W, Zakowski M, et al. Prospective trial with preoperative gefitinib to correlate lung cancer response with EGFR exon 19 and 21 mutations and to select patients for adjuvant therapy. Proc Am Soc Clin Onco. 2006;24:369s. [Google Scholar]
- 15.Miller VA, Kris MG, Shah N, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–9. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 16.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pao W, Wang TY, Riely GJ, et al. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the Epidermal Growth Factor Receptor and in KRAS Are Predictive and Prognostic Indicators in Patients With Non-Small-Cell Lung Cancer Treated With Chemotherapy Alone and in Combination With Erlotinib. J Clin Oncol. 2005 doi: 10.1200/JCO.2005.02.857. JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 20.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–6. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 21.Solomon SB, Zakowski MF, Pao W, et al. Core needle lung biopsy specimens: adequacy for EGFR and KRAS mutational analysis. AJR Am J Roentgenol. 2010;194:266–9. doi: 10.2214/AJR.09.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 23.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Pirker R, Pereira JR, Szczesna A, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009;373:1525–31. doi: 10.1016/S0140-6736(09)60569-9. [DOI] [PubMed] [Google Scholar]
- 25.Lynch TJ, Patel T, Dreisbach L, et al. Cetuximab and first-line taxane/carboplatin chemotherapy in advanced non-small-cell lung cancer: results of the randomized multicenter phase III trial BMS099. J Clin Oncol. 2010;28:911–7. doi: 10.1200/JCO.2009.21.9618. [DOI] [PubMed] [Google Scholar]
- 26.Khambata-Ford S, Harbison CT, Hart LL, et al. Analysis of potential predictive markers of cetuximab benefit in BMS099, a phase III study of cetuximab and first-line taxane/carboplatin in advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:918–27. doi: 10.1200/JCO.2009.25.2890. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch FR, Herbst RS, Olsen C, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol. 2008;26:3351–7. doi: 10.1200/JCO.2007.14.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabernero J, Cervantes A, Rivera F, et al. Pharmacogenomic and pharmacoproteomic studies of cetuximab in metastatic colorectal cancer: biomarker analysis of a phase I dose-escalation study. J Clin Oncol. 2010;28:1181–9. doi: 10.1200/JCO.2009.22.6043. [DOI] [PubMed] [Google Scholar]
- 29.Piessevaux H, Buyse M, De Roock W, et al. Radiological tumor size decrease at week 6 is a potent predictor of outcome in chemorefractory metastatic colorectal cancer treated with cetuximab (BOND trial) Ann Oncol. 2009;20:1375–82. doi: 10.1093/annonc/mdp011. [DOI] [PubMed] [Google Scholar]
- 30.Choi H, Charnsangavej C, Faria SC, et al. Correlation of computed tomography and positron emission tomography in patients with metastatic gastrointestinal stromal tumor treated at a single institution with imatinib mesylate: proposal of new computed tomography response criteria. J Clin Oncol. 2007;25:1753–9. doi: 10.1200/JCO.2006.07.3049. [DOI] [PubMed] [Google Scholar]
- 31.Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007;13:4954–5. doi: 10.1158/1078-0432.CCR-07-1387. [DOI] [PubMed] [Google Scholar]