Abstract
Chemokines are important regulators of directional cell migration and tumor metastasis. A genome-wide transcriptome array designed to uncover novel genes silenced by methylation in lung cancer identified the CXC-subfamily of chemokines. Expression of eleven of the sixteen known human CXC-chemokines was increased in lung adenocarcinoma cell lines after treatment with 5-aza-2deoxycytidine (DAC). Tumor-specific methylation leading to silencing of CXCL5, 12 and 14 was found in over 75% of primary lung adenocarcinomas and DAC treatment restored expression of each silenced gene. Forced expression of CXCL14 in H23 cells where this gene is silenced by methylation increased cell death in vitro and dramatically reduced in vivo growth of lung tumor xenografts through necrosis of up to 90% of the tumor mass. CXCL14 re-expression had a profound effect on the genome altering the transcription of over 1,000 genes, including increased expression of 30 cell cycle inhibitor and pro-apoptosis genes. In addition, CXCL14 methylation in sputum from asymptomatic early stage lung cancer cases was associated with a 2.9-fold elevated risk for this disease compared to controls, substantiating its potential as a biomarker for early detection of lung cancer. Together these findings identify CXCL14 as an important tumor suppressor gene epigenetically silenced during lung carcinogenesis.
Keywords: CXCL14, Chemokines, lung cancer, DNA methylation, CXCL5, CXCL12
Introduction
Aberrant DNA methylation has been established as one of the major mechanisms by which tumor suppressor genes are silenced in cancer (Baylin and Ohm, 2006). Several genome-wide methylation assays have identified a large number of abnormal gene methylation in various malignancies (Bennett et al., 2008; Jacinto et al., 2007; Kim et al., 2006; Meissner and Jaenisch, 2006; Shames et al., 2006). A genome-wide transcriptome based approach that can identify novel genes silenced by methylation in cancer has been developed (Schuebel et al., 2007). This approach relies on the differential response of densely methylated promoters to the demethylating agent DAC as compared to the histone deacetylase inhibitor trichostatin A (TSA). Using this approach we interrogated six lung tumor-derived cell lines and identified the CXC subfamily of chemokines as potential candidates for epigenetic silencing.
Chemokines are a superfamily of small chemotactic cytokines that direct the migration of leukocytes (Moser and Loetscher, 2001). In addition, they regulate cellular processes such as proliferation, migration, angiogenesis, and tumor related immunity (Muller et al., 2001; Shellenberger et al., 2004; Shurin et al., 2005; Strieter et al., 2004). Chemokines are classified into four subfamilies: C, CC, CXC, and CX3C chemokines based on the location of conserved cysteine residues (Strieter et al., 2004). The CXC subfamily in humans consists of sixteen members (CXCL1-14, 16, and 17) that are important regulators of tumor angiogenesis, immunity, and tissue-specific cancer metastasis (Darash-Yahana et al., 2009; Hromas et al., 1999; Mu et al., 2009; Strieter et al., 2004; Yuvaraj et al., 2009). CXC chemokines share four cysteine residues in a highly conserved location that determine the 3-dimensional structure of these heparin-binding proteins. A glutamate-leucine-arginine (ELR) motif near the NH2 terminus of the molecule determines the property of a specific chemokine. The ELR-positive chemokines, including CXCL1-3, 5-8, are proangiogenic, whereas members lacking the ELR motif such as CXCL4, 9-11 are interferon-inducible and are potential inhibitors of angiogenesis (Strieter et al., 2004).
CXCL14, also known as BRAK, is an ELR-negative chemokine abundantly expressed in most normal tissue including lung (Kurth et al., 2001; Ozawa et al., 2006; Parsanejad et al., 2008; Schwarze et al., 2005; Shurin et al., 2005). In contrast, the majority of established epithelial cancer cell lines and many primary carcinomas do not express CXCL14 suggesting a tumor suppressor function (McKinnon et al., 2008; Ozawa et al., 2006; Shellenberger et al., 2004; Shurin et al., 2005). Interestingly, CXCL14 expression was suppressed by epidermal growth factor (EGF) and restored by a EGF receptor tyrosine kinase inhibitor (gefitinib) in head and neck squamous cell carcinoma (HNSCC) cells (Ozawa et al., 2009). In addition, gefitinib-mediated re-expression of CXCL14 is strongly associated with the anti-tumor efficacy of this drug in HNSCC cell xenografts (Ozawa et al., 2009).
The purpose of this study was to evaluate primary lung adenocarcinomas from current, former, and never smokers for promoter hypermethylation of the CXC subfamily of chemokines identified by a genome-wide transcriptome array. Cancer specificity was determined by comparing methylation in lung cancer cell lines to normal human bronchial epithelial cells (NHBEC) from bronchoscopy of cancer free smokers and normal peripheral blood mononuclear cells (PBMC) from healthy donors. The potential use of aberrant methylation of these genes as biomarkers for early lung cancer detection was assessed using sputum samples from early stage lung cancer cases and controls. Finally, in vitro and in vivo studies were also conducted to evaluate the functional consequences associated with silencing of CXCL14.
Results
DNA methylation regulates transcription of CXC-chemokines in lung cancer
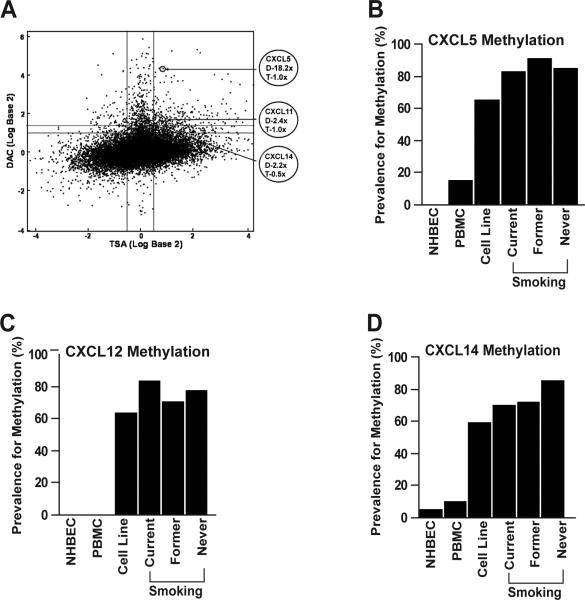
A genome-wide transcriptome array designed to discover novel aberrantly methylated genes identified 11 of the 16 known CXC genes (CXCL2-6, 8, 10-12, 14 and 16) in human lung cancer cell lines (Figure 1a and Table 1). Nine genes (CXCL1-6, 12, 14, and 16), including seven identified by the array, have promoter CpG islands (Table 1) (Gardiner-Garden and Frommer, 1987). The specificity of promoter methylation to tumor cells was validated by combined bisulfite restriction analysis (COBRA) using lung cancer cell lines, PBMC from healthy donors, and NHBEC obtained from bronchoscopy of cancer free smokers. Tumor-specific methylation (not detected in NHBEC) of CXCL5, CXCL6, CXCL12, and CXCL14 was found in 65, 65, 63 and 59% of lung cancer cell lines, respectively. In contrast, CXCL4 was methylated in 82% of the lung cancer cell lines, 90% of NHBEC and 100% of PBMC, and CXCL6 was methylated in 55% of PBMC. Although, CXCL4 and CXCL6 were methylated in lung cancer cell lines, the frequent methylation of these genes in PBMC negated their evaluation in primary adenocarcinomas. The promoter CpG islands of the remaining four genes (CXCL1, 2, 3, and 16) were unmethylated in all lung cancer cell lines (Table 1).
Figure 1.
Transcription of the CXC-chemokines in lung cancer is regulated by methylation. a) Genes that are most likely regulated by promoter hypermethylation are expected in the top tier group (2-fold or higher change with DAC treatment (Y-axis > 1.4) and little or no change with TSA treatment (X-axis = -0.5 to 0.5). Treatment of H23 cells with DAC led to an 18.2, 2.4, and 2.2-fold increase in expression of CXCL5, CXCL11, and CXCL14 respectively on Agilent 44K expression array. In contrast, H23 cells treated with trichostatin A (TSA) showed little or no change in the expression of these genes suggesting DNA methylation is the primary regulator of the transcription of these genes in H23. Primary lung adenocarcinomas from current, former and never smokers showed tumor specific and highly prevalent methylation in the promoter CpG island of CXCL5 (b), CXCL12 (c), and CXCL14 (d).
Table 1.
Promoter methylation of CXCL family genes in lung cancer
| No. | Gene | CpG island property |
Methylated (%) |
Array** positive (#) | |||||
|---|---|---|---|---|---|---|---|---|---|
| CpG* (#) | Size (bp) | GC (%) | Obs/exp | Cell lines | NHBEC | PBMC | |||
| 1 | CXCL1 | 54 | 626 | 69.3 | 0.74 | 0 | - | - | 0 |
| 2 | CXCL2 | 62 | 623 | 69.2 | 0.84 | 0 | - | - | 1 |
| 3 | CXCL3 | 62 | 636 | 69.0 | 0.84 | 0 | - | - | 1 |
| 4 | CXCL4 (PF4) | 22 | 302 | 65.6 | 0.68 | 82 | 90 | 100 | 2 |
| 5 | CXCL5 | 26 | 216 | 70.8 | 0.69 | 65 | 0 | 15 | 3 |
| 6 | CXCL6 | 23 | 207 | 73.4 | 0.85 | 65 | 0 | 55 | 1 |
| 7 | CXCL7 (PPBP) | 0 | - | - | - | - | - | - | 0 |
| 8 | CXCL8 (IL-8) | 0 | - | - | - | - | - | - | 1 |
| 9 | CXCL9 | 0 | - | - | - | - | - | - | 0 |
| 10 | CXCL10 | 0 | - | - | - | - | - | - | 1 |
| 11 | CXCL11 | 0 | - | - | - | - | - | - | 3 |
| 12 | CXCL12 | 223 | 2677 | 60.9 | 0.90 | 63 | 0 | 0 | 1 |
| 13 | CXCL13 | 0 | - | - | - | - | - | - | 0 |
| 14 | CXCL14 | 82 | 803 | 68.0 | 0.90 | 59 | 5 | 10 | 2 |
| 15 | CXCL16 | 134 | 1372 | 69.2 | 0.82 | 0 | - | - | 1 |
| 16 | CXCL17 | 0 | - | - | - | - | - | - | 0 |
When a gene has no CpG island the CpG number was shown as 0.
Array positive indicates the number of lung adenocarcinoma cell lines (out of 6 cell lines) that after DAC treatment showed a 2-fold or higher increase in expression.
Methylation of CXCL5, 12, and 14 is common in primary lung adenocarcinomas
The methylation status of CXCL5, 12, and 14 promoters was evaluated in primary lung adenocarcinomas using methylation-specific PCR (MSP) and methylation was found in 80, 75, and 78% of the tumors, respectively (Figures 1b-d). All three genes were methylated in 107 (61%) of the primary tumors whereas only 7 (4%) showed unmethylated promoter in all three genes. Although the prevalence for methylation of CXCL14 in never smokers is slightly higher than current and former smokers, the difference was not statistically significant (Figure 1d). Similarly, the prevalence for CXCL5 and CXCL12 methylation among the different smoking groups was similar (Figures 1b and 1c). There was no difference in prevalence for gene methylation by tumor stage and methylation of these chemokines alone or in combination was not prognostic for survival (not shown).
Methylation of CXCL5, 12, and 14 silences gene expression
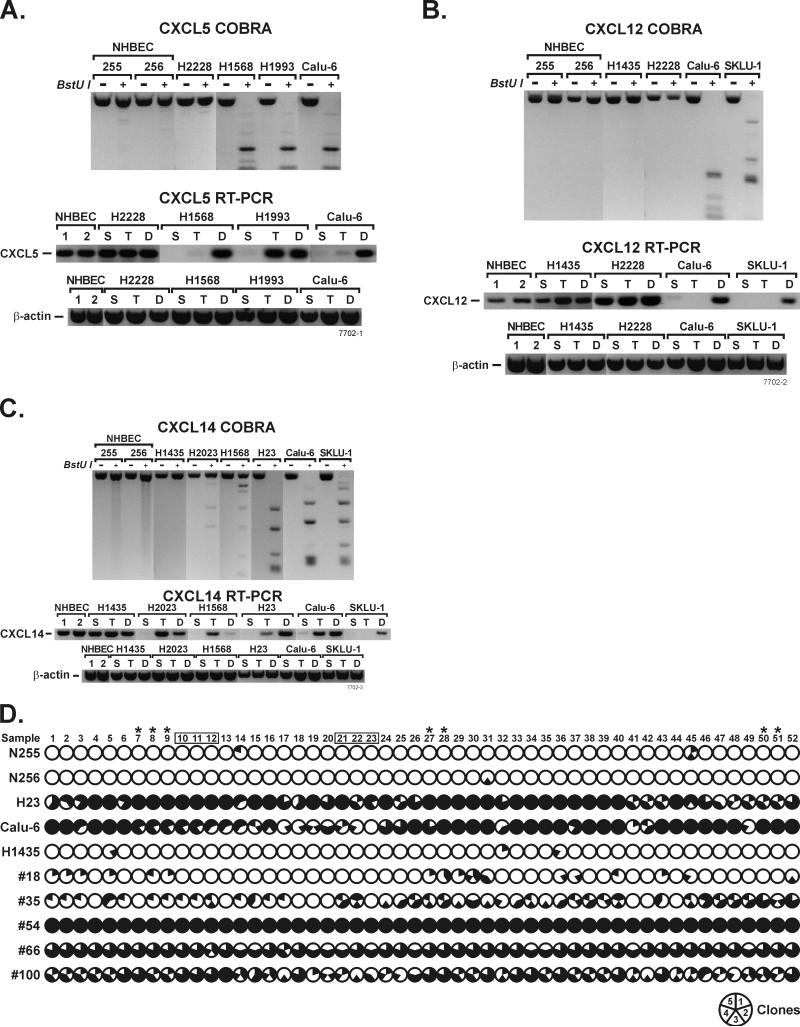
The effect of methylation on gene expression was compared between samples with and without methylation of CXCL5, 12 and 14 promoters using RT-PCR. Complete methylation of these genes (determined by a complete digestion of the multiple CGCG sites by the BstUI restriction enzyme) strongly correlated with loss of gene expression. In lung cancer cell lines with completely methylated CXCL5 (H1568, H1993, and Calu-6), CXCL12 (Calu-6 and SKLU-1) and CXCL14 (H23, Calu-6, and SKLU-1), transcription of these genes was absent (Figure 2a-c). In contrast, all three genes were readily transcribed in samples with unmethylated promoters such as NHBEC and H2228.
Figure 2.
DAC treatment restores expression of genes silenced by methylation. Expression of CXCL5 (a), CXCL12 (b), and CXCL14 (c) was silenced in untreated control (S) lung cancer cell lines with methylated promoter CpG islands and could be restored primarily with DAC (D) treatment. In some cell lines (CXCL5 in H1993 and CXCL14 in Calu-6) gene expression could also be restored by TSA (T) treatment. NHBEC and lung cancer cell lines with unmethylated promoter CpG islands, readily expressed these chemokines and their expression was not affected by treatment with either drug. CXCL14 expression in H2023 and H1568 cell lines was silenced in the absence of methylation and expression was primarily restored by TSA treatment. d) Dense methylation that corresponds with gene silencing was found in the promoter CpG island of CXCL14. Modified DNA was amplified using primers that do not discriminate between methylated and unmethylated alleles (the same used for COBRA), cloned, and five clones per sample were sequenced. The shaded area of each circle indicates the extent of methylation at that CpG and one clone represents 1/5th of the circle. CpGs 10-12 and 20-23 (placed in boxes) indicate the primer binding sites for the MSP assay and the CpGs recognized by BstU1 restriction enzyme used for COBRA are indicated by asterisks. Nearly all CpGs of samples that were unmethylated by COBRA and MSP assays (NHBEC 255, and 256, H1435, and primary lung adenocarcinoma sample #18) were free of methylation. Only 0-8% of the 260 CpGs (52 CpGs per clone and 5 clones per sample) were methylated in these samples. In contrast, 58-100% of the CpGs were methylated in samples that were strongly methylated by COBRA and MSP (H23, Calu-6 and primary tumor #54, #66, and #100). Primary adenocarcinoma #35 that was weakly methylated by the two assays was methylated for 33% of the CpGs.
DAC treatment restores expression of genes silenced by methylation
The causality of promoter hypermethylation and/or histone modification to gene silencing was evaluated using drugs to inhibit DNA methylation (DAC) and histone deacetylation (TSA). Lung cancer cell lines with or without methylation of CXCL5, 12, and 14 promoters were treated with vehicle (sham), TSA, or DAC and gene expression was evaluated by RT-PCR. DAC treatment restored the expression of CXCL5 (H1568, H1993, and Calu-6), CXCL12 (Calu-6 and SKLU-1), and CXCL14 (H23, Calu-6, and SKLU-1) to a level comparable to cell lines without methylation (Figure 2a-c). TSA was unable to restore expression of these genes in cell lines where dense methylation within the promoter CpG islands was detected by the COBRA assay. The only exceptions to this scenario were CXCL5 in H1993 and CXCL14 in Calu-6 where response to DAC and TSA was similar. Interestingly, CXCL14 was completely silenced in H2023 and H1568 where the promoter CpG island is unmethylated or weakly methylated, and expression was restored primarily by TSA suggesting histone modification is the predominant cause of CXCL14 silencing in these cell lines (Figure 2c).
Because epigenetic regulation of CXCL14 is unknown, unlike CXCL5 and CXCL12 (Speetjens et al., 2008; Wendt et al., 2008), studies on this cytokine were extended to map the distribution of methylation across the promoter CpG island. Out of 82 CpGs present within the promoter CpG island of CXCL14, 52 were analyzed using sodium bisulfite sequencing. Primary lung adenocarcinomas and lung cancer cell lines that were strongly positive for methylation in the MSP and COBRA (#54, #66, #100, H23 and Calu-6) were methylated for 58-100% of the 260 CpG islands evaluated (52 CpGs per clone and 5 clones per sample) (Figure 2d). In contrast, samples with no methylation (#18, H1435, and the NHBEC N255 and N256) or weak methylation (#35) in the MSP and COBRA assays were methylated in 0-8% and 33% of the CpGs, respectively (Figure 2d).
Methylation of CXCL14 is a diagnostic marker for early stage lung cancer
Detection of aberrant gene methylation in sputum is emerging as a promising early biomarker for lung cancer (Belinsky et al., 2006). Therefore, methylation of CXCL12 and CXCL14 was evaluated using sputum samples collected from asymptomatic stage I lung cancer cases (n = 40) and cancer-free smokers (control, n = 80) matched by age, gender, smoking status (current, former), and enrollment site. The low number of CpGs within CXCL5 promoter CpG island (Table 1) prohibited the development of a sensitive and specific nested, MSP assay required for interrogating DNA recovered from sputum samples. CXCL14 was methylated in 55% of cases compared to 33.8% in controls indicating a 2.9-fold increased risk (95% CI: 1.7, 7.3; p = 0.026) for lung cancer. Similarly, CXCL12 was methylated in 50% cases to 37.5% in controls suggesting an elevated, albeit not significant, 1.7-fold increased risk for lung cancer (95% CI: 0.7, 3.3; p = 0.24). As expected, the prevalence for methylation of these chemokines in sputum samples that contain cells from both the deep lung parenchyma and airways reflecting field cancerization was greater than seen in cytologically normal bronchial epithelial cells obtained by bronchoscopy.
CXCL14 expression increases cell death
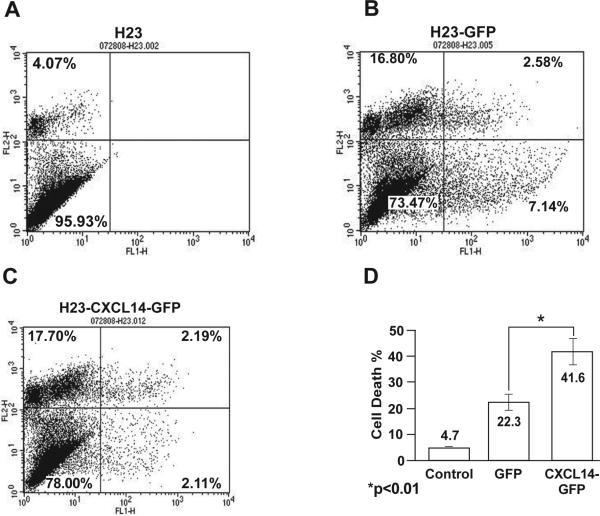
The effect of CXCL14 expression on cell survival and proliferation was evaluated using H23 and SKLU1 cells where this gene is completely silenced by methylation. Transient expression of CXCL14-GFP increased cell death in H23 by 20% (p < 0.01) (Figure 3a-d) and in SKLU1 by 40% (p < 0.001) (Figure S1) as compared to expression of GFP. Although expression of CXCL14-GFP also led to a 6% reduction in the number of actively cycling (G2-M phase) cells compared to the GFP control, the difference was not statistically significant (p = 0.28) (data not shown). In addition, migration of serum starved H23 cells with or without stable CXCL14 expression was compared using cell migration chambers with 8μm pores and H23 growth media containing 10% FBS as a chemo-attractant. Stable expression of CXCL14 increased cell migration by 40% (p < 0.01) compared to the parental cells (Figure S2).
Figure 3.
Transient re-expression of CXCL14 induces cell death. Cell survival was evaluated by flow cytometry using a GFP containing expression vector. The normal level of cell death which occurs in 4-5% of H23 cells (a) was increased to 15-20% due to exposure to the transfection reagent alone (data not shown). GFP expression in the transfected cells increased cell death by 20-25% which was only marginally greater than seen with the transfection media (b). In contrast, expression of CXCL14-GFP led to a 40-45% cell death (c). This indicates that re-expression of CXCL14 in H23 cells where the endogenous expression is silenced by methylation increased cell death by 19.3 % compared to the GFP control (d).
CXCL14 expression dramatically suppresses tumor growth in nude mice through induction of tumor necrosis
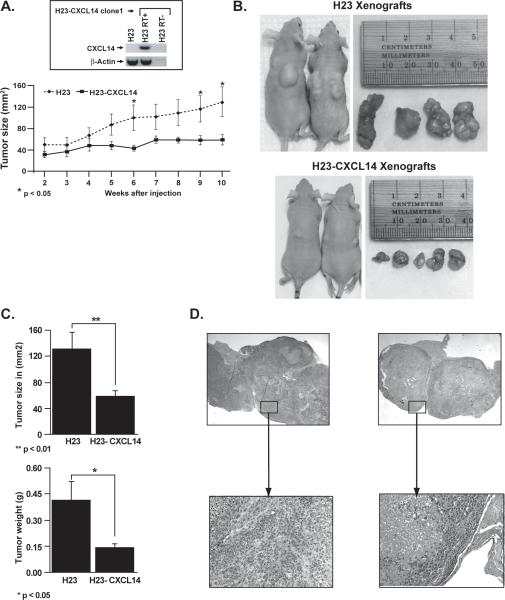
The candidate tumor suppressor function of CXCL14 was examined in vivo by comparing the tumorigenicity in nude mice of H23 cells with or without stable CXCL14 expression. Parental H23 cells with silenced CXCL14 and H23 clone (H23-CXCL14) stably expressing this chemokine (Figure 4a inset) were subcutaneously injected into nude mice and tumor growth was monitored for 10 wks. Tumor growth was comparable between the two cell lines for the first 4 wks. However, H23-CXCL14 tumors showed little additional increase in size while the parental line continued to grow (Figure 4a). The mice in both groups were sacrificed 10 wks post inoculation, the tumors were collected, and tumor volume and weight were measured. Tumors derived from the H23-CXCL14 cells were significantly smaller in size and weight than tumors from the parental H23 cell line (Figure 4a-c). Consistent with the flow cytometric cell death assay, H&E stained tumor sections revealed large areas of necrosis encompassing 50-90% of the CXCL14 expressing cell tumor mass compared to 10-30% tumor necrosis in CXCL14 negative tumors (Figure 4d).
Figure 4.
Stable re-expression of CXCL14 significantly reduced tumor growth and induced necrosis of H23 xenografts in nude mice. a) Both the parental and CXCL14 expressing H23 cells formed detectable tumors within two wks post inoculation and showed comparable rates of tumor growth in the first four wks. After wk four, tumors from the parental H23 kept growing while those from CXCL14 expressing cells barely increased in size. The asterisks at wk 6, 9, and 10 indicate significant differences in tumor size. b) The size differences between the tumors in the two groups was obvious under the skin when the mice were sacrificed ten wks post-injection. c) Similarly, the size and weight of the tumors harvested from the CXCL14 expressing cells were significantly smaller than tumors from the parental H23 cells. d) Histological examination of H&E stained slides revealed tumors from the CXCL14 expressing cells contain large necrotic foci that involved up to 90% of the tumor mass as compared to tumor necrosis in the range of 20-30% of the tumor mass from the parental H23 cells.
CXCL14 re-expression modulates pathways leading to cell cycle arrest and cell death
Genome-wide transcription patterns of H23 cells with or without stable CXCL14 expression were compared to identify pathways that might be altered by CXCL14. As compared to the parental H23, the CXCL14 expressing clone showed 659 and 445 genes with over 2-fold increased and decreased expression, respectively (data not shown). Because functional studies suggest a role for CXCL14 in cell cycle and cell death, we focused our evaluation of the microarray data on genes regulating these two pathways. Consistent with the in vitro and in vivo studies, expression of 30 genes that directly or indirectly inhibit cell cycle progression or promote apoptosis was increased in the CXCL14 expressing cells (Table 2). Moreover, expression of 41 genes that promote DNA replication, cell cycle progression and cytokinesis, or genes with anti-apoptosis and/or oncogenic properties was significantly reduced in the CXCL14 expressing cells. Fold expression changes and gene function are detailed in Table 2. Most notable, were the 4 – 7.6-fold increase in expression of caspases and the 20-fold increase in expression of TXNIP, an inducer of G1 cell cycle arrest. In contrast, expression of the cyclin family of genes (A2, A3, B1, D3, and E2) that promote cell cycle progression was reduced by 45 – 70% (Table 2).
Table 2.
Stable expression of CXCL14 in H23 cells promotes pathways that activate cell death and suppresses cell proliferation.
| Gene | Δ (fold) | Gene function |
|---|---|---|
| Inhibitors of cell cycle progression | ||
| CDKN1C | 2.89 | p57 (KIP2) is a tumor suppressor gene with reduced expression in many cancers |
| CEBPD | 8.14 | plays important role in IL-6/STAT3 mediated growth arrest |
| FOXN3 | 2.36 | S-phase checkpoint pathway gene arresting cell cycle in case of DNA damage |
| GAS1 | 3.18 | blocks entry to S phase and prevents cycling of normal and transformed cells |
| GAS5 | 4.30 | induces growth arrest and apoptosis and its expression is reduced in breast cancer |
| NDRG2 | 2.44 | leads to G1/S cell cycle arrest by attenuating AP-1 and downregulating of cyclin D1 |
| NEIL1 | 2.66 | damage sensor activating checkpoint control and involved in base excision repair. |
| NOTCH2 | 2.12 | potent inhibitor of NOTCH1 induced cell cycle progression and induces apoptosis |
| RBP1 | 4.30 | inhibits cell proliferation and induces expression of a senescence marker |
| RBP7 | 2.33 | internalization and degradation of nutrient transporters triggering nutrient starvation and induce cell death |
| SEPP1 | 4.33 | involved in hydrogen peroxide mediated oxidative stress induced cell cycle arrest |
| SPRY1 | 2.35 | suppresses proliferation and promote terminal differentiation via p21 and STAT1 upregulation and sustains ERK activation |
| SSBP2 | 2.48 | causes G1 arrest, partial differentiation and downregulation of c-MYC expression |
| Inducers of apoptosis | ||
| BAI1 | 2.13 | p53 regulated receptor for recognition and engulfment of apoptotic cells |
| BCL3 | 3.04 | increases apoptosis of multiple myeloma cells |
| CASP1 | 4.44 | caspase 1 promotes apoptosis |
| CASP4 | 7.68 | caspase 4 is effector of apoptosis |
| IFITM1 | 2.20 | promotes STAT1 and p53 crosstalk for the antiproliferative action of IFN-gamma |
| IFITM2 | 2.11 | interferon induced, p53 independent pro-apoptotic transmembrane protein |
| IFITM3 | 2.58 | human 1-8D gene is a novel p53 independent pro-apoptotic gene |
| GLTSCR2 | 2.92 | phosphorylates PTEN for caspase-independent PTEN-modulated apoptosis |
| NDRG1 | 5.30 | transcriptionally activated by p53 leading to caspase activation and apoptosis |
| OGT | 2.74 | modifies the anti-apoptotic Akt1 and induces apoptosis |
| PLAU | 3.11 | plasminogen activator induces apoptosis in brain endothelial cells (HBMEC). |
| PPARG | 2.60 | induces apoptotic cell death in NSCLC via ROS formation and POX induction |
| RHOBTB2 | 8.08 | required for CXCL14 expression and is a direct target of E2F1 with a short term increase in cell cycle but in the long term induces apoptosis |
| SALL2 | 2.30 | increase p21 and BAX for p53-independent regulation of growth and survival |
| TLE1 | 2.02 | promotes cell death through caspase-independent apoptosis |
| TRB2 | 4.38 | pro-apoptotic molecule leading to apoptosis of mainly hematopoietic cells |
| TXNIP | 20.02 | induces G1 cell cycle arrest via ASK1 activation, ER stress, p38 and JNK phosphorylation and stabilization of p27(kip1) |
| Promoters of cell cycle progression | ||
| CCNA2 | 0.31 | Cyclin A, critical for G2/M cell cycle progression |
| CCNB1 | 0.46 | Cyclin B1, required for the progression of M-phase of the cell cycle |
| CCND3 | 0.55 | Cyclin D3, important for G1/S cell cycle progression |
| CCNE2 | 0.46 | Cyclin E2, required for S/G2 cell cycle progression |
| CDCA3 | 0.50 | mediates the ubiquitination and degradation of the cdk1 inhibitor (WEE1) at G2/M leading to cdk1/cyclin B activation and mitotic entry |
| CDC2 | 0.45 | M-phase promoting factor (MPF) essential for G1/S and G2/M phase) |
| CDCA8 | 0.39 | key regulator of mitosis required for stability of the bipolar mitotic spindle |
| CDC20 | 0.48 | along with APC, required for cell cylce progression form metaphase to anaphase |
| CDC25A | 0.39 | required for progression from G1 to the S |
| CDC27 | 0.49 | component of APC, promotes ubiquitin-mediated degradation of B-type cyclins |
| PLK1 | 0.45 | promotes mitotic exit and cytokinesis via centrosome maturation, spindle assembly, removal of cohesins and inactivation of APC/C inhibitors |
| PLK2 | 0.49 | serine/threonine protein kinase that plays a role in normal cell division |
| Promoters of DNA replication | ||
| CDC6 | 0.41 | initiates DNA replication and ensures its completion before the initiation of mitosis |
| CDC45L | 0.48 | important for early steps of DNA replication in eukaryotes |
| ENDOG | 0.41 | generates the RNA primers for the initiation of mitochondrial DNA replication |
| MCM10 | 0.30 | helps form the pre-initiation complex for DNA replication and S to G2 transition |
| POLA2 | 0.49 | DNA primase that is a replicative polymerase |
| POLE4 | 0.48 | allows polymerase epsilon to carry out its replication and/or repair function |
| Promoters of Chromosomal assembly and cytokinesis | ||
| CDCA5 | 0.50 | Sororin, a regulator of sister chromatid cohesion in mitosis |
| CENPA | 0.48 | required for recruitment and assembly of kinetochore proteins, mitotic progression and chromosome segregation |
| CENPE | 0.41 | kinesin-like motor protein that accumulates in the G2 phase of the cell cycle |
| CENPO | 0.50 | component of the CENPA-CAD complex recruited to centromeres and involved in the assembly of kinetochore, mitotic progression, and chromosome segregation |
| CEP27 | 0.50 | centrosomal protein required for spindle assembly and completion of cytokinesis |
| CEP55 | 0.39 | centrosomal protein required for mitotic exit and cytokinesis |
| DSCC1 | 0.34 | involved in the establishment sister chromatid cohesion |
| NCAPG | 0.41 | helps to convert interphase chromatin into condensed mitotic chromosomes |
| NCAPH2 | 0.43 | play essential roles in mitotic chromosome assembly and segregation |
| NDC80 | 0.41 | expression peaks in mitosis and is required for the organization of microtubule binding sites, integrity of kinetochore, and chromosome segregation |
| STAG3 | 0.34 | cohesin complex required for the cohesion of sister chromatids. |
| Inhibitors of apoptosis | ||
| BIRC3 | 0.44 | suppresses apoptosis via interaction with TRAF1, TRAF2 and, TNFR2 |
| BIRC5 | 0.34 | Survivin, prevents apoptotic cell death and is overexpressed in many tumors including NSCLC and embryos, but is low in adult cells |
| IL7R | 0.37 | blocks apoptosis, differentiation, and activation of T lymphocytes |
| PTTG1 | 0.50 | suppresses transcriptional and apoptotic activity of TP53 and is tumorigenic |
| Oncogenes | ||
| ID1 | 0.42 | Ids are overexpressed in many tumors and correlate with advanced tumor stages. Id1 dimerizes with bHLH transcription factors, induces cell proliferation and inhibits differentiation and apoptosis. ID2 promotes cell survival and suppresses apoptosis of tumor cell by reducing the expression of p21 and the pro-apoptotic protein Bim/Bod, and preventing cleavage of caspase-7. Id1, 2, and 3 increase self-renewal and proliferation of cortical neural stem cells and inhibit neuronal differentiation. |
| ID2 | 0.09 | |
| ID3 | 0.34 | |
| ID4 | 0.05 | |
| EPGN | 0.27 | ligand for ERBB family receptors, promotes epithelial cell growth |
| MYB | 0.35 | oncogene in leukemia, breast and colon cancers |
| MYBL1 | 0.37 | Transcription factors required for cell proliferation and cell cycle progression. MYBL2 activates cell division cycle 2 and cyclin D1 |
| MYBL2 | 0.33 | |
Discussion
Chemokines regulate cell proliferation, apoptosis, angiogenesis, metastasis, and tumor immunity, pathways that are critical in carcinogenesis (Darash-Yahana et al., 2009; Hromas et al., 1999; Mu et al., 2009; Strieter et al., 2004). A genome-wide transcriptome array identified the CXCL5, CXCL12, and CXLC14 chemokines as common targets for silencing by promoter methylation in adenocarcinomas. Dense methylation that was reversible by treatment with a demethylating agent accounted for silencing of all three genes. Strong support for CXCL14 as a tumor suppressor gene was provided by its marked effect on growth of tumor xenografts, induction of tumor necrosis, and likely influence on many genes central to cell cycle control and apoptosis. The commonality and diverse function of the multitude of genes silenced by methylation in lung tumors has generated intense interest by our group and others for assessing their potential as biomarkers through detection of methylated genes in sputum from asymptomatic lung cancer patients (Belinsky et al., 2006; Belinsky et al., 1998; Cirincione et al., 2006; Hsu et al., 2007). CXCL14 methylation in sputum was associated with a 2.9-fold elevated risk for lung cancer, making it a potential marker for inclusion in our developing diagnostic gene panel (Belinsky et al., 2006).
The genome-wide transcriptome array developed to discover novel methylated genes in cancer identified eleven of the 16 CXC-chemokines as potential targets of DNA methylation in lung cancer. Five of the eight genes identified by the array that contained promoter CpG islands were methylated in lung cancer cell lines, a 62.5% accuracy for identifying genes regulated by methylation. However, two of the five methylated genes were also methylated in normal cells (CXCL4 in NHBEC and PBMC, and CXCL6 in PBMC) indicating that gene regulation through methylation can be cell-specific and related to state of differentiation. This conclusion is supported by our previous study identifying four genes within the lung cancer susceptibility locus 6q23-25 that were methylated in lymphocytes and two genes methylated in NHBEC (Tessema et al., 2008). Recently, a genome-wide screen for methylation also demonstrated that approximately 10% of all promoters were methylated in differentiated B cells (Rauch et al., 2009).
The expression of CXCL2, 3, and 16 that contain promoter CpG islands was significantly increased by DAC treatment; however, methylation of these genes was not seen in any lung cancer cell line. One explanation for this epigenetic regulation is re-expression of methylated genes whose proteins function as transcription factors to regulate expression of these chemokines. While detailed studies have not been conducted to map transcription factor binding sites within these chemokines, our own studies on regulation of the PAX5 β gene support this premise. PAX5 β is commonly silenced in lung cancer through methylation and encodes for the transcription factor B cell-specific activating protein that in turn, directly regulates CD19, a gene not containing a CpG island. A strong association was observed between PAX5 β methylation and loss of expression of CD19 and treatment with a demethylating agent restored expression of both genes (Palmisano et al., 2003). Unlike lung cancer, CXCL16 is commonly silenced by methylation in kidney tumors (Morris et al., 2008). In addition, RNA interference mediated knockdown of DNMT3a in melanoma cells resulted in CXCL9 and CXCL16 re-expression that was associated with suppression of tumor growth and metastasis suggesting methylation-mediated regulation of these chemokines in other tumors (Deng et al., 2009).
Among the CXC-chemokines with tumor-specific methylation in lung cancer, the role of CXCL12 in carcinogenesis is well established. Whereas CXCL12 is frequently methylated in various carcinomas, its cell surface receptor (CXCR4) is abundantly expressed in most tumors (Wendt et al., 2008; Wendt et al., 2006; Yoshino et al., 2009; Zhou et al., 2008). This limits the CXCL12-CXCR4 autocrine signaling and promotes directional migration of carcinoma cells toward organs/tissues with high CXCL12 expression. Lung is one of the organs expressing higher levels of CXCL12 and is a primary site for metastasis of carcinomas with low CXCL12 expression such as breast and colorectal cancers (Muller et al., 2001). Conversely, when CXCL12 is silenced in lung cancer, it may enhance local invasion as well as metastasis to other organs with high CXCL12 expression such as the adrenal glands, liver, and bone marrow (Phillips et al., 2003). Therefore, methylation-mediated silencing of CXCL12 in lung cancer could play a major role in deregulating the autocrine CXCL12-CXCR4 signaling pathway to promote tumor invasion and metastasis.
Unlike CXCL12, the role of CXCL5 in cancer is unclear and likely varies in different malignancies. ShRNA knockdown of CXCL5 in colon cancer increased tumor growth and metastasis that is reversed by restoring CXCL5 expression suggesting a tumor suppressor function (Speetjens et al., 2008). In contrast, expression of CXCL5 is increased in metastatic HNSCC and promotes cell proliferation, while shRNA knockdown of CXCL5 suppresses cell migration, proliferation, and tumorigenicity in nude mice suggesting an oncogenic role (Miyazaki et al., 2006). While CXCL5 is clearly silenced through methylation in lung cancer, its elevated expression in NSCLC could be from stromal cells within the tumor (Arenberg et al., 1998).
CXCL14 is a potent angiostatic chemokine that blocks CXCL8 (IL-8), VEGF and bFGF mediated angiogenesis (Shellenberger et al., 2004). It is also a chemo-attractant mediating infiltration of dendritic cells into tumors, which is critical for tumor immunity (Shurin et al., 2005). As shown in this study, re-expression of CXCL14 enhances cell death in vitro and induces marked necrosis in lung tumor xenografts, due in part through increased expression of multiple apoptosis effector proteins including the caspases. Moreover, re-expression of CXCL14 has a profound effect on the genome altering the expression of more than 1,000 genes. Taken together these results suggest CXCL14 expression is central for various anti-tumor mechanisms and its silencing could be critical for carcinogenesis.
Aberrant methylation may ultimately serve two vital roles in cancer: as biomarkers for detection and prognosis and targets for epigenetic therapy (Belinsky et al., 2003; Belinsky et al., 2006; Gore et al., 2006; Yang et al., 2006). CXCL14 may participate in both arenas. We are developing a gene panel whose methylation in sputum could identify people at high risk for cancer incidence. Previously, six of 14 genes evaluated in a lung cancer case-control study were associated with over 50% increase in lung cancer risk. Importantly, concomitant methylation of three or more of these six genes increased risk 6.5-fold with a sensitivity and specificity of 64% for classifying lung cancer cases. While, this sensitivity and specificity is not high enough for prospective population-based screening, additional genes such as CXLC14 whose methylation in sputum conferred a 190% increased risk, could contribute significantly to our gene panel.
Rather than targeting single pathway alterations in cancer, epigenetic therapy may circumvent the problem of tumor heterogeneity by inducing the re-expression of multiple tumor suppressor genes; several may be essential for abrogating cancer cell survival and proliferation. Demethylating agents used at doses far below their maximum tolerated dose have proven to be a potent therapy for a precursor state to acute myelogenous leukemia (AML) and myelodyplasia (MDS) and are now approved by the FDA for treatment of these cancers (Yang et al., 2006). Clinical trials with demethylating agents combined with histone deacetylase inhibitors also are showing promising responses in the treatment of myeloid malignancies (Gore et al., 2006). The extension of this targeted approach to solid tumors such as those in the lung may also hold promise as a therapy. Our recent work in which combined treatment with DAC and sodium phenylbutyrate reduced the number of developing lung tumors in a murine model by more than 50% support this supposition and Phase II trials in lung cancer are underway (Belinsky et al., 2003). The effective awakening of silenced genes such as CXCL14 that can affect the activity of many critical pathways could profoundly impact the growth and survival of tumor cells setting the stage for using epigenetic therapy in the management of lung cancer in some patients.
Materials and Methods
Samples
Frozen lung adenocarcinomas from current (n = 37), former (n = 59) and never (n=75) smokers were obtained from tumor banks at Johns Hopkins and the Mayo Clinic. Demographic variables including age, gender, and stage of lung cancer and selection criteria for suitable adenocarcinomas has been described previously (Tessema et al., 2008). NHBEC isolated from bronchoscopy of cancer-free smokers (n = 20) and PBMC from healthy donors (n = 20) were used as controls. Seventeen lung cancer-derived cell lines (H23, H358, H1435, H1568, H1993, H2023, H2085, H2228, H2009, Calu-3, Calu-6, SKLU-1, SKMES1, H1299, H1975, HCC827, and HCC4006) were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). Sputum samples collected from stage I NSCLC cases prior to surgery (n = 40) and cancer free smokers (n = 80) and were matched on age (+/- 5 years), gender, smoking status (current, former) and location of hospital as described (Chin et al., 2008).
Treatment and Genome-Wide Transcriptome Array
Three lung adenocarcinoma cell lines (H23, H1568, and H1993) from smokers and three (H2023, H2085, and H2228) from never smokers were used for the transcriptome array as described (Tessema et al., 2008). Briefly, cells at log phase of growth were treated as follows: Control (culture medium), TSA (culture medium containing 300 nM of trichostatin A, [Sigma, St. Louis, MO; stock solution 5 μM in ethanol] for 18 h), or DAC (500nM 5-aza-2-deoxycytidine [Sigma; stock solution 10 mM in PBS] for 96 h with fresh medium containing the drug changed every 24 h). Cells were harvested in TRI Reagent (Sigma-Aldrich, Steinheim, Germany) and genome-wide transcriptome array analysis was conducted using the Agilent 44K expression array as described (Tessema et al., 2008).
DNA methylation analysis
DNA was extracted and modified as described (Tessema et al., 2009) and 40 ng of modified DNA was used per PCR. Methylation in NHBEC, PBMC, primary lung tumors and cancer cell lines was studied using COBRA and MSP as described (Tessema et al., 2009). Methylation of DNA isolated from sputum was assessed using a nested, MSP assay developed as described (Belinsky et al., 2006). Primer sequences and PCR conditions are described in Tables S1 and S2. Bisulfite sequencing of CXCL14 promoter CpG island was conducted using modified DNA and COBRA primers (Table S1) that do not discriminate between methylated and unmethylated DNA. PCR products were cloned into pCR II cloning vector (Invitrogen, Santa Clara CA) and five clones were sequenced per sample.
Gene Expression Analysis
Total RNA (3 μg) isolated from sham (vehicle), TSA, and DAC treated cells as described (Tessema et al., 2009) was reverse transcribed using the SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen) according to the protocol from Invitrogen®. Transcription of CXCL5, 12 and 14 was evaluated using RT-PCR and electrophoresis in 3% agarose gels. To avoid PCR products from DNA, RNA was treated with DNase and PCR primers were located in exons separated by a large intron. For CXCL14 transcripts originating from plasmid vectors (no intron), RT-negative (RT-) PCR was done in parallel using cDNA synthesized in the absence of Superscriptase-II. RT-PCR primers and amplification conditions are described in Table S3.
Cloning, transfection, and establishment of stable CXCL14 expressing clones
Full-length CXCL14 transcript amplified by PCR using cDNA from NHBEC 255 CXCL14 F2/R2 primer pairs (Table S3) was directly ligated into pcDNA3.1/NT-GFP-TOPO® vector (Invitrogen) for transient transfection and into pTARGET™ Mammalian Expression Vector System (Promega, Madison, WI) for stable expression and cloned by TA cloning strategy. The sense orientation and sequences of the cloned CXCL14 cDNA was confirmed by DNA sequencing. H23 cells (1 × 105/well in 6-well plate) were transfected with CXCL14-GFP or the GFP expression vector (Mock) using Lipofectamine™ LTX (Invitrogen). Stable transformants were selected from CXCL14 and Mock pTARGET™ vector transfected H23 cells using 400 μg/ml Geneticin (Geneticin® Liquid (G-418 sulfate), Invitrogen).
Cell death, cell cycle, and cell migration assays
H23 cells were plated at a density of 1 × 105/well in 6-well plates and transfected 24 hours later with Mock (GFP) or CXCL14-GFP vectors. For cell death analysis, cells were harvested (48 hours post transfection) with trypsin, washed once with PBS, stained in the dark with 10 μg/ml Propidium iodide (PI) (Sigma-Aldrich) at 37°C for 1 hr, washed, resuspended in 1 ml PBS and the PI and GFP fluorescence were evaluated using flow cytometry (Becton Dickinson FACScalibur Flow Cytometer). The proportion of PI and/or GFP positive cells was calculated from 10,000 events. For cell cycle analysis, cells were plated, transfected, harvested, and washed as described above and resuspended in 500 μl ice-cold PBS, fixed by adding 500 μl of ice cold 2% buffered paraformaldehyde, and incubated at 4°C for 30 min. The cells were then washed, permeablized with 1 ml ethanol at 4°C overnight, stained with 1 ml PI solution containing 40 μg/ml PI, 100 μg/ml RNase in PBS at 37°C for 1 hr in the dark, washed, resuspended in PBS and cells were analyzed with flow cytometry as described above. For cell migration, H23 cells with or without stable CXCL14 expression were serum starved for 48 hrs and cell migration was measured using CytoSelect™ 24-Well Cell Migration Assay (8 μm, Colorimetric Format) (Cell Biolabs, San Diego, CA) and 10% serum containing growth medium as a chemo-attractant for 24 hrs according to the protocol.
In vivo tumor growth
Matrigel™ Basement Membrane Matrix (BD Biosciences, San Jose, CA) was mixed 1:1 with H23-CXCL14 clone stably expressing CXCL14 and the parental H23 cell line and subcutaneously injected (5 × 106/site) into both sides of the dorso-lateral region of four female athymic nude mice (Athymic NCr-nu/nu, Frederick, MD) per group. Tumor size was quantified once a wk from 2nd to 10th wk post injection and tumor volume was calculated as: (a × b2)/2, where a and b represent the longer and shorter dimensions respectively. On the 10th wk, all the mice were sacrificed, tumors collected, and weighed. Tumors were formalin fixed, paraffin embedded, sectioned, and stained with hematoxylin and eosin.
Supplementary Material
Figure S1 Transient re-expression of CXCL14 in SKLU1 cells increases cell death by 40% compared to the GFP control.
Figure S2 CXCL14 expression increases cell migration. H23 cells with or without stable CXCL14 expression were serum starved for 48h and their migration potential was compared using cell migration chambers with 8μm pores and 10% FBS containing H23 growth medium as chemo-attractant for 24h.
Acknowledgments
The authors thank Elizabeth Burki, Ph.D. for recruitment of lung cancer patients, Maria Picchi, MPH and Chris Stidley, Ph.D. for statistical analysis and Wayne Yu, Ph.D. at Johns Hopkins for running Agilant expression arrays.
Grant support: Supported largely by NIH grant R01 ES008801 and in part by P50 CA58184.
Footnotes
Conflict of interest
S.A. Belinsky is a consultant to Oncomethylome Sciences. Under a licensing agreement between Lovelace Respiratory Research Institute and Oncomethylome Sciences, nested MSP was licensed to Oncomethylome Sciences, and the author is entitled to a share of the royalties received by the Institute from sales of the licensed technology. The Institute, in accordance with its conflict-of-interest policies, is managing the terms of these arrangements.
References
- Arenberg DA, Keane MP, DiGiovine B, Kunkel SL, Morris SB, Xue YY, et al. Epithelial-neutrophil activating peptide (ENA-78) is an important angiogenic factor in non-small cell lung cancer. J Clin Invest. 1998;102:465–72. doi: 10.1172/JCI3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–93. [PubMed] [Google Scholar]
- Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95:11891–6. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KL, Karpenko M, Lin MT, Claus R, Arab K, Dyckhoff G, et al. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008;68:4494–9. doi: 10.1158/0008-5472.CAN-07-6509. [DOI] [PubMed] [Google Scholar]
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirincione R, Lintas C, Conte D, Mariani L, Roz L, Vignola AM, et al. Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: a nested case-control study. Int J Cancer. 2006;118:1248–53. doi: 10.1002/ijc.21473. [DOI] [PubMed] [Google Scholar]
- Darash-Yahana M, Gillespie JW, Hewitt SM, Chen YY, Maeda S, Stein I, et al. The chemokine CXCL16 and its receptor, CXCR6, as markers and promoters of inflammation-associated cancers. PLoS One. 2009;4:e6695. doi: 10.1371/journal.pone.0006695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Kuang Y, Wang L, Li J, Wang Z, Fei J. An essential role for DNA methyltransferase 3a in melanoma tumorigenesis. Biochem Biophys Res Commun. 2009;387:611–6. doi: 10.1016/j.bbrc.2009.07.093. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–9. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- Hromas R, Broxmeyer HE, Kim C, Nakshatri H, Christopherson K, 2nd, Azam M, et al. Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem Biophys Res Commun. 1999;255:703–6. doi: 10.1006/bbrc.1999.0257. [DOI] [PubMed] [Google Scholar]
- Hsu HS, Chen TP, Wen CK, Hung CH, Chen CY, Chen JT, et al. Multiple genetic and epigenetic biomarkers for lung cancer detection in cytologically negative sputum and a nested case-control study for risk assessment. J Pathol. 2007;213:412–9. doi: 10.1002/path.2246. [DOI] [PubMed] [Google Scholar]
- Jacinto FV, Ballestar E, Ropero S, Esteller M. Discovery of epigenetically silenced genes by methylated DNA immunoprecipitation in colon cancer cells. Cancer Res. 2007;67:11481–6. doi: 10.1158/0008-5472.CAN-07-2687. [DOI] [PubMed] [Google Scholar]
- Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- Kurth I, Willimann K, Schaerli P, Hunziker T, Clark-Lewis I, Moser B. Monocyte selectivity and tissue localization suggests a role for breast and kidney-expressed chemokine (BRAK) in macrophage development. J Exp Med. 2001;194:855–61. doi: 10.1084/jem.194.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon CM, Lygoe KA, Skelton L, Mitter R, Mellor H. The atypical Rho GTPase RhoBTB2 is required for expression of the chemokine CXCL14 in normal and cancerous epithelial cells. Oncogene. 2008;27:6856–65. doi: 10.1038/onc.2008.317. [DOI] [PubMed] [Google Scholar]
- Meissner A, Jaenisch R. Generation of nuclear transfer-derived pluripotent ES cells from cloned Cdx2-deficient blastocysts. Nature. 2006;439:212–5. doi: 10.1038/nature04257. [DOI] [PubMed] [Google Scholar]
- Miyazaki H, Patel V, Wang H, Edmunds RK, Gutkind JS, Yeudall WA. Down-regulation of CXCL5 inhibits squamous carcinogenesis. Cancer Res. 2006;66:4279–84. doi: 10.1158/0008-5472.CAN-05-4398. [DOI] [PubMed] [Google Scholar]
- Morris MR, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, et al. Functional epigenomics approach to identify methylated candidate tumour suppressor genes in renal cell carcinoma. Br J Cancer. 2008;98:496–501. doi: 10.1038/sj.bjc.6604180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2:123–8. doi: 10.1038/84219. [DOI] [PubMed] [Google Scholar]
- Mu X, Chen Y, Wang S, Huang X, Pan H, Li M. Overexpression of VCC-1 gene in human hepatocellular carcinoma cells promotes cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2009;41:631–7. doi: 10.1093/abbs/gmp051. [DOI] [PubMed] [Google Scholar]
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Ozawa S, Kato Y, Ito S, Komori R, Shiiki N, Tsukinoki K, et al. Restoration of BRAK / CXCL14 gene expression by gefitinib is associated with antitumor efficacy of the drug in head and neck squamous cell carcinoma. Cancer Sci. 2009;100:2202–9. doi: 10.1111/j.1349-7006.2009.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa S, Kato Y, Komori R, Maehata Y, Kubota E, Hata R. BRAK/CXCL14 expression suppresses tumor growth in vivo in human oral carcinoma cells. Biochem Biophys Res Commun. 2006;348:406–12. doi: 10.1016/j.bbrc.2006.07.070. [DOI] [PubMed] [Google Scholar]
- Palmisano WA, Crume KP, Grimes MJ, Winters SA, Toyota M, Esteller M, et al. Aberrant promoter methylation of the transcription factor genes PAX5 alpha and beta in human cancers. Cancer Res. 2003;63:4620–5. [PubMed] [Google Scholar]
- Parsanejad R, Fields WR, Steichen TJ, Bombick BR, Doolittle DJ. Distinct regulatory profiles of interleukins and chemokines in response to cigarette smoke condensate in normal human bronchial epithelial (NHBE) cells. J Interferon Cytokine Res. 2008;28:703–12. doi: 10.1089/jir.2008.0139. [DOI] [PubMed] [Google Scholar]
- Phillips RJ, Burdick MD, Lutz M, Belperio JA, Keane MP, Strieter RM. The stromal derived factor-1/CXCL12-CXC chemokine receptor 4 biological axis in non-small cell lung cancer metastases. Am J Respir Crit Care Med. 2003;167:1676–86. doi: 10.1164/rccm.200301-071OC. [DOI] [PubMed] [Google Scholar]
- Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–8. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA Hypermethylome with Gene Mutations in Human Colorectal Cancer. PLoS Genet. 2007;3:e157. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze SR, Luo J, Isaacs WB, Jarrard DF. Modulation of CXCL14 (BRAK) expression in prostate cancer. Prostate. 2005;64:67–74. doi: 10.1002/pros.20215. [DOI] [PubMed] [Google Scholar]
- Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3:e486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shellenberger TD, Wang M, Gujrati M, Jayakumar A, Strieter RM, Burdick MD, et al. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Res. 2004;64:8262–70. doi: 10.1158/0008-5472.CAN-04-2056. [DOI] [PubMed] [Google Scholar]
- Shurin GV, Ferris RL, Tourkova IL, Perez L, Lokshin A, Balkir L, et al. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. J Immunol. 2005;174:5490–8. doi: 10.4049/jimmunol.174.9.5490. [DOI] [PubMed] [Google Scholar]
- Speetjens FM, Kuppen PJ, Sandel MH, Menon AG, Burg D, van de Velde CJ, et al. Disrupted expression of CXCL5 in colorectal cancer is associated with rapid tumor formation in rats and poor prognosis in patients. Clin Cancer Res. 2008;14:2276–84. doi: 10.1158/1078-0432.CCR-07-4045. [DOI] [PubMed] [Google Scholar]
- Strieter RM, Belperio JA, Burdick MD, Sharma S, Dubinett SM, Keane MP. CXC chemokines: angiogenesis, immunoangiostasis, and metastases in lung cancer. Ann N Y Acad Sci. 2004;1028:351–60. doi: 10.1196/annals.1322.041. [DOI] [PubMed] [Google Scholar]
- Tessema M, Willink R, Do K, Yu YY, Yu W, Machida EO, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707–14. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- Tessema M, Yu YY, Stidley CA, Machida EO, Schuebel KE, Baylin SB, et al. Concomitant promoter methylation of multiple genes in lung adenocarcinomas from current, former and never smokers. Carcinogenesis. 2009;30:1132–8. doi: 10.1093/carcin/bgp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt MK, Cooper AN, Dwinell MB. Epigenetic silencing of CXCL12 increases the metastatic potential of mammary carcinoma cells. Oncogene. 2008;27:1461–71. doi: 10.1038/sj.onc.1210751. [DOI] [PubMed] [Google Scholar]
- Wendt MK, Johanesen PA, Kang-Decker N, Binion DG, Shah V, Dwinell MB. Silencing of epithelial CXCL12 expression by DNA hypermethylation promotes colonic carcinoma metastasis. Oncogene. 2006;25:4986–97. doi: 10.1038/sj.onc.1209505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, et al. DNA methylation changes after 5-aza-2'-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- Yoshino M, Suzuki M, Tian L, Moriya Y, Hoshino H, Okamoto T, et al. Promoter hypermethylation of the p16 and Wif-1 genes as an independent prognostic marker in stage IA non-small cell lung cancers. Int J Oncol. 2009;35:1201–9. doi: 10.3892/ijo_00000437. [DOI] [PubMed] [Google Scholar]
- Yuvaraj S, Griffin AC, Sundaram K, Kirkwood KL, Norris JS, Reddy SV. A novel function of CXCL13 to stimulate RANK ligand expression in oral squamous cell carcinoma cells. Mol Cancer Res. 2009;7:1399–407. doi: 10.1158/1541-7786.MCR-08-0589. [DOI] [PubMed] [Google Scholar]
- Zhou W, Jiang Z, Song X, Liu Y, Wen P, Guo Y, et al. Promoter hypermethylation-mediated down-regulation of CXCL12 in human astrocytoma. J Neurosci Res. 2008;86:3002–10. doi: 10.1002/jnr.21746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Transient re-expression of CXCL14 in SKLU1 cells increases cell death by 40% compared to the GFP control.
Figure S2 CXCL14 expression increases cell migration. H23 cells with or without stable CXCL14 expression were serum starved for 48h and their migration potential was compared using cell migration chambers with 8μm pores and 10% FBS containing H23 growth medium as chemo-attractant for 24h.