Abstract
Epidermal Growth Factor Receptor (EGFR) is frequently over-expressed in head and neck squamous cell carcinoma (HNSCC) where aberrant signaling downstream of this receptor contributes to tumor growth. EGFR variant III (EGFRvIII) is the most commonly altered form of EGFR and contains a truncated ligand-binding domain. We previously reported that EGFRvIII is expressed in up to 40% of HNSCC tumors where it is associated with increased proliferation, tumor growth and chemoresistance to anti-tumor drugs including the EGFR targeting monoclonal antibody cetuximab. Cetuximab was FDA-approved in 2006 for HNSCC but has not been shown to prevent invasion or metastasis. The present study was undertaken to evaluate the mechanisms of EGFRvIII-mediated cell motility and invasion in HNSCC. We found that EGFRvIII induced HNSCC cell migration and invasion in conjunction with increased STAT3 activation, which was not abrogated by cetuximab treatment. Further investigation demonstrated that EGF-induced expression of the STAT3 target gene HIF1-α, was abolished by cetuximab in HNSCC cells expressing wild-type EGFR under hypoxic conditions, but not in EGFRvIII-expressing HNSCC cells. These results suggest that EGFRvIII mediates HNSCC cell migration and invasion via increased STAT3 activation and induction of HIF1-α, which contribute to cetuximab resistance in EGFRvIII-expressing HNSCC tumors.
Keywords: head and neck cancer, epidermal growth factor receptor, EGFRvIII, STAT3 and cancer cell invasion
Introduction
The epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that regulates crucial cellular signaling pathways contributing to tumor progression. EGFR is frequently amplified and over-expressed in several human solid tumors including in a high percentage of head and neck squamous cell carcinomas (HNSCC). EGFR overexpression in HNSCC has been correlated with tumor progression, resistance to conventional therapy and poor prognosis (Grandis and Tweardy, 1993). Preclinical studies demonstrated the anti-tumor effects of EGFR targeting and the FDA approved the EGFR monoclonal antibody cetuximab for clinical use in HNSCC based on the results of a phase III trial (Bonner et al., 2006). However, while combining EGFR targeting with radiation prolonged overall survival, it did not reduce the incidence of metastasis. Despite the nearly ubiquitous expression of EGFR in HNSCC, there is only a 13% response rate when cetuximab is administered as a single agent (Vermorken et al., 2007). The tumor features that contribute to resistance to EGFR targeting are incompletely understood.
Receptor alterations that influence ligand and antibody binding may play a role in therapeutic resistance. The most common EGFR alteration in several cancers, including HNSCC, consists of a truncation in the extracellular domain known as EGFR variant III (EGFRvIII). This mutation eliminates exons 2-7 resulting in a distorted ligand-binding region (Bigner et al., 1990; Sugawa et al., 1990). EGFRvIII does not bind ligand but is constitutively activated in a ligand-independent manner. The presence of EGFRvIII in human tumors has been associated with tumor growth, metastasis, and survival in several malignancies including glioma, carcinomas of the breast, lung and HNSCC (Pedersen et al., 2001). Furthermore, EGFRvIII has been reported to increase resistance to anti-tumor agents including EGFR inhibitors (Sok et al., 2006).
We previously reported the expression of EGFRvIII in up to 42% of HNSCC where co-expression of EGFRvIII with wild-type EGFR increased HNSCC cell proliferation in vitro and tumor volume in vivo. Moreover, EGFRvIII decreased HNSCC cell apoptosis in response to cisplatin and decreased growth inhibition following treatment with cetuximab (Sok et al., 2006). While these results support the role of EGFRvIII in mediating tumor growth in response to EGFR targeting, the contribution of EGFRvIII to invasion and the precise downstream pathways that are induced by EGFRvIII are incompletely understood. EGFRvIII expression in glioma has been reported to correlate with expression of phosphotyrosine STAT3 (Mizoguchi et al., 2006). The lethality of HNSCC is associated with the tendency of these cancers to invade surrounding structures and metastasize. The present study was undertaken to test the hypothesis that EGFRvIII induces HNSCC invasion and subsequently, metastasis via activation of STAT3 signaling.
Results
Expression of EGFRvIII in engineered HNSCC cells
We previously reported that EGFRvIII is expressed in approximately 40% of HNSCC tumors (Sok et al., 2006). For reasons that are incompletely understood, expression of EGFRvIII in human tumors is routinely lost in tissue culture (Bigner et al., 1990). Therefore, to study the consequences of EGFRvIII in HNSCC, we stably transfected EGFR vIII into a representative HNSCC cell line (UM-22B) as described previously (Nishikawa et al., 1994; Sok et al., 2006). Four independent clones that stably expressed EGFRvIII were isolated (vIII-1 to 4) as well as four vector-transfected control clones (control-1 to 4). Due to the high level of wild-type EGFR in HNSCC cell lines and tissues, commercially available EGFR antibodies that are reported to detect both EGFRvIII and wild-type EGFR in other tumor systems, are unable to identify EGFRvIII expression in HNSCC. Expression of the EGFRvIII transcript was therefore determined by RT-PCR where the rodent fibroblasts stably expressing wild-type EGFR (NR6W) or EGFRvIII (NR6M) were used as controls for EGFR and EGFRvIII, respectively. EGFRvIII gene expression was detected in all four vIII clones but not in vector-transfected control clones while wild-type EGFR gene expression was observed in both control and vIII clones (Figure 1A). Flow cytometry was also performed to measure the degree of expression of EGFRvIII in these clones. FACS analyses of the cell surface EGFRvIII receptor expression revealed no EGFRvIII in the vector transfected control cells and approximately 104 EGFRvIII receptors per cell in each of the HNSCC clones (Figure 1B). In addition, using flow cytometry we determined the number of EGFR receptors to be 1.61 × 105 receptors per cell (Supplemental Figure 1). Thus, the numbers of wild-type EGF receptors are 10-fold higher than EGFRvIII in the EGFRvIII- transfected clones. These results are consistent with findings in human HNSCC where all HNSCC tumors that express EGFRvIII also express wild-type EGFR, with a higher level of wild-type EGFR compared to EGFRvIII (Sok et al., 2006).
Figure 1. EGFRvIII expression in HNSCC-transfected cells.
(A) EGFRvIII mRNA levels were examined in HNSCC-transfected cells (UM-22B) by RT-PCR. Representative ethidium bromide-stained gel showing EGFR and EGFRvIII PCR products from four vector-transfected control clones (control 1-4) and EGFRvIII-expressing clones (vIII-1 to vIII-4). The EGFR-transfected NR6W cell line was used as positive control for EGFR and EGFRvIII-transfected NR6M cells served as positive control for EGFRvIII. Experiments were repeated 3 times with similar results. (B) Flow cytometry results for UM-22B clones stably transfected with EGFRvIII. Surface EGFRvIII on four HNSCC-transfected clones (vIII-1 to vIII-4) were detected by flow cytometry. A vector-transfected control clone (control-1) was used as negative control. Data is represented as Mean Fluorescent Intensity. Experiments were repeated 3 times with similar results.
EGFRvIII increases HNSCC motility and invasion
We previously reported that EGFRvIII induces HNSCC cell proliferation in vitro and tumor growth in vivo (Sok et al., 2006). EGFRvIII has been shown to induce motility in murine fibroblasts (Pedersen et al., 2004). To determine the consequences of EGFRvIII on directional HNSCC cell motility, cell migration (wound-healing) assays were performed. Briefly, a wound was created in a confluent monolayer of HNSCC cells. Images captured at the creation of the wound and at the end of the assay were analyzed to measure the distance traveled by the leading edge of the cells in the wound. As shown in Figure 2A, HNSCC cell migration was increased in all four EGFRvIII-expressing clones compared to a vector-transfected control (p=0.03). In order to validate these findings in other HNSCC cell lines, we transiently transfected 1483 and PCI-37A cells with an EGFR vIII expression plasmid and tested the migration of the cells in a transwell migration assay. EGFR vIII expressing cells demonstrated increased migration (Supplemental Figure 2A). We next assessed the consequences of EGFRvIII on HNSCC cell invasion through Matrigel, controlling for proliferation. As shown in Figure 2B, EGFRvIII-expressing HNSCC cells were significantly more invasive than vector transfected controls (p=0.03).
Figure 2. EGFRvIII increases HNSCC migration and invasion.
(A) The denuded area of vector control-transfected cells (control-1) and EGFRvIII-expressing cells (vIII-1 to 4) was measured 0 h and 6 h after a single scrape (wound) was made in the confluent monolayer using a sterile pipette tip. The area covered by the migrating cells was measured using computer-assisted image analysis. The decreased area was determined by subtracting the values obtained a 6 hr from the 0 hr time point. The fold-decreased area in the EGFRvIII-transfected cells was compared with vector-transfected controls. Cumulative results are shown from four independent experiments (*p=0.03). (B) The invasive capacity of EGFRvIII-expressing cells (vIII-1 to -4) compared with a vector-transfected control (control-1) was determined using a Boyden chamber Matrigel assay. The fold increase in invasion relative to vector-transfected control is shown. Cumulative results are shown from four independent experiments (*p=0.03). (C) EGFRvIII-expressing cells (vIII-4) and vector transfected control cells (control-1) were subjected to an invasion assay in the presence of EGF (10 ng/ml) and/or C225 (7 μg/ml) for 24 hours. The fold-increase in invasion relative to untreated vector-transfected control (control-1 NoTx) is shown. The invasion of EGFRvIII-expressing HNSCC cells was not abrogated by C225 treatment compared with vector-transfected control cells (*p=0.03).
Cetuximab was FDA-approved for the treatment of HNSCC in 2006. We previously reported that HNSCC cells expressing EGFRvIII are relatively resistant to the growth inhibitory effects of cetuximab in vitro and in vivo (Sok et al., 2006). Since the addition of cetuximab to radiation did not prevent metastasis in HNSCC patients, we next determined the effects of cetuximab on EGFRvIII-mediated invasion (Bonner et al., 2006). As shown in Figure 2C, while cetuximab abrogated EGF-induced invasion of vector control-transfected HNSCC cells, treatment of EGFRvIII-expressing HNSCC cells with cetuximab failed to decrease invasion. While EGF induced invasion of vector-transfected control cells, EGF treatment had no significant effect on the invasive capacity of EGFRvIII cells, which are more invasive than controls in the absence of growth factor stimulation. These results suggest that EGFRvIII induces HNSCC cell motility and invasion in vitro, which are not abrogated by treatment with the only clinically approved EGFR targeting strategy in HNSCC.
EGFRvIII increases STAT3 activation
The precise signaling pathways induced by EGFRvIII in the setting of cancer cells that also express wild-type EGFR are incompletely understood. STAT3 is activated downstream of several receptor and non-receptor tyrosine kinases including EGFR. A correlation between EGFRvIII expression and expression of phosphotyrosine STAT3 has been noted in glioblastomas (Mizoguchi et al., 2006). We previously reported that STAT3 is activated downstream of wild-type EGFR in HNSCC (Grandis et al., 1998). To determine whether STAT3 is differentially activated by EGFRvIII, we analyzed expression of tyrosine phosphorylated STAT3 by immunoblotting in addition to STAT3 transcriptional activity in the EGFRvIII-expressing HNSCC cells compared with vector-controls. As shown in Figure 3A, phosphotyrosine STAT3 was expressed at higher levels in HNSCC cells that contain EGFRvIII compared to vector-transfected controls. Transiently transfected HNSCC cells 1483 and PCI-37A also demonstrated higher levels of phosphorylated STAT3 (Supplemental Figure 2B). In addition, EGFRvIII-expressing HNSCC cells demonstrated increased STAT3 transcriptional activity using an hSIE luciferase reporter assay compared with controls (p=0.03) (Figure 3B). To determine the relative expression levels of phosphorylated and total STAT3 in EGFRvIII-expressing HNSCC tumors, xenografts derived from EGFRvIII-expressing cells were analyzed. As shown in Figure 3C, EGFRvIII-expressing tumors contained higher levels of phosphorylated STAT3 compared to tumors derived from vector-transfected control cells (p=0.03). Further, EGFRvIII expression was associated with increased tumor growth in a xenograft model, thus confirming our previous findings (Sok et al., 2006) (data not shown). To determine the effects of cetuximab on EGFRvIII-mediated STAT3 activation, HNSCC cells expressing EGFRvIII were treated with cetuximab followed by STAT3 promoter assays. While cetuximab decreased STAT3 promoter activity in vector-transfected control cells cetuximab was unable to abrogate STAT3 activation in HNSCC cells expressing EGFRvIII (Figure 3D). Similarly, EGF increased STAT3 promoter activity in vector-transfected controls, while EGF was unable to augment the already elevated levels of STAT3 promoter activation in HNSCC cells expressing EGFRvIII. These results indicate that EGFRvIII enhances STAT3 transcription and phosphorylation in HNSCC, effects that are resistant to treatment with cetuximab.
Figure 3. EGFRvIII increases STAT3 activation.
(A) After serum starvation for 24 hours, cell extracts from vector-transfected control cells (control-1), EGFRvIII-transfected cells (vIII-1 and vIII-4) were analyzed by Western blot analysis. The blot was incubated with phosphotyrosine-STAT3 antibody, stripped and probed for total STAT3 and β-actin to ensure equivalent loading. The experiment was performed 3 times with similar results. (B) Vector-transfected control cells (control-1) and EGFRvIII cells (vIII-1 and vIII-4) were transiently co-transfected with a luciferase construct under the control of STAT3 responsive promoter and a Renilla luciferase construct, incubated in complete media for 24h and assayed for luciferase activity. The fire-fly luciferase activity units (RLU) were normalized to Renilla luciferase RLU and micrograms of total protein and expressed as a fold of the activity of the control in each experiment. Cumulative results from two independent experiments indicate a significant increase in STAT3 promoter activity in HNSCC cells expressing EGFR vIII (*p=0.05). (C) HNSCC xenografts derived from EGFRvIII-expressing cells and xenografts derived from vector-transfected control cells were analyzed for phosphorylated and total STAT3 expression by immunoblotting. A representative immunoblot of pSTAT3 and STAT3 levels in control and vIII xenograft tumors are depicted. Densitometry analysis was performed on immunoblots from all tumors (n=8) and phosphorylated STAT3 expression levels relative to total STAT3 are shown as mean ± SE. EGFRvIII-expressing HNSCC cell xenograft tumor (n=8) expressed 2-fold higher levels of phosphorylated STAT3 compared to control HNSCC xenografts (n=8) (*p=0.027). (D) EGFRvIII-transfected cells (vIII-4) and vector-transfected control cells (control-1) were transiently co-transfected with a hSIE-luciferase construct and Renilla luciferase construct for 4 h. Cells were incubated in serum free media +/− EGF (10ng/ml) and/or cetuximab (C225) (0.7μg/ml ) for 24h and assayed for luciferase activity. The luciferase RLU were normalized to Renilla luciferase-RLU and to micrograms of total protein and expressed as a fold of the activity of the vector control (control-1, NoTx) in each experiment. Cumulative results are shown from two independent experiments. The hSIE promoter activity of EGFRvIII-expressing cells was not stimulated by EGF or abrogated by cetuximab treatment. In contrast, cetuximab effectively abrogated EGF induced STAT3 promoter activity in HNSCC cells (*p=0.05).
STAT3 is required for EGFRvIII-mediated motility and invasion
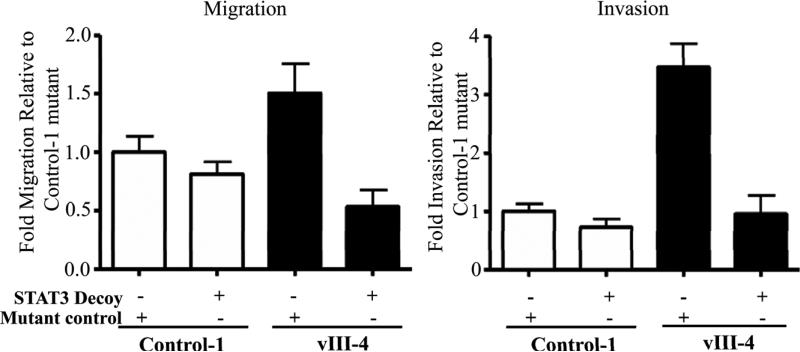
STAT3 has been implicated in several oncogenic processes including proliferation, survival, and invasion and may represent a therapeutic target for cancer (Germain and Frank, 2007). To determine whether STAT3 is required for EGFRvIII-mediated cell motility and invasion, we performed wound healing and invasion assays in the presence or absence of siRNA targeting STAT3, under conditions where siRNA did not modulate proliferation. In order to examine the phenotypic effects of EGFR vIII signaling via STAT3, we assessed cell invasion and migration in the absence of EGFR ligand. STAT3 siRNA effectively abrogated STAT3 levels in vector control and EGFR vIII expressing cells (Figure 4A). As shown in Figures 4B and 4C, knockdown of STAT3 (and phosphotyrosine STAT3) reduced the motility and invasion of EGFRvIII-expressing HNSCC cells, under conditions controlling for proliferation (p=0.03). In fact, on STAT3 knockdown, the degree of wound healing and invasion in the EGFRvIII cells were comparable to levels in vector-transfected controls. In addition to downmodulation of STAT3 expression using siRNA, we also blocked STAT3 in the cells with a transcription factor decoy directed against STAT3 as described previously (Leong et al., 2003). The STAT3 decoy interferes with STAT3-mediated DNA binding and abrogates STAT3 target gene expression. As shown in Figures 4D and 4E, treatment with the STAT3 decoy resulted in reduction of both migration and invasion in vIII-4 cells compared to treatment with a mutant control decoy, under conditions where the decoy did not affect cell growth. In the absence of EGFR ligand there was no significant reduction in the migration or invasion of control cells treated with the STAT3 decoy (p=0.19). However, the STAT3 decoy significantly abrogated the migration and invasion of EGFRvIII expressing HNSCC cells compared to the control cells (p=0.03 and p=0.048, respectively).
Figure 4. STAT3 plays a critical role in EGFRvIII-mediated migration and invasion.
(A) EGFRvIII-expressing cells (vIII-4) and vector transfected control cells (control-1) were transfected with non-targeting siRNA or STAT3 siRNA for 4 h. Untreated cells (NoTx) and transfected cells were collected at hours 48 for the analysis of total STAT3 protein levels by immunoblotting. B-tubulin levels demonstrate equal loading of protein in all lanes. The experiment was repeated 4 times with similar results. (B) STAT3 siRNA decreases HNSCC migration and invasion. Forty eight hours after non-targeting or STAT3 siRNA transfection, EGFRvIII-transfected HNSCC cells (vIII-4) and vector-transfected controls (control-1) were subjected to migration (left panel) and invasion assays (right panel) with the same methods described in materials and methods. Both control cells and EGFRvIII expressing cells decreased their migration and invasion when cells are treated with STAT3 siRNA compared to non-targeting siRNA treated condition. (C) Percent reduction of migration and invasion with STAT3 siRNA in both control-1 and vIII-4 was calculated from the results shown in panel B. The degree of both migration (left panel) and invasion (right panel) with STAT3 siRNA was greater in EGFRvIII expressing cells than in control cells (*p=0.03). (D) STAT3 decoy decreases HNSCC migration and invasion. After mutant or STAT3 decoy treatment, EGFRvIII-expressing HNSCC cells (vIII-4) and vector-transfected controls (control-1) were subjected to migration (left panel) and invasion assays (right panel). Both control cells and EGFRvIII-expressing cells demonstrated decreased migration and invasion when treated with STAT3 decoy compared to mutant control decoy treatment. (E) Percent reduction of migration and invasion with STAT3 decoy in both control-1 and vIII-4 was calculated from the results shown in Figure 4D. HNSCC cells expressing EGFR vIII had significantly reduced migration (*p=0.03) and invasion (*p=0.048) compared to control cells in the presence of STAT3 decoy.
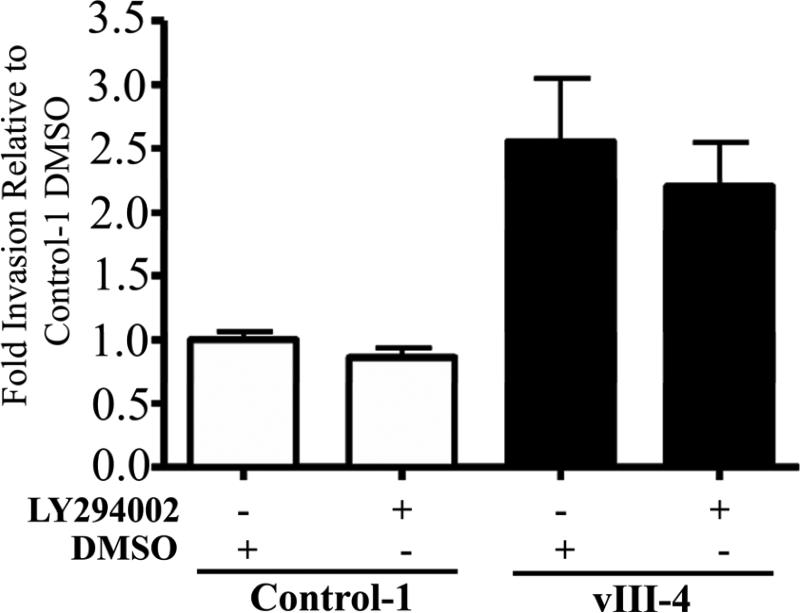
Others have reported that PI3K/AKT is activated downstream of EGFRvIII in glioma (Antonyak et al., 1998; Li et al., 2004). To determine whether motility and invasion are also mediated by this pathway, in addition to STAT3, we examined the expression of AKT phosphorylation in EGFRvIII and vector-transfected control HNSCC cells and found that similar to results in previous reports, EGFRvIII-expressing HNSCC cells expressed increased levels of pAKT (Figure 5A). However, in contrast to our observations with STAT3 targeting, blockade of PI3K/AKT using the pharmacologic inhibitor LY294002 abrogated cell growth but not invasion, in HNSCC cells expressing EGFRvIII (Figures 5B-C). Similar results were also obtained using siRNA directed against the p85 subunit of PI3K (data not shown). These results suggest that STAT3 is specifically required for the EGFRvIII-mediated enhancement of HNSCC cell motility and invasion.
Figure 5. Increased PI3K/AKT signaling in EGFRvIII-expressing HNSCC cells mediates proliferation, but not invasion.
(A) EGFRvIII-expressing HNSCC cells express increased levels of phosporylated AKT. After serum starvation for 24 hours, cell extracts from vector-transfected control cells (control-1) and EGFRvIII-transfected cells (vIII-1 and vIII-4) were analyzed by Western blot analysis. The blot was incubated with phosphoserine-AKT antibody (Ser473), stripped and immunoblotted for total AKT and β-actin to ensure equivalent loading. The experiment was performed 3 times with similar results. (B) Blockade of PI3K/AKT abrogates cell growth in HNSCC cells expressing EGFRvIII. HNSCC cells expressing EGFRvIII and vector-transfected control cells were plated onto 6 well plates with presence (◆, vIII-4), (◇, control-1) or absence (■, vIII-4), (□, control-1) of 20μM of LY294002. Growth curve of the transfected cell lines were obtained by cell counting using vital dye exclusion at several time points for 6 days. HNSCC cells expressing EGFRvIII showed increased growth rates compared with the vector- transfected control cells where cell growth was abrogated by PI3K/AKT blockade in both cell lines. Representative results from 3 independent experiments are shown. (C) Cell invasion was not abrogated by PI3K/AKT blockade. Cell invasion assay was performed with HNSCC cells expressing EGFRvIII and vector-transfected control cells with or without PI3K/AKT inhibition using 20μM of LY294002. HNSCC cells expressing EGFRvIII show increased cell invasion compared with the vector-transfected control cells but cell invasive capacity was not decreased by PI3K/AKT blockade in either cell line.
Cetuximab does not abrogate EGFRvIII-induced HIF-1α expression under hypoxic conditions
Solid tumors, including HNSCC, contain large regions of low oxygen concentrations (hypoxic) regions, which contribute to resistance to treatment with standard approaches including chemotherapy and radiation. Hypoxia potently induces expression of hypoxia inducible factor (HIF-1α), which has been shown to be a STAT3 target gene (Niu et al., 2008). EGFRvIII has been reported to contribute to hypoxia-mediated tumor growth in conjunction with radiation therapy but has not been previously linked to HIF-1α expression (Weppler et al., 2007). We therefore examined the expression of HIF-1α following treatment of HNSCC cells expressing EGFRvIII (or vector-transfected controls) with EGF and/or cetuximab. As shown in Figure 6, hypoxia-induced expression of HIF-1α was abrogated by cetuximab in vector-transfected control cells but not in HNSCC cells expressing EGFRvIII. These results suggest that STAT3 signaling via HIF-1α may contribute to cetuximab resistance in EGFRvIII-expressing HNSCC tumors under hypoxia.
Figure 6. EGFRvIII induces HIF-1α expression under hypoxic conditions, which is not abrogated by ceutximab.
EGFRvIII-expressing cells (vIII-4) and vector transfected control cells (control-1) were treated with (or without) EGF (10 ng/ml) and/or cetuximab (C225, 7 μg/ml) in under normoxic or hypoxic conditions for 24 h. Under hypoxic conditions and EGF treatment, EGFRvIII-expressing cells maintained expression of HIF-1α with cetuximab treatment whereas the expression of HIF-1α in vector-transfected control cells was significantly decreased by cetuximab (p=0.014). The experiment was performed 3 times with similar results.
Discussion
We previously reported that EGFRvIII is expressed in up to 40% of HNSCC tumors where expression of this altered receptor mediates growth and resistance to chemotherapy or EGFR targeting using cetuximab (Sok et al., 2006). Patients with HNSCC succumb to their disease due to invasion into surrounding tissues and regional and distant metastasis. Although the addition of cetuximab to radiation was shown to improve survival, it did not decrease metastasis (Bonner et al., 2006). The present study was undertaken to determine the effects of EGFRvIII on the migration and invasion of HNSCC cells and the signaling pathways that mediate these properties. Our results suggest that EGFRvIII increases HNSCC motility and invasion, at least in part, through activation of STAT3.
EGFRvIII is the most common EGFR alteration in human cancers. Deletion of exons 2-7 gives rise to a receptor that lacks a ligand binding site and is constitutively activated in a ligand-independent manner. EGFRvIII has not been observed in normal tissue, but it has been detected in carcinomas of the brain, breast, ovary (Moscatello et al., 1995), lung (Garcia de Palazzo et al., 1993), prostate (Olapade-Olaopa et al., 2000) and head and neck (Sok et al., 2006). Expression of EGFRvIII has been correlated with poor prognosis in brain tumors (Diedrich et al., 1995). Further, EGFRvIII can to transform fibroblasts in vitro (Pedersen et al., 2004) and enhance the tumorigenicity of cancer cells in vivo, supporting its oncogenic function (Tang et al., 2000).
The effect of EGFRvIII on tumor cell behavior is incompletely understood. Nagane et al. reported that EGFRvIII-expressing cells demonstrated less apoptosis in response to cisplatin treatment (Nagane et al., 1998). Others have reported that EGFRvIII induced glioma cell migration and invasion via induction of metalloproteases and extracellular matrix components (Lal et al., 2002); (Cai et al., 2005). The precise signaling events that mediate EGFRvIII-induced migration and invasion need further investigation. Moscatello et al. reported that EGFRvIII activates PI3-K pathway instead of the Ras-Raf-MEK pathway, which is preferentially activated by wild-type EGFR (Moscatello et al., 1998). Further investigation suggested that constitutive PI3-K/AKT activation by EGFRvIII may contribute to chemoresistance and radioresistance in these cells (Li et al., 2004; Narita et al., 2002). Antonyak et al. showed that c-Jun N-terminal Kinase (JNK) was constitutively activated by EGFRvIII and was down-regulated by PI3-K inhibition (Antonyak et al., 1998). To date, signaling through PI3-K/AKT has not been correlated with tumor cell migration or invasion mediated by EGFRvIII. We found that although EGFRvIII-expressing HNSCC cell expressed increased levels of phosphorylated AKT, abrogation of PI3-K/AKT using either LY294002 or p85 siRNA, did not abrogate invasion in EGFRvIII-expressing HNSCC cells. LY294002 treatment decreased the proliferation of EGFRvIII-expressing HNSCC cells, suggesting that activated PI3K/AKT by EGFRvIII contributes to EGFRvIII-induced HNSCC cell proliferation, but not invasion (Figure 5).
There are few, but conflicting, reports linking EGFRvIII to STAT3 signaling. While Mizoguchi et al. reported a correlation of expression levels of EGFRvIII and phosphotyrosine STAT3 in glioblastoma (Mizoguchi et al., 2006), Andersen et al. recently reported that glioma cells that express EGFRvIII fail to induce IRF-1 via STAT3 phosphorylation (Andersen et al., 2008). Cumulative evidence has implicated STAT3 as an critical oncogene where elevated expression levels of tyrosine phosphorylated STAT3 are detected in numerous human cancers (Bowman et al., 2000). Studies in HNSCC demonstrate that STAT3 is activated downstream of receptor and non-receptor tyrosine kinases including EGFR and Src family kinases as well as IL-6/gp130 (Kijima et al., 2002), (Xi et al., 2003), (Sriuranpong et al., 2003). Targeting STAT3 in HNSCC preclinical HNSCC models inhibited tumor growth but not the growth of normal epithelial cells (Leong et al., 2003). Expression of tyrosine phosphorylated STAT3 in the primary HNSCC tumor has been correlated with nodal metastasis, advanced tumor stage and decreased survival (Masuda et al., 2002). The results of the present study suggest that activation of STAT3 downstream of EGFRvIII in HNSCC contributes to the increased migration and invasion.
STAT3 target genes include cell cycle regulators (Sinibaldi et al., 2000), anti-apoptotic genes (Oritani et al., 1999) and pro-angiogenic factors (Huang et al., 2002), each of which has been implicated in tumorigenic processes including invasion and metastasis. STAT3 has been shown to contribute to cancer migration and invasion thorough regulation of genes that stimulate these processes including matrix metalloproteinases (e.g. MMP-2 and MMP-9), VEGF and/or bFGF (Dechow et al., 2004); (Qiu et al., 2007). In addition to the transcriptionally mediated effects of STAT3 on cell migration and invasion, transcription-independent pathways have also been described for the effects of STAT3 on cell motility. Specifically, STAT3 has been found to directly interact with cell motility components such as focal adhesion components, FAK, paxillin (Silver et al., 2004), p130CAS (Kira et al., 2002), or cytoskeltal microtubles (Ng et al., 2006). We did not detect increased expression of MMP-2, MMP-9 or VEGF in association with the EGFRvIII-mediated migration and invasion observed in these cells (data not shown), suggesting that other STAT3 target genes or STAT3 interacting proteins may be playing a role. Additionally, the lack of HIF-1α decrease in EGFRvIII cells treated with cetuximab under hypoxic conditions suggests a more complex regulatory balance between oxygen-dependent and –independent factors that influence HIF-1α.
Aberrant activation of STAT3 has been shown to contribute to tumor progression making STAT3 an attractive therapeutic target. To date, no STAT3 targeting strategies have undergone clinical evaluation. We have developed a highly specific transcription factor decoy approach to block STAT3 signaling and demonstrated that it inhibits tumor growth in vitro and in vivo in HNSCC preclinical models (Leong et al., 2003). Using the same STAT3 decoy, others have reported antitumor effects in a murine model of cutaneous squamous cell carcinoma (Sano et al., 2005). Toxicology studies in non-human primates were recently completed and demonstrated no evidence of toxicity (Sen et al., 2008). In the present study, treatment of EGFRvIII-expressing HNSCC cells with the STAT3 decoy abrogated cell motility and invasion. These results suggest that selective activation by EGFRvIII in HNSCC contributes to the invasive phenotype, which can potentially be targeted with therapeutic strategies that inhibit STAT3. In addition to the decoy, others have reported the use of siRNA, peptidomimetic strategies, and G-quartet oligonucleotides to inhibit STAT3 in cancer models (Leeman et al., 2006).
These cumulative results suggest that STAT3 activation is critical for cancer progression mediated by both wild-type EGFR and EGFRvIII, which are often co-expressed in HNSCC tumors (Sok et al., 2006). Knockdown or blockade of STAT3 preferentially abrogated the migration and invasion of HNSCC cells that expressed EGFRvIII implicating STAT3 as a critical pathway in mediating HNSCC invasion in tumors that express this altered receptor. HNSCC cells co-express the wild-type and mutant EGFRvIII receptors with an estimated 1.6 × 105 wild-type receptors (Supplemental Figure 1) and ~104 mutant EGFRvIII receptors per cell. Thus, in the presence of EGF, EGFRvIII-expressing HNSCC cells invade despite STAT3 knockdown (Supplemental Figure 3). We have previously demonstrated that both PLCγ-1 and c-Src mediate HNSCC invasion downstream of EGFR (Thomas 2003, Zhang 2004). Thus EGF stimulation in cells expressing both EGFR and EGFR vIII likely results in invasion via multiple downstream signaling molecules including STAT3.
Therapeutic agents with selective activity against EGFRvIII are presently under clinical investigation including the immunotoxin MR1-1, the chimeric antibody 806 (Li et al., 2007) and irreversible HER1/HER2 inhibitors (Ji et al., 2006) that appear to have selective activity against EGFRvIII. The EGFR monoclonal antibody cetuximab is the only FDA-approved EGFR targeting strategy for HNSCC. We previously reported that HNSCC xenografts expressing EGFRvIII were resistant to the growth inhibitory effects of cetuximab (Sok et al., 2006). Here we demonstrate that EGFRvIII cells are resistant to anti-invasive effects of cetuximab in HNSCC. Further, EGFRvIII expression results in an increase in phosphorylation of STAT3 in HNSCC cells. These results suggest that HNSCC tumors that express EGFRvIII may be best treated with strategies that selectively block EGFRvIII, or its downstream signaling pathways, in addition to targeting wild-type EGFR.
Materials and Methods
Cell lines, reagents, and cell culture
C225/cetuximab was purchased from the research pharmacy at the University of Pittsburgh Cancer Institute. For in vitro cell stimulation, recombinant human EGF (Sigma Chemical Co.) was used. Phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 was obtained from Calbiochem (San Diego, CA). NR6 (Swiss 3T3 murine fibroblasts) cells expressing EGFRwt (NR6W) were a generous gift from Dr. Alan Wells (University of Pittsburgh School of Medicine). NR6 cells expressing human EGFRvIII (NR6M) were generated as described previously (Batra et al., 1995). EGFRvIII-transfected HNSCC cells (vIII-1 to 4) and vector control transfected HNSCC cells (control-1 to 4) were generated as descried previously (Sok et al., 2006). All cells were maintained in DMEM (Mediatech, Inc., Herndon, VA) with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA) and incubated at 37°C in the presence of 5% CO2. To establish hypoxic conditions (1% O2), cells were placed in an InVivo300 hypoxia workstation (Ruskinn Life Sciences Ltd, UK).
RT-PCR analysis and cDNA sequencing of EGFRvIII
Total RNA was isolated from HNSCC cell lines (5 × 106 cells) using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Total RNA (1 μg) was reverse-transcribed with the first-strand cDNA synthesis using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). To detect the deleted region of EGFRvIII, standard RT-PCR was performed as described previously (Sok et al., 2006).
Flow cytometry
Indirect analytic flow cytometry was done on a Becton Dickinson FACS calibur equipped with CellQuest Pro software (Becton Dickinson, San Jose, CA ). Assays were done at 4°C; all washes were done with iced medium to facilitate the detection of cell surface receptors without allowing internalization to occur. All profiles were obtained with cells maintained in ice-cold 1% bovine serum albumin/PBS. The percentage of a population designated as positive was arbitrarily defined as that region in which only the highest fluorescing 10% of the isotype-control stained cells graphed, corrected for background; this is a conservative estimate of the total positive staining population. In order to examine the cell surface expression of EGFRvIII proteins, target cultured cells were stained with anti-EGFRvIII monoclonal antibody L8A4 under nonpermeabilized conditions. Subconfluent cells were detached from culture flasks by incubation with 0.02% EDTA/PBS; 106 cells were maintained in 0.5% paraformaldehyde/ PBS for 10 minutes at 4°C, washed, resuspended in 150 mL PBS containing 10% fetal bovine serum, and blocked for 20 minutes at 4°C. After two washes, the samples were reacted with L8A4 monoclonal antibody (10 mg/mL, black line) and irrelevant mouse IgG1k (10 mg/mL, solid gray) in PBS for 60 min. After two additional washes, cells were incubated with FITC-labeled secondary antibody for 30 minutes at 4°C and analyzed on a Becton Dickinson FACS calibur instrument (Becton Dickinson).
Western blotting
Cell lines were lysed in detergent containing 1% NP40, 0.1 mM phenylmethylsulfonyl fluoride, 1 mg/ml leupeptin, and 1 mg/ml aprotinin, and protein levels were determined using the Bio-Rad protein assay method (Bio-Rad Laboratories, Hercules, CA). Forty μg of total protein were separated on 8% SDS-PAGE gels and transferred to nitrocellulose membranes using the semi-dry transfer machine (BioRad Laboratories, Hercules, CA). Membranes were blocked with 5% skim milk/Tris-buffered saline with Tween 20 () solution for 2 hour at room temperature, and incubated with primary antibodies in 5% skim milk in TBS-T overnight at 4°C. After washing with TBS-T three times, membranes were incubated for 1 hour with HRP-conjugated secondary antibody (Bio-Rad Laboratories, Hercules, CA) 1:3000 diluted in 5% skim milk in TBS-T. The filters were rinsed with TBS-T three times, and the blot was developed using Luminol Regent (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) by autoradiography. Antibodies used for blotting included β-actin (Oncogene Research Products, Boston, MA), β-tubulin (Abcam, Cambridge, UK), HIF-1α (BD Transduction Laboratories, San Jose, CA), phospho-AKT (Ser473), AKT, phospho-STAT3 (Tyr705) and STAT3 (Cell Signaling Technology, Beverly, MA).
Luciferase reporter assay
HNSCC cells (4 × 105/ml) were plated onto 6-well tissue culture plates. After sixteen hours incubation, cells were transiently co-transfected with pSTAT3TALuc, a generous gift from Dr. Jacqueline Bromberg (Memorial Sloan-Kettering Cancer Center, New York, NY) and pRL-TK (Promega, Madison, WI) a Renilla luciferase construct, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions (Besser et al., 1999). The transfection media was replaced to complete DMEM after 4 h of transfection. Cells were lysed and luciferase assays were performed 24h after the transfection using the Dual-Luciferase Reporter Assay System kit (Promega, Madison, WI). Cell lysates were subjected to protein estimation using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA). Relative light units (RLU) from luciferase was normalized to RLU from renilla luciferase (to account for differences in transfection efficiency) and to micrograms of protein (to account for differences in protein concentrations between samples).
Matrigel invasion assay
Cell invasion was evaluated in vitro using Matrigel-coated semi-permeable modified Boyden inserts with a pore size of 8 μm (Becton Dickinson/Biocoat, Bedford, MA). Cells were plated in triplicate at a density of 5 × 103 cells per well in DMEM in the insert. At the same time cells were plated in 96-well plates to serve as loading controls. Both the insert and the holding well were subjected to the same medium composition with the exception of serum. The insert contained no serum, whereas the lower well contained 10% fetal bovine serum (FBS) that served as a chemo-attractant. After 24 h of treatment at 37°C in a 5% CO2 incubator, the cells in the insert were removed by wiping gently with a cotton swab. Cells on the reverse side of the insert were fixed and stained with Hema 3 (Fisher Scientific, Hampton, NH) according to the manufacturer's instructions. Cells plated in 96-well plates were subjected to MTT assays and the cell numbers across the groups were normalized. The number of invading cells was adjusted accordingly.
Wound healing assay
HNSCC cells were grown to confluence on 6-well tissue culture dishes and a single scrape was made in the confluent monolayer using a sterile pipette tip. The monolayer was washed with PBS and complete medium was added. Photographs were taken at 0 and 6 h, and the relative denuded area at the cellular front was determined by computer-assisted image analysis; markings on the plate ensured measurement of the same site for the photographs. The decreased area was then expressed as a percentage within each experiment, allowing direct comparisons between experiments.
STAT3 siRNA and STAT3 decoy transfection
The STAT3 decoy and the mutant control decoy sequences (double-stranded deoxyribonucleotides with phosphorothioate modifications in the first three bases and last three bases of the sequences) were generated as described previously (Leong et al., 2003). The mutant control decoy, carrying a single base mutation, that does not abrogate STAT3 DNA binding activity, was used as a control as in previous studies (Leong et al., 2003; Xi et al., 2005). The siRNA sequences targeting STAT3 human mRNA (D-003544-01, sense 5’-CCAACGACCUGCAGCAAUAUU-3’, and antisense 5’-PUAUUGCUGCAGGUCGUUGGUU-3’; Dharmacon, Lafayette, CO) were transfected into HNSCC cells for STAT3 silencing. The nontargeting siRNA (D-001210-01, sense 5'-UAGCGACUAAACACAUCAAUU-3', antisense 5-UUGAUGUGUUUAGUCGCUAUU-3'; Dharmacon) was used as a control. The siRNA or decoy transfections were performed using the Lipofectamine 2000 (Life Technologies Inc). In brief, HNSCC cells were plated (1.8 × 105/well for siRNA or 1.0 × 106/well for decoy transfection in a six-well tissue culture plate). Sixteen hours after plating, cells were transfected with 200 pmol of STAT3 siRNA or non-targeting control siRNA, or 12.6 pmol of STAT3 decoy or mutant control decoy. The transfection medium was replaced with complete DMEM after 4 h of transfection.
In vitro growth assay
To determine if the sensitivity of HNSCC cells to PI3K/AKT inhibition was affected by EGFRvIII expression, vector-transfected and EGFRvIII-expressing HNSCC cells (3 × 104) were seeded onto 6 well plates and treated with a PI3K inhibitor LY294002. Each cell population was then harvested in triplicate, transferred to a hemocytometer, and counted every other day over a period of 6 days.
In vivo growth of HNSCC cell expressing EGFRvIII and analysis of HNSCC xenografts
HNSCC cells [UM-22B-expressing EGFRvIII (vIII-4) or empty vector-transfected parental cells (control-1)] were cultured in DMEM containing 10% fetal bovine serum. Cells were trypsinized and washed 3 times with HBSS (Life Technologies, Carlsbad, CA). Cell number and viability were determined using trypan blue dye exclusion. A suspension of 3 × 106 HNSCC cells in 50 μL HBSS was injected into floor of mouth of nu/nu athymic nude mice (n = 8 per cell line; Harlan Sprague-Dawley, Indianapolis, IN) transcutaneously. Tumor volumes were measured in two dimensions with vernier calipers and calculated using the formula: (length × width2) × 0.52. At the end of the study, mice were sacrificed by cervical dislocation under anesthesia the tumors surgically excised and snap frozen in dry ice. To evaluate the expression of phosphorylated and total STAT3 in HNSCC xenografts, tumors homogenized, sonicated in detergent containing 1% NP40, 0.1 mM phenylmethylsulfonyl fluoride, 1 mg/ml leupeptin, and 1 mg/ml aprotinin. Forty μg of total protein were separated on 8% SDS-PAGE gels and immunoblotted for phosphorylated and total STAT3 and β-tubulin. Animal use and care was in strict compliance with institutional guidelines established by the Institutional Animal Care and Use Committee at the University of Pittsburgh.
Statistical analysis
For wound healing and invasion studies, the statistical significance of differences in the number of invading cells or migrated area were assessed by use of Wilcoxon-Mann-Whitney two-tailed exact test.
Supplementary Material
Supplemental methods
Quantitative flow cytometry
QFACS procedures and labeling of monoclonal antibodies have been previously described (Wikstrand et al., 1997) using the Quantum Simply Cellular kit from Bangs Laboratories (Fishers, IN). Appropriate isotype control monoclonal antibody, murine IgG2b grown and purified in house was labeled and used under identical conditions as the specific EGFR antibody. Anti-EGFR (wild type) monoclonal antibody EGFR1 (IgG2b) specific for the extracellular domain of wild-type EGFR was from BD Biosciences (San Jose, CA). Cells for analysis were grown in Richter's Zinc Option MEM plus 10% FBS (Invitrogen, Carlsbad, CA), harvested and washed in medium plus 5% FBS. Five micrograms of EGFR antibody or control immunoglobulin (IgG2b) were added to 1 × 106 cells resuspended in 0.1 ml of medium with 5% FBS. Cells were incubated for 1 hour on ice and then washed twice with medium plus 5% FBS. Live cells were suspended in PBS and were analyzed by using a Becton Dickinson FACSort flow cytometer with CellQuest software (Becton Dickinson, San Jose, CA).
Transient transfection of HNSCC cells with EGFR vIII
HNSCC cell lines 1483 and PCI-37A were transiently transfected using Lipofectamine 2000 (Invitrogen; Carlsbad,CA) with the pLERN vector control or pLERN EGFRvIII construct. Cells were allowed to grow in serum containing medium overnight.
Supplemental Figure 1: UM-22B cells express 1.61 × 105 EGFR per cell. UM-22B cells were analyzed by quantitative flow cytometry (QFC). Monoclonal antibody to EGFR was used to determine levels of wild-type EGFR. Background binding was determined with fluoresceinated irrelevant isotype control (IgG2b).
Supplemental Figure 2: EGFR vIII expression increase cellular migration and pSTAT3 levels. A) After 24 hours of serum starvation cells were plated in BD Biocoat control inserts (BD Biosciences; Bedford, MA) in triplicate in serum free DMEM (1.2×104 cells per insert) with 10% FBS media in the well. The cells were incubated for 24 hours and then stained following manufacturer's protocol and 6 fields counted and averaged. HNSCC cells transiently transfected with EGFRvIII have increased migration compared to vector controls. B) After 24 hours of serum starvation, transiently transfected cells were lysed and the protein fractionated on an 8% PAGE gel. HNSCC cells transiently transfected with EGFRvIII have higher levels of phosphorylated STAT3 compared to the vector control cells.
Supplemental Figure 3: Inhibition of STAT3 does not abrogate EGF-induced invasion in EGFR vIII expressing HNSCC cells. UM-22B cells stably expressing EGFRvIII or vector control where transfected with STAT3 siRNA or non-targeting siRNA and stimulated with EGF (10 ng/ml) or vehicle. STAT3 downregulation did not abrogate EGF-stimulated invasion in vector control or EGFRvIII transfected cells.
Acknowledgement
This work was supported by grants RO1 CA77308, RO1 CA101840, P50 CA097190 and an American Cancer Society Clinical Research professorship CRP-08-229-01 (to JRG) and the NIH core grant EY08098.
References
- Andersen P, Pedersen MW, Woetmann A, Villingshoj M, Stockhausen MT, Odum N, et al. EGFR induces expression of IRF-1 via STAT1 and STAT3 activation leading to growth arrest of human cancer cells. Int J Cancer. 2008;122:342–9. doi: 10.1002/ijc.23109. [DOI] [PubMed] [Google Scholar]
- Antonyak MA, Moscatello DK, Wong AJ. Constitutive activation of c-Jun N-terminal kinase by a mutant epidermal growth factor receptor. J Biol Chem. 1998;273:2817–22. doi: 10.1074/jbc.273.5.2817. [DOI] [PubMed] [Google Scholar]
- Batra SK, Castelino-Prabhu S, Wikstrand CJ, Zhu X, Humphrey PA, Friedman HS, et al. Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell Growth Differ. 1995;6:1251–9. [PubMed] [Google Scholar]
- Besser D, Bromberg JF, Darnell JE, Jr., Hanafusa H. A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation. Mol Cell Biol. 1999;19:1401–9. doi: 10.1128/mcb.19.2.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigner SH, Humphrey PA, Wong AJ, Vogelstein B, Mark J, Friedman HS, et al. Characterization of the epidermal growth factor receptor in human glioma cell lines and xenografts. Cancer Res. 1990;50:8017–22. [PubMed] [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- Cai XM, Tao BB, Wang LY, Liang YL, Jin JW, Yang Y, et al. Protein phosphatase activity of PTEN inhibited the invasion of glioma cells with epidermal growth factor receptor mutation type III expression. Int J Cancer. 2005;117:905–12. doi: 10.1002/ijc.21251. [DOI] [PubMed] [Google Scholar]
- Dechow TN, Pedranzini L, Leitch A, Leslie K, Gerald WL, Linkov I, et al. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc Natl Acad Sci U S A. 2004;101:10602–7. doi: 10.1073/pnas.0404100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diedrich U, Lucius J, Baron E, Behnke J, Pabst B, Zoll B. Distribution of epidermal growth factor receptor gene amplification in brain tumours and correlation to prognosis. J Neurol. 1995;242:683–8. doi: 10.1007/BF00866920. [DOI] [PubMed] [Google Scholar]
- Garcia de Palazzo IE, Adams GP, Sundareshan P, Wong AJ, Testa JR, Bigner DD, et al. Expression of mutated epidermal growth factor receptor by non-small cell lung carcinomas. Cancer Res. 1993;53:3217–20. [PubMed] [Google Scholar]
- Germain D, Frank DA. Targeting the cytoplasmic and nuclear functions of signal transducers and activators of transcription 3 for cancer therapy. Clin Cancer Res. 2007;13:5665–9. doi: 10.1158/1078-0432.CCR-06-2491. [DOI] [PubMed] [Google Scholar]
- Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, et al. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor- mediated cell growth in vitro. J Clin Invest. 1998;102:1385–92. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandis JR, Tweardy DJ. TGF-alpha and EGFR in head and neck cancer. J Cell Biochem Suppl. 1993:188–91. doi: 10.1002/jcb.240531027. [DOI] [PubMed] [Google Scholar]
- Huang M, Dorsey JF, Epling-Burnette PK, Nimmanapalli R, Landowski TH, Mora LB, et al. Inhibition of Bcr-Abl kinase activity by PD180970 blocks constitutive activation of Stat5 and growth of CML cells. Oncogene. 2002;21:8804–16. doi: 10.1038/sj.onc.1206028. [DOI] [PubMed] [Google Scholar]
- Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006;103:7817–22. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima T, Niwa H, Steinman RA, Drenning SD, Gooding WE, Wentzel AL, et al. Stat3-mediated EGFR-independent growth in SCCHN. Cell Growth Differ. 2002;13:355–362. [PubMed] [Google Scholar]
- Kira M, Sano S, Takagi S, Yoshikawa K, Takeda J, Itami S. STAT3 deficiency in keratinocytes leads to compromised cell migration through hyperphosphorylation of p130(cas). J Biol Chem. 2002;277:12931–6. doi: 10.1074/jbc.M110795200. [DOI] [PubMed] [Google Scholar]
- Lal A, Glazer CA, Martinson HM, Friedman HS, Archer GE, Sampson JH, et al. Mutant epidermal growth factor receptor up-regulates molecular effectors of tumor invasion. Cancer Res. 2002;62:3335–9. [PubMed] [Google Scholar]
- Leeman RJ, Lui VW, Grandis JR. STAT3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6:231–41. doi: 10.1517/14712598.6.3.231. [DOI] [PubMed] [Google Scholar]
- Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci U S A. 2003;100:4138–43. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Yuan M, Kim IA, Chang CM, Bernhard EJ, Shu HK. Mutant epidermal growth factor receptor displays increased signaling through the phosphatidylinositol-3 kinase/AKT pathway and promotes radioresistance in cells of astrocytic origin. Oncogene. 2004;23:4594–602. doi: 10.1038/sj.onc.1207602. [DOI] [PubMed] [Google Scholar]
- Li D, Ji H, Zaghlul S, McNamara K, Liang MC, Shimamura T, et al. Therapeutic anti-EGFR antibody 806 generates responses in murine de novo EGFR mutant-dependent lung carcinomas. J Clin Invest. 2007;117:346–52. doi: 10.1172/JCI30446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, et al. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–5. [PubMed] [Google Scholar]
- Mizoguchi M, Betensky RA, Batchelor TT, Bernay DC, Louis DN, Nutt CL. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol. 2006;65:1181–8. doi: 10.1097/01.jnen.0000248549.14962.b2. [DOI] [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J Biol Chem. 1998;273:200–6. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, Zoltick PW, et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55:5536–9. [PubMed] [Google Scholar]
- Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci U S A. 1998;95:5724–9. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK. Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Cancer Res. 2002;62:6764–9. [PubMed] [Google Scholar]
- Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, et al. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol. 2006;172:245–57. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–31. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu G, Briggs J, Deng J, Ma Y, Lee H, Kortylewski M, et al. Signal transducer and activator of transcription 3 is required for hypoxia-inducible factor-1alpha RNA expression in both tumor cells and tumor-associated myeloid cells. Mol Cancer Res. 2008;6:1099–105. doi: 10.1158/1541-7786.MCR-07-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olapade-Olaopa EO, Moscatello DK, MacKay EH, Horsburgh T, Sandhu DP, Terry TR, et al. Evidence for the differential expression of a variant EGF receptor protein in human prostate cancer. Br J Cancer. 2000;82:186–94. doi: 10.1054/bjoc.1999.0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oritani K, Tomiyama Y, Kincade PW, Aoyama K, Yokota T, Matsumura I, et al. Both Stat3-activation and Stat3-independent BCL2 downregulation are important for interleukin-6-induced apoptosis of 1A9-M cells. Blood. 1999;93:1346–54. [PubMed] [Google Scholar]
- Pedersen MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001;12:745–60. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- Pedersen MW, Tkach V, Pedersen N, Berezin V, Poulsen HS. Expression of a naturally occurring constitutively active variant of the epidermal growth factor receptor in mouse fibroblasts increases motility. Int J Cancer. 2004;108:643–53. doi: 10.1002/ijc.11566. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Huang C, Sun J, Qiu W, Zhang J, Li H, et al. RNA interference-mediated signal transducers and activators of transcription 3 gene silencing inhibits invasion and metastasis of human pancreatic cancer cells. Cancer Sci. 2007;98:1099–106. doi: 10.1111/j.1349-7006.2007.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano S, Chan KS, Carbajal S, Clifford J, Peavey M, Kiguchi K, et al. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat Med. 2005;11:43–9. doi: 10.1038/nm1162. [DOI] [PubMed] [Google Scholar]
- Sen M, Tosca PJ, Zwayer C, Ryan MJ, Johnson JD, Knostman KA, et al. Lack of toxicity of a STAT3 decoy oligonucleotide. Cancer Chemother Pharmacol. 2008 doi: 10.1007/s00280-008-0823-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–8. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- Sinibaldi D, Wharton W, Turkson J, Bowman T, Pledger WJ, Jove R. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene. 2000;19:5419–27. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, Hunt JL, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12:5064–73. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res. 2003;63:2948–56. [PubMed] [Google Scholar]
- Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87:8602–6. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Shou M, Mei Q, Rushmore TH, Rodrigues AD. Major role of human liver microsomal cytochrome P450 2C9 (CYP2C9) in the oxidative metabolism of celecoxib, a novel cyclooxygenase-II inhibitor. J Pharmacol Exp Ther. 2000;293:453–9. [PubMed] [Google Scholar]
- Vermorken JB, Trigo J, Hitt R, Koralewski P, Diaz-Rubio E, Rolland F, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. J Clin Oncol. 2007;25:2171–7. doi: 10.1200/JCO.2006.06.7447. [DOI] [PubMed] [Google Scholar]
- Weppler SA, Li Y, Dubois L, Lieuwes N, Jutten B, Lambin P, et al. Expression of EGFR variant vIII promotes both radiation resistance and hypoxia tolerance. Radiother Oncol. 2007;83:333–9. doi: 10.1016/j.radonc.2007.04.025. [DOI] [PubMed] [Google Scholar]
- Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer Res. 1997;57:4130–40. [PubMed] [Google Scholar]
- Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications for cancer therapy. Oncogene. 2005;24:970–9. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- Xi S, Zhang Q, Dyer KF, Lerner EC, Smithgall TE, Gooding WE, et al. Src kinases mediate STAT growth pathways in squamous cell carcinoma of the head and neck. J Biol Chem. 2003;278:31574–83. doi: 10.1074/jbc.M303499200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods
Quantitative flow cytometry
QFACS procedures and labeling of monoclonal antibodies have been previously described (Wikstrand et al., 1997) using the Quantum Simply Cellular kit from Bangs Laboratories (Fishers, IN). Appropriate isotype control monoclonal antibody, murine IgG2b grown and purified in house was labeled and used under identical conditions as the specific EGFR antibody. Anti-EGFR (wild type) monoclonal antibody EGFR1 (IgG2b) specific for the extracellular domain of wild-type EGFR was from BD Biosciences (San Jose, CA). Cells for analysis were grown in Richter's Zinc Option MEM plus 10% FBS (Invitrogen, Carlsbad, CA), harvested and washed in medium plus 5% FBS. Five micrograms of EGFR antibody or control immunoglobulin (IgG2b) were added to 1 × 106 cells resuspended in 0.1 ml of medium with 5% FBS. Cells were incubated for 1 hour on ice and then washed twice with medium plus 5% FBS. Live cells were suspended in PBS and were analyzed by using a Becton Dickinson FACSort flow cytometer with CellQuest software (Becton Dickinson, San Jose, CA).
Transient transfection of HNSCC cells with EGFR vIII
HNSCC cell lines 1483 and PCI-37A were transiently transfected using Lipofectamine 2000 (Invitrogen; Carlsbad,CA) with the pLERN vector control or pLERN EGFRvIII construct. Cells were allowed to grow in serum containing medium overnight.
Supplemental Figure 1: UM-22B cells express 1.61 × 105 EGFR per cell. UM-22B cells were analyzed by quantitative flow cytometry (QFC). Monoclonal antibody to EGFR was used to determine levels of wild-type EGFR. Background binding was determined with fluoresceinated irrelevant isotype control (IgG2b).
Supplemental Figure 2: EGFR vIII expression increase cellular migration and pSTAT3 levels. A) After 24 hours of serum starvation cells were plated in BD Biocoat control inserts (BD Biosciences; Bedford, MA) in triplicate in serum free DMEM (1.2×104 cells per insert) with 10% FBS media in the well. The cells were incubated for 24 hours and then stained following manufacturer's protocol and 6 fields counted and averaged. HNSCC cells transiently transfected with EGFRvIII have increased migration compared to vector controls. B) After 24 hours of serum starvation, transiently transfected cells were lysed and the protein fractionated on an 8% PAGE gel. HNSCC cells transiently transfected with EGFRvIII have higher levels of phosphorylated STAT3 compared to the vector control cells.
Supplemental Figure 3: Inhibition of STAT3 does not abrogate EGF-induced invasion in EGFR vIII expressing HNSCC cells. UM-22B cells stably expressing EGFRvIII or vector control where transfected with STAT3 siRNA or non-targeting siRNA and stimulated with EGF (10 ng/ml) or vehicle. STAT3 downregulation did not abrogate EGF-stimulated invasion in vector control or EGFRvIII transfected cells.