Abstract
Tumor cells are often deficient in DNA repair and damage response (DDR) pathways, and anticancer therapies are commonly based on genotoxic treatments using radiation and/or drugs that damage DNA directly or interfere with DNA metabolism leading to the formation of DNA double-strand breaks (DSBs), and ultimately to cell death. Since DSBs induce the phosphorylation of histone H2AX (γH2AX) in the chromatin flanking the break site, an antibody directed against γH2AX can be employed to measure DNA damage levels before and after patient treatment. Poly(ADP-ribose) polymerases (PARP1 and PARP2) are also activated by DNA damage and PARP inhibitors show promising activity in cancers with defective homologous recombination (HR) pathways for DSB repair. Ongoing clinical trials are testing combinations of PARP inhibitors with DNA damaging agents. Poly(ADP-ribosylation) (PAR) can be measured in clinical samples and used to determine the efficiency of PARP inhibitors. This review summarizes the roles of γH2AX and PAR in the DDR and their use as biomarkers to monitor drug response and guide clinical trials, especially Phase 0 clinical trials. We will also discuss the choices of relevant samples for γH2AX and PAR analyses.
Keywords: γH2AX, pharmacodynamics, indenoisoquinolines, poly(ADP-ribose), PARP, DNA repair
Background
There is a critical need in the field of cancer treatment to accelerate the validation of candidate drugs while ultimately reducing costs. One approach promising to improve the efficiency and speed of clinical trials utilizes biological markers to measure pharmacodynamic parameters in samples taken from the cancer patients themselves during drug protocols (1). Because many anticancer agents target DNA and the DDR pathways, biomarkers based on DNA damage endpoints. Biomarkers may also be useful for identifying individuals hypersensitive to radiotherapy in order to tailor treatments to minimize undesired side effects. The most desirable biomarkers would be those able to determine early in the course of treatment whether a drug is reaching its target and acting as intended. The last few years has seen the development of new biomarkers for clinical trials (2), especially in the context of Phase 0 trials (1). Here, we discuss two pharmacodynamic biomarkers, γH2AX and PAR, currently being developed at the NIH medical center (Bethesda, MD, USA) by the National Cancer Institute.
γH2AX
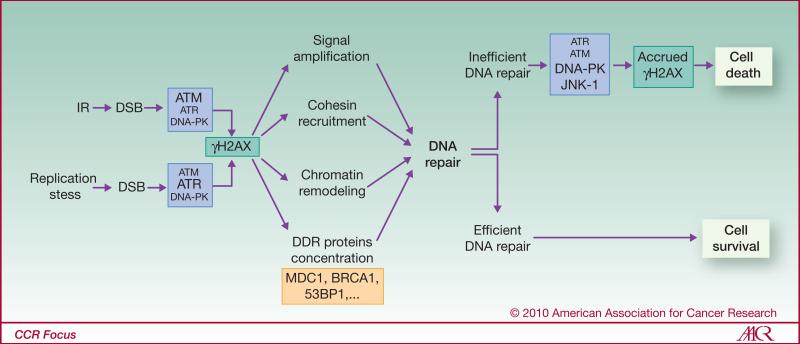
H2AX belongs to the H2A family of histones, one of four families present in the nucleosomes that package eukaryotic DNA into chromatin (3). Upon DSB formation, hundreds of H2AX molecules in the chromatin are rapidly phosphorylated by members of the phosphatidylinositol-3 kinase (PI3K) family (4, 5) forming a focus at the DSB site (see Fig. 1 for details). H2AX-deficient cells exhibit cell cycle checkpoint deficiency, increased genomic instability and sensitivity to genotoxic agents (6), observations corroborating findings of roles for γH2AX in multiple processes necessary to return the broken chromatin to its original state (see Fig. 1 for details). After completion of DNA repair and chromatin remodeling, dephosphorylation of γH2AX is carried out by several phosphatases including the p53-inducible phosphatase Wip1 (7, 8), protein phosphatases 6 (9), 4 (10) and 2A (11).
Figure 1. γH2AX following DSB formation.
H2AX is phosphorylated by members of the phosphatidylinositol-3 kinase (PI3K) family; which one is involved depends on the type of genotoxic stress (4). While ATM and ATR are primarily involved in H2AX phosphorylation following IR and replication stress respectively, DNA-PK and JNK-1 were shown to be responsible for γH2AX formation during apoptosis (97, 98). γH2AX foci are known to be involved in the recruitment and stabilization of DDR proteins including Mre11, Rad50, Nbs1 (the MRN complex), MDC1, 53BP1, BRCA1, ATM, and RNF8 (6, 99-101). DSB repair is performed by the homologous recombination (HR) and non-homologous end joining (NHEJ) pathways. HR, driven by the BRCA2, RAD51 and RAD52/54 genes, is the more accurate because it utilizes a homologous DNA segment to act as a template for the damaged DNA region. Repair is also performed by sister chromatid-dependent recombination repair via cohesin recruitment (102, 103). In contrast, NHEJ is faster, does not require a homologous DNA segment, and can operate in non-replicating cells. However, it is error-prone. The classical effectors of NHEJ are the end-binding proteins Ku70/80, DNA-dependent protein kinase (DNA-PKcs), the nuclease Artemis, the scaffolding protein XRCC4 and ligase IV. Recently, a slow DNA-PK-independent NHEJ pathway involving PARP1, histone H1, XRCC1 and ligase III has been proposed (24, 25). The γH2AX foci are also involved in chromatin alteration via recruitment of remodeling complexes and in signal transduction (accrued ATM activation, G2/M cell cycle checkpoint). In addition, γH2AX foci, through their recruitment of the cohesions and the MRN complexes, are involved in binding and tethering the broken DNA ends may help prevent the dissociation of the broken chromosome ends (104).
Since its discovery in 1998 at the NCI (5), γH2AX has been a topic of basic research analyzing the DDR and of translational studies as a biodosimeter to measure the genotoxic effects of drugs and/or radiation exposure (4, 5). γH2AX foci are found in almost all cell types after exposure to agents that directly induce DSBs--radiation therapy, and classical chemotherapy with DNA alkylating and radiomimetic agents. Importantly however, treatment with inhibitors of DNA topoisomerases and replication also generate a variety of DNA lesions that may result in the formation of DSBs (12), primarily in S-phase cells. It is important to note that γH2AX foci, usually in small numbers, may be present in cells even in the absence of exogenous damage (4) due to DNA damage that can occur during many common cellular processes, including replication, senescence, viral infection, exposure to endogenous reactive oxygen species (ROS), and carcinogenic adducts. DSBs may form during these processes or during DNA repair, producing genomic instability and increasing cancer risk, as exemplified by the increased cancer incidence in individuals with genetic defects in DNA repair genes, such as BRCA1, BRCA2, the FANC genes, ATM and CHEK2.
Poly(ADP-ribose) polymerases (PARPs) and Poly(ADP-ribose) polymers (PAR)
Poly (ADP-ribose) polymerases (PARPs) belong to a family of 17 structurally related mammalian enzymes [reviewed in (13-16)]. Only three PARPs have demonstrable poly(ADP-ribose) polymerase activity: PARP1, PARP2 (PARP1/2) and tankyrase 1. Other proteins containing all the structural features of functional PARP have been identified (i.e. PARP3, PARP4) but their ability to form poly(ADP-ribose) (PAR) chains still has to be demonstrated. PARP1 is the most abundant PARP and is normally associated with chromatin. Binding of non-PARylated PARP1 to chromatin increases chromatin compaction (14). PARP1 activation and PARylation are associated with transcription activation, in some cases in association with DNA cleavage by topoisomerase II beta (17, 18). PARP1/2 are both strongly activated by DNA damage including DNA single- and double-strand breaks. The N-terminal region of PARP1 contains zinc binding domains that bind DNA (13-15). PARP1/2 complement each other. Single PARP1 or PARP2 knockout mice are viable though hypersensitive to radiation and DNA damaging drugs, whereas the double PARP1/2 knockout is an early embryonic lethal (15).
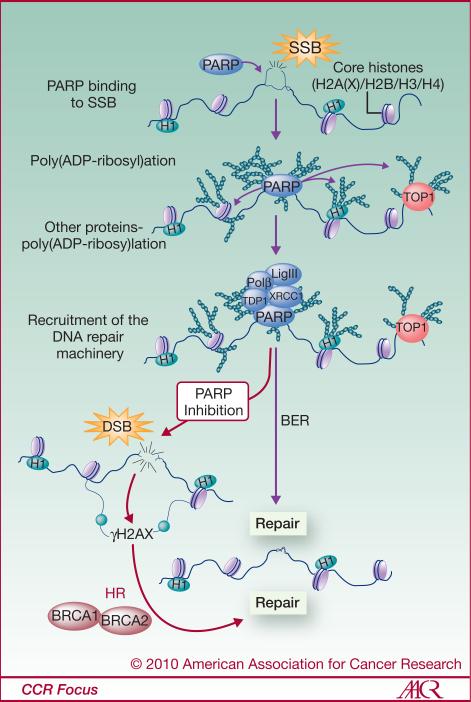
PAR was discovered approximately 50 years ago by Chambon and coworkers (19). Its formation is catalyzed by PARP1/2 in several steps (20). PARP is activated upon DNA binding by intramolecular folding and dimerization. The PARP homodimer then catalyzes the transfer of an ADP-ribose moiety from nicotinamide adenine dinucleotide (NAD+) to a lysine (or glutamate) residue of an acceptor protein, followed by the sequential addition of multiple ADP-ribose units to the preceding ones forming linear and branched PAR chains containing up to 200 units. In response to DNA damage, PARP1 itself is the main PARylation acceptor (automodification) and over 90% of PAR is found on PARP1 (13). PARylation is a reversible reaction. Poly(ADP-ribose) glycosylase (PARG) acts as an endo- and exo-glycosidase and hydrolyzes the glycosidic linkages between ADP-ribose units of PAR producing free ADP-ribose, and ADP-ribosyl protein lyase hydrolyzes the remaining protein-proximal ADP-ribose monomers (14). Poly(ADP-ribose) hydrolysis via endo- and exo-glycosidase activities would restore PARP1's ability to identify DNA single strand breaks and activate a new DNA damage response if necessary. Another consequence of poly(ADP-ribose) hydrolysis would be the production of high cellular levels of ADP-ribose, which in turn, may be hydrolyzed to phosphoribose and AMP, increasing the AMP:ATP ratio and inducing an autophagic state (16). PAR functions as a docking polymer for a variety of chromatin and DNA repair proteins including histones, XRCC1, Ku, DNA-PKcs, condensins and DNA topoisomerases (13, 15). PARylation acts as an early DDR post-translational modification that recruits repair proteins to the DNA damage site, including scaffolding protein, XRCC1, a factor involved in base excision repair (BER), DNA ligase III and DNA polymeraseβ (21-23) (see Fig. 2 for details). PARP along with XRCC1 has also been implicated in the alternative-NHEJ pathway of DSB repair (24, 25), and in DSB repair during spermatogenesis (26). Because of PARP's involvement in SSB repair, its inhibition has been proposed to lead to DSB formation.
Figure 2. Schematic representation of PARP1's role in single-strand break repair.
Upon detection of the DNA single-strand break (SSB) lesion, PARP1 is activated and, in turn, synthesizes PAR polymers attached to itself and other acceptor proteins at the DNA lesion site (Histone H1, other core histones, TOP1,...). These accrued post-translational modifications favor the recruitment of other factors involved in DNA repair, especially those involved in the base excision repair (XRCC1, Tdp1, Ligase III, Polβ). If left unrepaired, for example with PARP inhibition (red arrows), SSBs can lead to DSBs and γH2AX foci formation. DSB repair requires BRCA1/2, proteins that are deficient in many, including breast and ovarian, cancers.
Since BRCA1 and BRCA2 are defective in many cancers and necessary for DSB repair, inhibition of PARP1 in such cancer cells results in enhanced and selective killing (27, 28) (Fig. 2). PARP inhibitors provide a novel way to treat BRCA-deficient cells in combination with chemotherapy and radiation while sparing normal cells. Therefore, PARP is a valuable target for pharmacological strategies (16, 29) and PARP inhibitors are now used in an increasing number of clinical trials (30).
Biosampling for γH2AX and PAR measurements
Ideally, the efficiency of a cancer therapy based on PARP inhibitors would be to measure both γH2AX formation and PARP activity (i.e. PAR levels) in tumor biopsies (31, 32). However, issues of tumor accessibility, patient discomfort, and risk of infection prevent the widespread acquisition of tumor biopsies. Even if tumor biopsies were routinely available, other confounding issues might prevent straightforward interpretations of any results obtained from them. One issue stems from tumor heterogeneity (33), due to differing replicating fractions and to vascularization anomalies affecting oxygen, nutrient and drug delivery. Therefore, γH2AX and PAR amounts may differ among multiple biopsies of the same tumor. Perhaps comparability of tumor responses might be improved by characterizing the extent of hypoxia and/or replication status along with γH2AX levels in the cells being analyzed. Another confounding factor involves decisions to repeatedly sample the same nodule with consequent influence on the biomarker or to sample a different nodule with questions of biological variability. One unique solution to obtain multiple tumor samples that may yield useful information involves the isolation of circulating tumor cells (CTC) from the patient's blood (34), which is much more feasible than obtaining multiple tumor biopsies (35).
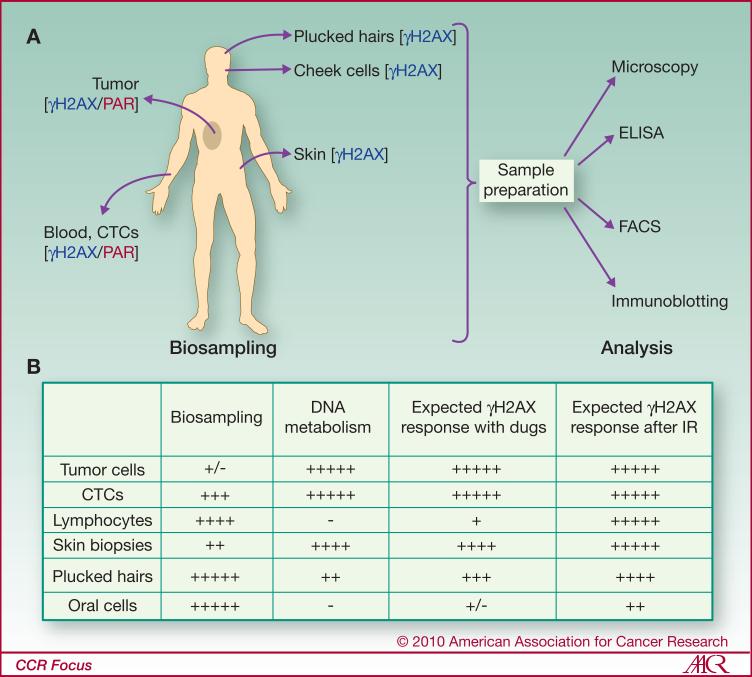
Because of these confounding issues concerning tumor biopsies, procedures to assess other more accessible, surrogate, patient tissues by less invasive means are being developed. The advantages and inconveniences of different tumor and surrogate tissues for γH2AX detection are summarized in Figure 3. The most suitable surrogate sample should be selected considering the type of treatment received by the patient (see Fig. 3 for details).
Figure 3. Human biopsy analysis.
(A) Biopsy sites and methods used to analyze γH2AX levels and PARP activities. (B) Biosample accessibility, state of DNA metabolism, and expected γH2AX response. Note that the most accessible tissues for analysis are not always the most appropriate for measuring a drug response. For example, lymphocyte and oral cells, two highly differentiated cell types, may exhibit poor responses to chemotherapeutic drugs targeting DNA replication. In contrast, tumor cells, while clearly appropriate, are often poorly accessible particularly for repetitive sampling. γH2AX formation is independent of the cell cycle state, occurring in cancer cells as well as in lymphocytes and oral cells after irradiation (37, 40, 70). CTCs: circulating tumor cells.
Circulating blood lymphocytes and leucocytes are terminally differentiated (post-mitotic) cells which respond consistently to irradiation with robust γH2AX signals proportional to the radiation doses (36, 37); however, their use for assessing the action of anticancer drugs that interfere with DNA replication is problematic. In contrast, PAR measurements in lymphocytes can be performed and reduction in PAR levels can readily be observed following PARP inhibitor treatment in most patients (32).
Buccal cells can be collected very simply by swabbing the patient's inner cheeks, but collection of numbers of living cells sufficient for testing can be problematical. In addition, buccal cells, a type of terminally differentiated stratified squamous epithelium, can present a challenge in procedures involving cell lysis or permeabilization when using microscopy (38). In addition, buccal cells appear to have very high background levels of DNA damage (38, 39). Nevertheless, significantly increased γH2AX formation was detected in patients undergoing routine dental radiographic examination, indicating that the use of γH2AX in oral cells could serve as sensitive indicators of low-dose radiation exposure (40).
Another minimally invasive procedure consists of collecting hairs plucked from the scalp or eyebrows. A substantial portion of the hair bulb may stay attached to the hair shaft and contain dividing and stem cells (41) which can be used to measure γH2AX levels and PARP inhibition (42). However, considering that a patient's hair follicles are in various stages of growth, plucked hairs containing replicating cells (i.e. in anagen phase) should be favored for chemotherapy assessments (Redon C. and Bonner W.M., unpublished).
Finally, since skin contains proliferative cells, such a tissue would be valuable to determine DNA damage (i.e. γH2AX levels) following treatment with drugs that interfere with DNA replication. However, like tumor biopsies, multiple sampling would result in patient discomfort and may lead to possible complications, such as infections.
A major issue is the relationship of the responses of γH2AX and PAR in surrogate tissues vs. the tumors and to the relationship between those responses and the treatment outcome. While performing these comparisons for solid tumors is challenging because of the difficulty of biopsy collection, blood malignancies may present a more accessible situation. γH2AX and PAR levels could be directly measured in malignant cells collected in blood samples. However, the use of γH2AX in surrogate tissues can help determine both a drug's genotoxicity and its pharmacokinetics in vivo. Such comparisons could be extended to multiple drug combinations.
Assays for γH2AX and PAR detection
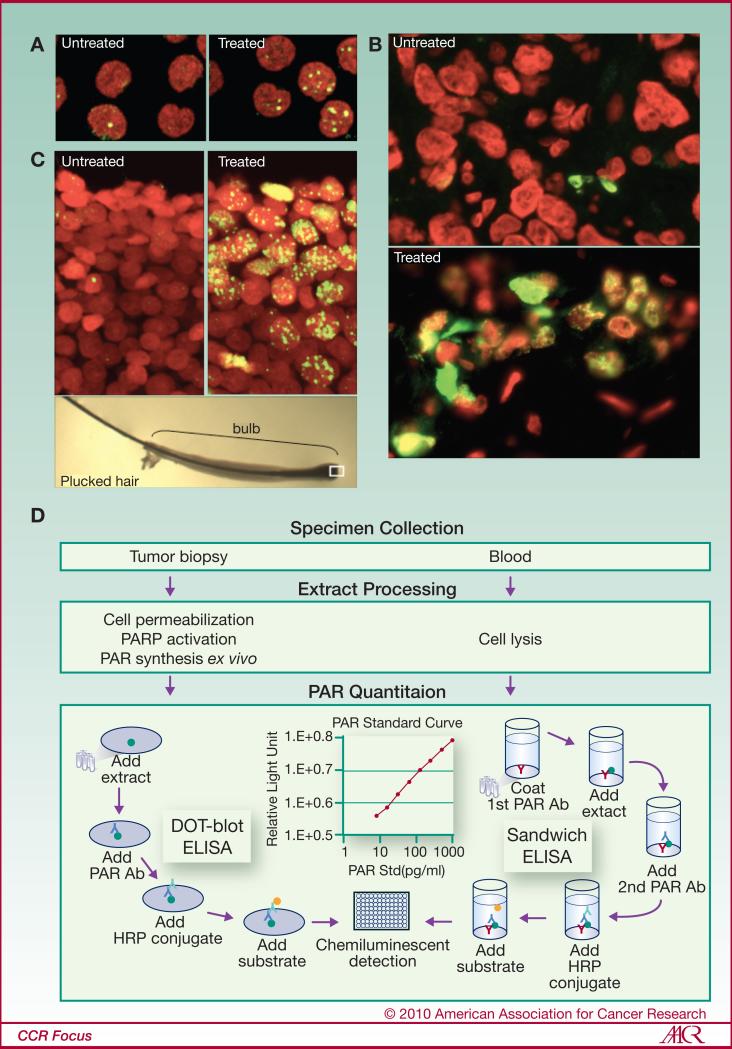
Several immunological techniques can be used to quantify γH2AX and PAR (PARP activity is deduced from changes in PAR levels). They may consist of counting antigen intensities in cells and tissues [i.e. microscopy and fluorescence-activated cell sorting (FACS)] or quantifying their overall levels (i.e. Western blotting and ELISA) (4, 32, 42-44) (Figs. 3, 4). In recent years, the Pharmacodynamic Assay Development and Implementation Section (PADIS - National Cancer Institute (NCI)), in collaboration with Abbott Laboratories and the National Clinical Target Validation Laboratory at the NCI has developed an ELISA for PAR levels in tissue samples. With this assay, the in vivo effect of a PARP inhibitor could be measured (45) (Fig. 4). Finally, a method using an electrochemoluminescent detection system (an assay derived from the Meso Scale Discovery Technology or MSD assay) was recently reported. This ELISA-based methodology allows γH2AX measurements in tumors after irradiation (46).
Figure 4. γH2AX and PAR detection.
γH2AX detection in lymphocytes (A), tumor needle biopsies (B), and plucked hairs (C) from patients undergoing chemotherapy. The white box in the lower panel (C) marks the region of active γH2AX formation in a typical plucked hair. Green, γH2AX; red, DNA. (D) Pharmacodynamic assay developed at the National Cancer Institute to measure PAR as a biomarker for PARP inhibition in both tumor biopsies and peripheral blood mononuclear cells.
Although each of these methods can be effective and gives important information, they are not currently usable for high throughput screening (HTS) and still involve extensive human labor. Recently, Garty et al. reported the development of HTS system as a radiation biodosimetry tool for radiation triage and using γH2AX immunofluorescence assay, called as the RABIT (Rapid Automated Biodosimetry Tool) that would allow the screening of 6,500 samples a day (47).
Both γH2AX and PAR assays are specific and very sensitive. A single DSB (corresponding to one γH2AX focus) can be visualized by microscopy and responses to radiation doses as low as 1.2 mGy and 100 mGy can be detected by microscopy and FACS respectively (4). The ELISA-based measurements of PAR has a lower limit of detection (LLD) <6 pg/ml with a dynamic range of 7.8 to 1000 pg/ml (Dr. J. Ji, personal communication).
γH2AX in clinical oncology
γH2AX in cancer chemotherapy
Many cancer therapies rely on agents that preferentially kill tumor cells by generating DNA damage (48) including DSBs [reviewed in (4)] (Table 1). Methods to detect DSB levels directly in patient material cells could be invaluable for optimizing treatment, particularly with chemotherapeutic agents whose efficacies could vary among individuals with different genetic backgrounds. Because biosampling tumors is not always possible, alternate surrogate tissues have been developed depending on availability (i.e. hair sampling could be problematic in patients with alopecia due to prior treatments) or on the agent (i.e. terminally differentiated surrogate cells may not respond well when using a drug interfering with replication). For this reason, biosampling of several tissues would be preferable during drug development.
Table 1. Anticancer drugs that produce γH2AX.
Note that all the anticancer drugs listed can also induce delayed γH2AX activation by apoptosis.
| DRUGS | MECHANISM OF INDUCTION | REFERENCES |
|---|---|---|
| Bleomycin | Direct DSB – iron mediated oxidative cleavage | (70) |
| Camptothecins and Indenoisoquinolines | Indirect: conversion of SSB to DSB by replication | (12, 71, 72) |
| Doxorubicin, Etoposide, Mitoxantrone Batracylin | Direct DSB by trapping topoisomerase II cleavage complexes. Also indirect: ROS formation | (73-75) |
| Cytarabine, Gemcitabine, Hydroxyurea | Indirect: replication fork collapse (chain termination; deoxyribonucleotide pool depletion) | (76-78) |
| Cisplatin, Temozolomide Aminoflavone Trabectedin | Indirect: DNA alkylation | (79-82) |
| Imatinib (Gleevec®) | Indirect: apoptosis induced by Kit/PDGF tyrosine kinase inhibition | (83) |
| 5-azacytidine SAHA (vironostat) | Indirect: epigenetic modifications | (84, 85) |
| PARP and DNA-PK inhibitors) (see ref. 1 for PARPi) | Indirect: interference with SSB and DSB repair | (27, 86, 87) |
| SJG-136 | Indirect: DNA alkylation | (53) |
| Tirapazamine | Indirect: ROS production in hypoxic cells | (73, 88) |
| TRAIL | Indirect: Death receptors-mediated activation of DNA-PK | (89, 90) |
| UCN-01 and AZD7762 | Indirect: Checkpoint inhibitor potentiating IR- and replication-induced DNA damage by topoisomerase I inhibitors, cytarabine and gemcitabine. | (77, 91, 92) |
Clinical studies with different methods of γH2AX analysis are described in Table 2. In a phase I clinical study for patients with refractory leukemia, flow cytometry revealed increased γH2AX levels in circulating leukemia cells in 12 of 13 patients after treatment with a novel deoxyadenosine analog (clofarabine) combined with an alkylating agent (cyclophosphamide) (49). The same group also reported that the peripheral blood mononuclear cells from 23 patients with myelodysplasia (MDS), and high-risk acute myelogenous leukemia (AML) exhibited increased γH2AX levels after sequential administration of a DNA methyltransferase inhibitor (5-azacytidine) and a histone deacetylase (HDAC) inhibitor (Ectinostat) (50). However the latter data did not show any correlation between the patients’ responses and γH2AX induction. Another clinical study demonstrated the ability of the minor groove binding agent SJG-136 to increase γH2AX levels in both the patients’ lymphocytes and tumor biopsies (51). Fong et al. described the use of plucked-eyebrow hairs to follow patients treated with a PARP inhibitor. Their study showed a correlation between γH2AX levels and PARP inhibition (42). Finally, a trial testing the combination of a farnesyltransferase inhibitor (Tipifarnib) and a topoisomerase II inhibitor (etoposide) for individuals diagnosed with AML used a γH2AX assay on AML marrow blasts (52). This drug combination demonstrated genotoxicity by both the use of γH2AX and increased subdiploid DNA content.
Table 2. Some published Examples of clinical studies using γH2AX detection in vivo.
Studies are listed in three separate groups: A. γH2AX used in clinical trials, B. γH2AX used for radiation biodosimetry, C. γH2AX used for diagnostics.
| APPLICATION | TISSUE | STUDY DETAILS | METHOD | REF. |
|---|---|---|---|---|
| (A) γH2AX used in clinical trials | ||||
| Chemotherapy | PBMCs | Phase 1 study of clofarabine followed by cyclophosphamide for adults with refractory acute leukemias | FACS | (49) |
| Chemotherapy | PBMCs / tumor biopsies | A phase 1 study of SJG-136 | Microscopy | (51) |
| Chemotherapy | AML marrow blasts | A phase 1 study of the combination of tipifarnib and etoposide for patients with AML | FACS | (52) |
| Chemotherapy | PBMCs | A phase 1 study of 5-azacytidine and entinostata for patients with MDS, chronic myelomonocytic leukemia, and AML | Immunoblotting | (50) |
| Chemotherapy | Plucked eye-brows | A phase 1 study of olaparib | Microscopy | (42) |
| (B) γH2AX used for radiation biodosimetry | ||||
| Computed tomography | PBMCs | DNA damage measured after multi-detector row CT | Microscopy | (93) |
| radiotherapy | PBMCs | DNA damage measured in cancer patients after local radiotherapy to different sites of the body | Microscopy | (54) |
| radiotherapy | Skin | DNA damage measured in prostate cancer patients undergoing radiotherapy with curative intent | Microscopy | (53) |
| X-ray examination | PBMCs | DNA damage measured after cardiac catheterization | Microscopy | (94) |
| X-ray examination | PBMCs | DNA damage measured after coronary CT angiographic procedure | Microscopy | (95) |
| X-ray examination | PBMCs | DNA damage measured after angiographic procedure | Microscopy | (56) |
| X-ray examination | PBMCs | DNA damage measured after percutaneous transluminal angioplasty | Microscopy | (55) |
| (C) γH2AX used for diagnostics | ||||
| Diagnosis | Tumor biopsies | Diagnosis of metastatic Renal Cell Carcinoma | Microscopy | (64) |
| Diagnosis | Tissue biopsies | Monitoring DNA damage in Ulcerative Colitis | Immunoblotting | (96) |
| Radiosensitivity diagnosis | T-cells and lymphoblast oid cell lines and/or PBMCs | Confirmation of radiosensitive A-T patients | FACS | (58) |
γH2AX use in radiotherapy
In contrast to chemotherapeutic agents, ionizing radiation can induce DSBs regardless of cell cycle phase, thus, radiation induced-γH2AX could in principle be detectible in all tissues or cells including G0 lymphocytes and oral cells. Counting γH2AX foci has been used successfully for biodosimetry (36, 53-56). After exposure γH2AX foci levels in patient lymphocytes and skin biopsies exhibited a good correlation with the radiation doses.
Additionally, γH2AX measurements might provide information useful in improving patient outcome during radiotherapy. Although treatment with radiation is commonly used, some patients develop severe, possibly lethal, side effects. Patients with DSB repair deficiencies, such as ataxia telangiectasia caused by mutation in ATM gene, are highly sensitive to radiation exposure (57). Thus, γH2AX measurements may allow the identification of hypersensitive individuals in order to improve the efficacy of radiotherapy and to avoid therapeutic accidents (58, 59).
It is important to note that therapeutic and accidental radiation exposures are rarely uniform and therefore can result in subpopulations of lymphocytes with different exposures, a phenomenon that should be taken into account for dose estimation. Moreover, because hypoxic microenvironments in solid tumors result in the decrease of the number of DSBs by radiotherapy (33), variations in γH2AX levels could occur in different tumor samples for a same irradiation dose.
γH2AX use in diagnostics
γH2AX also has potential uses in medical diagnosis (Table 2). Since both pre-cancerous and cancerous cells were found to exhibit increased genomic instability (60, 61), a biomarker able to identify such cells in patients would be a useful diagnostic tool. In fact, it was shown that γH2AX can be used as a biomarker for cancer as tumor biopsies show increased γH2AX levels (62, 63). γH2AX measurements also established high levels of DNA damage in colon biopsies of patients with ulcerative colitis, an inflammation disease linked to higher risk for colorectal cancer. A recent study showed the dependability and accuracy of γH2AX in the diagnosis of metastatic renal cell carcinoma (64).
PAR in clinical oncology
Several PARP inhibitors (at least eight: Iniparib/BSI-201, Olaparib/AZ2281, Veliparib/ABT-888, AG014699, MK-4827, CEP-8933/CEP-9722, INO-1001, GPI 21016) have been developed and are now used alone or combined with other drugs in several clinical trials (30). Because of the high homology between PARP1 and PARP2, most PARP inhibitors can target both enzymes and future studies will probably look for specific inhibitors (65). The first report of a PARP clinical trial using the drug BSI-201 alone in a phase I study came out in 2008 (66). Since, this drug has been used in more phase I trials in combination with other drugs, in phase II clinical studies and is now entering phase III study (67, 68). Olaparib and veliparib/ABT-888 are also undergoing clinical development in several phase I and phase II trials. The National Cancer Institute conducted the first clinical pharmacodynamic trial (Phase 0) of ABT-888, an orally available small-molecule inhibitor of PARP in patients with advanced malignancies. PAR levels in tumor biopsies and peripheral blood mononuclear cells were measured by using a validated ELISA (32). The data obtained could be used to guide the design of several phase I combination studies of veliparib, including ongoing trials with topotecan and cyclophosphamide, each of which include measurement of PAR as a pharmacodynamic endpoint.
In these studies, it is crucial to take measurements at the in vivo cellular level, both for the drug effect on PARP activity (i.e. PAR levels) and the consequences of this inhibition (i.e. DNA damage induction via γH2AX detection). Recent years have seen the development of ELISA for PAR detection that can now replace the less reliable immunoassays (32, 69) (Fig. 4D). Thus, PAR levels can be measured in both peripheral blood mononuclear cells (PBMCs) (mostly lymphocytes) and tumor biopsies. These measurements show a significant correlation between the effects of the PARP inhibitor in PBMCs and the tumor samples (32) raising the possibility that blood samples could be used as tumor surrogates to follow PARP inhibition.
Conclusions
The availability of biomarkers of DNA damage offers the opportunity to evaluate clinical cancer samples and determine their DDR status prior and during therapy. The systematic use of γH2AX and PAR in tumor samples in the absence of treatment may identify groups of patients with a particular prognosis. γH2AX and PAR may also be useful to evaluate the activity of novel drugs in the tumor and normal tissues in response to treatment, which could accelerate the drug triage process -- aiding go - no-go decisions, or lead compound selection, for example. Hopefully, incorporation of such biomarkers will eventually eliminate a substantial fraction of drugs that fail in Phase II-III clinical trials.
The related question is whether γH2AX and PAR could be developed beyond that of a tool for clinical trials, to a marker with utility in cancer therapy. Following treatment, assessment of γH2AX and PAR could allow identification of patients who had insufficient evidence of DNA damage so that different drugs could be selected, or strategies to increase drug exposure, change schedule, or improve activity could be identified. It is likely that γH2AX and PAR only represent the first generation of DDR biomarkers and that more sensitive and convenient biomarkers will be developed in the near future. Ultimately, one of the major challenges remains to sample tumors and to develop non-invasive detection procedures. In the meantime, it will be interesting to take advantage of analyses in circulating tumor cells.
Acknowledgements
This work was supported by the Center for Cancer Research, Intramural Program, National Cancer Institute, National Institutes of Health, Bethesda, Maryland. We thank Dr. Susan Bates, Christine Bryla, Robin Frye, Kathryn Compton, Merrill Goldsmith, Gareth Peters and William Yutzy for their help in the biomarker studies.
References
- 1.Kummar S, Kinders R, Rubinstein L, et al. Compressing drug development timelines in oncology using phase ‘0’ trials. Nat Rev Cancer. 2007;7:131–9. doi: 10.1038/nrc2066. [DOI] [PubMed] [Google Scholar]
- 2.Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nat Rev Drug Discov. 2003;2:566–80. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- 3.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–9. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 4.Bonner WM, Redon CE, Dickey JS, et al. gammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 6.Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha H, Lowe JM, Li H, et al. Wip1 directly dephosphorylates gamma-H2AX and attenuates the DNA damage response. Cancer Res. 2010;70:4112–22. doi: 10.1158/0008-5472.CAN-09-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moon SH, Nguyen TA, Darlington Y, Lu X, Donehower LA. Dephosphorylation of gammaH2AX by WIP1: An important homeostatic regulatory event in DNA repair and cell cycle control. Cell Cycle. 2010;9 doi: 10.4161/cc.9.11.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas P, Zhong J, Ye R, Moorhead GB, Xu X, Lees-Miller SP. Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol Cell Biol. 30:1368–81. doi: 10.1128/MCB.00741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakada S, Chen GI, Gingras AC, Durocher D. PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 2008;9:1019–26. doi: 10.1038/embor.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–9. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 13.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–68. [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–67. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 16.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ju BG, Lunyak VV, Perissi V, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 18.Lis JT, Kraus WL. Promoter cleavage: a topoIIbeta and PARP-1 collaboration. Cell. 2006;125:1225–7. doi: 10.1016/j.cell.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 20.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–38. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–71. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–69. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 23.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–33. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–26. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 25.Iliakis G. Backup pathways of NHEJ in cells of higher eukaryotes: cell cycle dependence. Radiother Oncol. 2009;92:310–5. doi: 10.1016/j.radonc.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed EA, de Boer P, Philippens ME, Kal HB, de Rooij DG. Parp1-XRCC1 and the repair of DNA double strand breaks in mouse round spermatids. Mutat Res. 2010;683:84–90. doi: 10.1016/j.mrfmmm.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 28.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 29.Drew Y, Plummer R. PARP inhibitors in cancer therapy: two modes of attack on the cancer cell widening the clinical applications. Drug Resist Updat. 2009;12:153–6. doi: 10.1016/j.drup.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Annunziata CM, O'Shaughnessy J. Poly(ADP-Ribose) Polymerase as a novel therapeutic target in cancer. Clin Cancer Res. 2010;16 doi: 10.1158/1078-0432.CCR-10-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oka K, Tanaka T, Enoki T, et al. DNA damage signaling is activated during cancer progression in human colorectal carcinoma. Cancer Biol Ther. 9:246–52. doi: 10.4161/cbt.9.3.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kummar S, Kinders R, Gutierrez ME, et al. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009;27:2705–11. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan N, Bristow RG. “Contextual” synthetic lethality / loss of heterozygoty: tumor hypoxia and modification of DNA repair. Clin Cancer Res. 2010;16 doi: 10.1158/1078-0432.CCR-10-0527. [DOI] [PubMed] [Google Scholar]
- 34.Wang LH, Pfister TD, Parchment RE, et al. Monitoring drug-induced gammaH2AX as a pharmacodynamic biomarker in individual circulating tumor cells. Clin Cancer Res. 16:1073–84. doi: 10.1158/1078-0432.CCR-09-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maheswaran S, Haber DA. Circulating tumor cells: a window into cancer biology and metastasis. Curr Opin Genet Dev. 2010;20:96–9. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lobrich M, Rief N, Kuhne M, et al. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci U S A. 2005;102:8984–9. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redon CE, Dickey JS, Bonner WM, Sedelnikova OA. gamma-H2AX as a biomarker of DNA damage induced by ionizing radiation in human peripheral blood lymphocytes and artificial skin. Adv Space Res. 2009;43:1171–8. doi: 10.1016/j.asr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szeto YT, Benzie IF, Collins AR, et al. A buccal cell model comet assay: development and evaluation for human biomonitoring and nutritional studies. Mutat Res. 2005;578:371–81. doi: 10.1016/j.mrfmmm.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Rothkamm K, Horn S. gamma-H2AX as protein biomarker for radiation exposure. Ann Ist Super Sanita. 2009;45:265–71. [PubMed] [Google Scholar]
- 40.Yoon AJ, Shen J, Wu HC, et al. Expression of activated checkpoint kinase 2 and histone 2AX in exfoliative oral cells after exposure to ionizing radiation. Radiat Res. 2009;171:771–5. doi: 10.1667/RR1560.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gho CG, Braun JE, Tilli CM, Neumann HA, Ramaekers FC. Human follicular stem cells: their presence in plucked hair and follicular cell culture. Br J Dermatol. 2004;150:860–8. doi: 10.1111/j.1365-2133.2004.05862.x. [DOI] [PubMed] [Google Scholar]
- 42.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 43.Kunzmann A, Liu D, Annett K, et al. Flow-cytometric assessment of cellular poly(ADP-ribosyl)ation capacity in peripheral blood lymphocytes. Immun Ageing. 2006;3:8. doi: 10.1186/1742-4933-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang SX, Kummar S, Steinberg SM, et al. Immunohistochemical detection of poly(ADP-ribose) polymerase inhibition by ABT-888 in patients with refractory solid tumors and lymphomas. Cancer Biol Ther. 2009;8:2004–9. doi: 10.4161/cbt.8.21.9917. [DOI] [PubMed] [Google Scholar]
- 45.Kinders RJ, Hollingshead M, Khin S, et al. Preclinical modeling of a phase 0 clinical trial: qualification of a pharmacodynamic assay of poly (ADP-ribose) polymerase in tumor biopsies of mouse xenografts. Clin Cancer Res. 2008;14:6877–85. doi: 10.1158/1078-0432.CCR-08-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avondoglio D, Scott T, Kil WJ, Sproull M, Tofilon PJ, Camphausen K. High throughput evaluation of gamma-H2AX. Radiat Oncol. 2009;4:31. doi: 10.1186/1748-717X-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garty G, Chen Y, Salerno A, et al. The RABIT: a rapid automated biodosimetry tool for radiological triage. Health Phys. 98:209–17. doi: 10.1097/HP.0b013e3181ab3cb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plummer R. Perspective on the pipeline of drugs being developed with modulation of DNA damage as a target. Clin Cancer Res. 2010;16 doi: 10.1158/1078-0432.CCR-10-0984. [DOI] [PubMed] [Google Scholar]
- 49.Karp JE, Ricklis RM, Balakrishnan K, et al. A phase 1 clinical-laboratory study of clofarabine followed by cyclophosphamide for adults with refractory acute leukemias. Blood. 2007;110:1762–9. doi: 10.1182/blood-2007-03-081364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fandy TE, Herman JG, Kerns P, et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood. 2009;114:2764–73. doi: 10.1182/blood-2009-02-203547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hochhauser D, Meyer T, Spanswick VJ, et al. Phase I study of sequence-selective minor groove DNA binding agent SJG-136 in patients with advanced solid tumors. Clin Cancer Res. 2009;15:2140–7. doi: 10.1158/1078-0432.CCR-08-1315. [DOI] [PubMed] [Google Scholar]
- 52.Karp JE, Flatten K, Feldman EJ, et al. Active oral regimen for elderly adults with newly diagnosed acute myelogenous leukemia: a preclinical and phase 1 trial of the farnesyltransferase inhibitor tipifarnib (R115777, Zarnestra) combined with etoposide. Blood. 2009;113:4841–52. doi: 10.1182/blood-2008-08-172726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qvarnstrom OF, Simonsson M, Johansson KA, Nyman J, Turesson I. DNA double strand break quantification in skin biopsies. Radiother Oncol. 2004;72:311–7. doi: 10.1016/j.radonc.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 54.Sak A, Grehl S, Erichsen P, et al. gamma-H2AX foci formation in peripheral blood lymphocytes of tumor patients after local radiotherapy to different sites of the body: dependence on the dose-distribution, irradiated site and time from start of treatment. Int J Radiat Biol. 2007;83:639–52. doi: 10.1080/09553000701596118. [DOI] [PubMed] [Google Scholar]
- 55.Geisel D, Heverhagen JT, Kalinowski M, Wagner HJ. DNA double-strand breaks after percutaneous transluminal angioplasty. Radiology. 2008;248:852–9. doi: 10.1148/radiol.2483071686. [DOI] [PubMed] [Google Scholar]
- 56.Kuefner MA, Grudzenski S, Schwab SA, et al. DNA double-strand breaks and their repair in blood lymphocytes of patients undergoing angiographic procedures. Invest Radiol. 2009;44:440–6. doi: 10.1097/RLI.0b013e3181a654a5. [DOI] [PubMed] [Google Scholar]
- 57.Taylor AM, Byrd PJ. Molecular pathology of ataxia telangiectasia. J Clin Pathol. 2005;58:1009–15. doi: 10.1136/jcp.2005.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porcedda P, Turinetto V, Brusco A, et al. A rapid flow cytometry test based on histone H2AX phosphorylation for the sensitive and specific diagnosis of ataxia telangiectasia. Cytometry A. 2008;73:508–16. doi: 10.1002/cyto.a.20566. [DOI] [PubMed] [Google Scholar]
- 59.Porcedda P, Turinetto V, Orlando L, et al. Two-tier analysis of histone H2AX phosphorylation allows the identification of Ataxia Telangiectasia heterozygotes. Radiother Oncol. 2009;92:133–7. doi: 10.1016/j.radonc.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 60.Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 61.Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 62.Sedelnikova OA, Bonner WM. GammaH2AX in cancer cells: a potential biomarker for cancer diagnostics, prediction and recurrence. Cell Cycle. 2006;5:2909–13. doi: 10.4161/cc.5.24.3569. [DOI] [PubMed] [Google Scholar]
- 63.Yu T, MacPhail SH, Banath JP, Klokov D, Olive PL. Endogenous expression of phosphorylated histone H2AX in tumors in relation to DNA double-strand breaks and genomic instability. DNA Repair (Amst) 2006;5:935–46. doi: 10.1016/j.dnarep.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 64.Wasco MJ, Pu RT. Utility of antiphosphorylated H2AX antibody (gamma-H2AX) in diagnosing metastatic renal cell carcinoma. Appl Immunohistochem Mol Morphol. 2008;16:349–56. doi: 10.1097/PAI.0b013e3181577993. [DOI] [PubMed] [Google Scholar]
- 65.Ishida J, Yamamoto H, Kido Y, et al. Discovery of potent and selective PARP-1 and PARP-2 inhibitors: SBDD analysis via a combination of X-ray structural study and homology modeling. Bioorg Med Chem. 2006;14:1378–90. doi: 10.1016/j.bmc.2005.09.061. [DOI] [PubMed] [Google Scholar]
- 66.Kopetz SMM, Mok I, et al. First in human phase I study of BSI-201, a small molecule inhibitor of poly ADP-ribose (PARP) in subjects with advanced solid tumors.. ASCO Annual meeting proceedings; 2008; p. Abstract 3577. [Google Scholar]
- 67.Mahani JJ, Lewis N, Heath EI, et al. A phase IB study evaluating BSI-201 in combination with chemotherapy in subjects with advanced solid tumors.. ASCO Annual meeting; 2008; 2008. p. Abstract 3579. [Google Scholar]
- 68.O'Shaugnessy J, Osborne C, Pippen J, et al. Efficacy of BSI-201, a poly (ADP-ribose) polymerase-I (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomized phase II trial.. ASCO annual meeting; 2009; 2009. [Google Scholar]
- 69.Liu X, Palma J, Kinders R, et al. An enzyme-linked immunosorbent poly(ADP-ribose) polymerase biomarker assay for clinical trials of PARP inhibitors. Anal Biochem. 2008;381:240–7. doi: 10.1016/j.ab.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 70.Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–9. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 71.Furuta T, Takemura H, Liao ZY, et al. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J Biol Chem. 2003;278:20303–12. doi: 10.1074/jbc.M300198200. [DOI] [PubMed] [Google Scholar]
- 72.Antony S, Agama KK, Miao ZH, et al. Novel indenoisoquinolines NSC 725776 and NSC 724998 produce persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 2007;67:10397–405. doi: 10.1158/0008-5472.CAN-07-0938. [DOI] [PubMed] [Google Scholar]
- 73.Olive PL, Banath JP, Sinnott LT. Phosphorylated histone H2AX in spheroids, tumors, and tissues of mice exposed to etoposide and 3-amino-1,2,4-benzotriazine-1,3-dioxide. Cancer Res. 2004;64:5363–9. doi: 10.1158/0008-5472.CAN-04-0729. [DOI] [PubMed] [Google Scholar]
- 74.Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst) 2004;3:889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 75.Rao VA, Agama K, Holbeck S, Pommier Y. Batracylin (NSC 320846), a dual inhibitor of DNA topoisomerases I and II induces histone gamma-H2AX as a biomarker of DNA damage. Cancer Res. 2007;67:9971–9. doi: 10.1158/0008-5472.CAN-07-0804. [DOI] [PubMed] [Google Scholar]
- 76.Krynetskaia N, Xie H, Vucetic S, Obradovic Z, Krynetskiy E. High mobility group protein B1 is an activator of apoptotic response to antimetabolite drugs. Mol Pharmacol. 2008;73:260–9. doi: 10.1124/mol.107.041764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ewald B, Sampath D, Plunkett W. H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther. 2007;6:1239–48. doi: 10.1158/1535-7163.MCT-06-0633. [DOI] [PubMed] [Google Scholar]
- 78.Kurose A, Tanaka T, Huang X, Traganos F, Dai W, Darzynkiewicz Z. Effects of hydroxyurea and aphidicolin on phosphorylation of ataxia telangiectasia mutated on Ser 1981 and histone H2AX on Ser 139 in relation to cell cycle phase and induction of apoptosis. Cytometry A. 2006;69:212–21. doi: 10.1002/cyto.a.20241. [DOI] [PubMed] [Google Scholar]
- 79.Pabla N, Huang S, Mi QS, Daniel R, Dong Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem. 2008;283:6572–83. doi: 10.1074/jbc.M707568200. [DOI] [PubMed] [Google Scholar]
- 80.Mirzoeva OK, Kawaguchi T, Pieper RO. The Mre11/Rad50/Nbs1 complex interacts with the mismatch repair system and contributes to temozolomide-induced G2 arrest and cytotoxicity. Mol Cancer Ther. 2006;5:2757–66. doi: 10.1158/1535-7163.MCT-06-0183. [DOI] [PubMed] [Google Scholar]
- 81.Meng LH, Kohlhagen G, Liao ZY, Antony S, Sausville E, Pommier Y. DNA-protein cross-links and replication-dependent histone H2AX phosphorylation induced by aminoflavone (NSC 686288), a novel anticancer agent active against human breast cancer cells. Cancer Res. 2005;65:5337–43. doi: 10.1158/0008-5472.CAN-05-0003. [DOI] [PubMed] [Google Scholar]
- 82.Guirouilh-Barbat J, Redon C, Pommier Y. Transcription-coupled DNA double-strand breaks are mediated via the nucleotide excision repair and the Mre11-Rad50-Nbs1 complex. Mol Biol Cell. 2008;19:3969–81. doi: 10.1091/mbc.E08-02-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y, Tseng M, Perdreau SA, et al. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Res. 2007;67:2685–92. doi: 10.1158/0008-5472.CAN-06-3497. [DOI] [PubMed] [Google Scholar]
- 84.Kiziltepe T, Hideshima T, Catley L, et al. 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Mol Cancer Ther. 2007;6:1718–27. doi: 10.1158/1535-7163.MCT-07-0010. [DOI] [PubMed] [Google Scholar]
- 85.Conti C, Leo E, Eichler GS, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–80. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Albert JM, Cao C, Kim KW, et al. Inhibition of poly(ADP-ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models. Clin Cancer Res. 2007;13:3033–42. doi: 10.1158/1078-0432.CCR-06-2872. [DOI] [PubMed] [Google Scholar]
- 87.Zhao Y, Thomas HD, Batey MA, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–62. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 88.Evans JW, Chernikova SB, Kachnic LA, et al. Homologous recombination is the principal pathway for the repair of DNA damage induced by tirapazamine in mammalian cells. Cancer Res. 2008;68:257–65. doi: 10.1158/0008-5472.CAN-06-4497. [DOI] [PubMed] [Google Scholar]
- 89.Solier S, Pommier Y. The apoptotic ring: a novel entity with phosphorylated histones H2AX and H2B and activated DNA damage response kinases. Cell Cycle. 2009;8:1853–9. doi: 10.4161/cc.8.12.8865. [DOI] [PubMed] [Google Scholar]
- 90.Solier S, Sordet O, Kohn KW, Pommier Y. Death receptor-induced activation of the Chk2- and histone H2AX-associated DNA damage response pathways. Mol Cell Biol. 2009;29:68–82. doi: 10.1128/MCB.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Furuta T, Hayward RL, Meng LH, et al. p21CDKN1A allows the repair of replication-mediated DNA double-strand breaks induced by topoisomerase I and is inactivated by the checkpoint kinase inhibitor 7-hydroxystaurosporine. Oncogene. 2006;25:2839–49. doi: 10.1038/sj.onc.1209313. [DOI] [PubMed] [Google Scholar]
- 92.Mitchell JB, Choudhuri R, Fabre K, et al. In vitro and in vivo radiation sensitization of human tumor cells by a novel checkpoint kinase inhibitor, AZD7762. Clin Cancer Res. 2010;16:2076–84. doi: 10.1158/1078-0432.CCR-09-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rothkamm K, Balroop S, Shekhdar J, Fernie P, Goh V. Leukocyte DNA damage after multi-detector row CT: a quantitative biomarker of low-level radiation exposure. Radiology. 2007;242:244–51. doi: 10.1148/radiol.2421060171. [DOI] [PubMed] [Google Scholar]
- 94.Beels L, Bacher K, De Wolf D, Werbrouck J, Thierens H. gamma-H2AX foci as a biomarker for patient X-ray exposure in pediatric cardiac catheterization: are we underestimating radiation risks? Circulation. 2009;120:1903–9. doi: 10.1161/CIRCULATIONAHA.109.880385. [DOI] [PubMed] [Google Scholar]
- 95.Kuefner MA, Hinkmann FM, Alibek S, et al. Reduction of X-ray induced DNA double-strand breaks in blood lymphocytes during coronary CT angiography using high-pitch spiral data acquisition with prospective ECG-triggering. Invest Radiol. 45:182–7. doi: 10.1097/RLI.0b013e3181d3eddf. [DOI] [PubMed] [Google Scholar]
- 96.Risques RA, Lai LA, Brentnall TA, et al. Is Ulcerative Colitis a Disease of Accelerated Colon Aging? Evidence From Telomere Attrition and DNA Damage. Gastroenterology. 2008 doi: 10.1053/j.gastro.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu C, Zhu F, Cho YY, et al. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell. 2006;23:121–32. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mukherjee B, Kessinger C, Kobayashi J, et al. DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair (Amst) 2006;5:575–90. doi: 10.1016/j.dnarep.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 99.Rossetto D, Truman AW, Kron SJ, Côté J. Epigenetic modification in double-strand break DNA damage signaling and repair. Clin Cancer Res. 2010;16 doi: 10.1158/1078-0432.CCR-10-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Downs JA, Allard S, Jobin-Robitaille O, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–90. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 101.van Attikum H, Gasser SM. ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle. 2005;4:1011–4. doi: 10.4161/cc.4.8.1887. [DOI] [PubMed] [Google Scholar]
- 102.Unal E, Arbel-Eden A, Sattler U, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 103.Xie A, Puget N, Shim I, et al. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16:1017–25. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–53. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]