Abstract
Significant emphasis has recently been placed on the characterization of the human cancer genome. This effort has been assisted by the development of new DNA sequencing technologies that allow the genomes of individual tumors to be analyzed in much greater detail. However, the genetic complexity of human cancer has complicated the identification of driver mutations among the more abundant passenger mutations found in tumors. Recently, the Sleeping Beauty (SB) transposon system has been engineered to model cancer in mice. SB-induced tumors are produced by transposon insertional mutagenesis, thus the tagged mutations facilitate the identification of novel cancer genes. This review provides a brief summary of the SB system and its use in modeling cancer in mice.
Introduction
While human cancer is caused by the cumulative effects of several factors (e.g. genetic, environmental), the acquisition of somatic gene mutations plays a key role in tumor initiation and progression. Many years of cancer research have identified a large number of human tumor suppressors and oncogenes that are frequently targeted by somatic mutation in a wide variety of tumor types. In addition, it is well established that cellular transformation requires the stepwise acquisition of somatic mutations in multiple genes. Thus each tumor is the product of many genetic alterations that cooperate to convert a cell from a normal to a malignant state.
Recently, significant resources have been committed to cataloging the human cancer genome as part of an ongoing effort to better understand the frequency and distribution of somatic mutations found within tumors of the lung, breast, colon and brain [1-3]. The goal of each study was to identify mutations that directly contribute to the process of cellular transformation (i.e. driver mutations). While the conclusions of each study vary depending on the number of genes and tumors analyzed, the general indication is that many more genes are mutated in human cancer than was previously appreciated.
Animal models have long been used to gain insight into the molecular and cellular events that contribute to cancer. More recently, cancer geneticists have taken advantage of high throughput analyses, such as comparative genomic hybridization, to identify genetic lesions common to similar tumor types from animals and humans. The basic assumption is that the genetic mechanisms that drive cellular transformation are shared among different species, while the spontaneous mutations will be different in each species. Thus, this type of comparative oncogenomic approach provides another avenue to identify driver mutations within human tumors.
Insertional mutagenesis has been used in mouse models of cancer for decades to simplify the genetic analysis of tumors. Mouse models of spontaneous lymphoma and mammary cancer were initially identified in the 1930's [4-5]. Subsequent work with these strains revealed that tumors in these mice were induced by endogenous retroviruses — the murine leukemia virus (MuLV) or the mouse mammary tumor virus (MMTV). These viruses are often referred to as “slow transforming retroviruses” because they do not encode any oncogenic peptides, but instead cause tumors by inducing insertional mutations when integrated into the host cell genome as a provirus [6]. The use of the inserted provirus as a sequence tag has greatly accelerated the identification of driver mutations within tumors induced by retroviral insertional mutagenesis, and this approach has made significant contributions to our understanding of the oncogenic networks that drive mouse lymphoma [7] and mammary cancer [8]. Unfortunately, the application of retroviral insertional mutagenesis is limited to the study of these specific forms of cancer due to the biology of the retroviruses (e.g. cellular tropism). However, other mouse models of cancer that are induced by insertional mutagenesis would be extremely valuable for comparative oncogenomic studies with more common forms of human cancer.
Transposons have been used extensively to perform insertional mutagenesis screens in invertebrate organisms such as yeast, Caenorhabditis elegans and Drosophila melanogaster [9-11]. These studies have co-opted an active endogenous transposon found in each species to perform mutagenesis. Unfortunately, most mammalian species, including mice, do not have active endogenous transposons, and thus transposon insertional mutagenesis in the mouse was not possible.
This changed in 1997 when Ivics et al. described the molecular reconstruction of an active transposon, called Sleeping Beauty [12]. Subsequent work showed that the Sleeping Beauty (SB) transposon functions in vivo in the mouse and could be used to perform insertional mutagenesis [13-15]. Unlike endogenous retroviruses, SB showed activity in many tissues in the mouse [15]. This suggested that SB could be used to induce tumors in mice, but without the limitations in tissue specificity caused by retroviruses. This chapter summarizes the development of the SB system to achieve this goal, as well as the current applications of SB in generating novel mouse models of human cancer.
Adapting Sleeping Beauty to model cancer
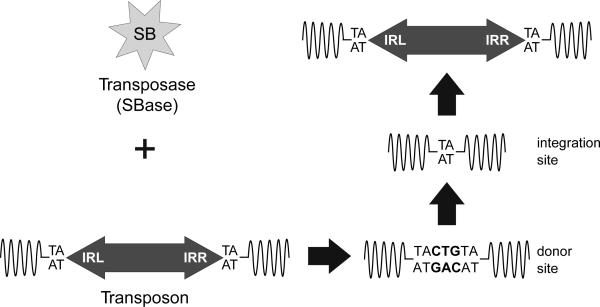
The SB transposon is a member of the Tc1/mariner family of class II transposable elements that use a cut-and-paste transposition mechanism. The basic requirements to drive SB transposition are quite simple: the transposase enzyme and a transposon vector. When both components are present within the same nucleus, the transposase can bind to specific sites within the inverted repeats of the transposon vector, mediate excision of the transposon from the donor site and integration into another target site in the host cell genome (Figure 1). The target sequence for SB is a TA dinucleotide pair — a simple sequence that is found within the mouse genome in more than 340 million sites. Therefore, the SB system has the potential to produce a wide variety of insertional mutations in virtually every gene in the mouse genome.
Figure 1.
The Sleeping Beauty (SB) transposon system. Like all cut-and-paste transposons, the SB system requires two functional parts: the transposase enzyme (SBase) and the transposon vector. When these two elements are found within the same host cell nucleus, the SBase can bind to the inverted repeats (IRL and IRR) at the ends of the transposon, mediate excision of the transposon from the donor site and insertion into a new TA dinucleotide site. The TA site is duplicated and flanks each end of the transposon at the insertion site. The DNA breaks generated by the SBase at the donor site are repaired by the host cell. The repair often leaves behind a footprint (TACTGTA) at the donor site.
The SB transposase (SBase) has several conserved domains that are critical for its function. First, a bipartite DNA-binding domain is found at the N-terminus of the SBase protein [16]. This domain confers the specificity with which the SBase is able to recognize and bind its target sequences within the transposon [17]. The catalytic function of the SBase is found in the C-terminal DDE motif — a domain that is common to transposase, recombinase and viral integrase enzymes. This DDE domain mediates the cleavage and joining reactions of the target DNA. The initial version of the SBase engineered by Ivics and colleagues is referred to as SB10 [12]. However, several improved versions of the SBase have since been described that show increased transposition rates in some assays [18-20].
The SB transposon, the second part of the SB system, is essentially a DNA fragment flanked by the inverted repeats (IRs) (Figure 1). A specific left (IRL) and right (IRR) inverted repeat, each having a distinct sequence, are required for efficient transposition [21]. Each IR contains two direct repeats that function as the binding sites for the SBase. The DNA cargo for SB transposons varies depending on the application. However, the transposition rate is inversely correlated to transposon size, with an optimal transposon size of roughly 2 kilobases [19]. As with the SBase enzyme, improvements to the SB transposon have also been made to improve the efficiency of transposition [21].
The initial studies to use the SB system in mice generated transgenic strains to express the SBase ubiquitously [13, 15] or specifically in the mouse germ line [14]. These strains were then bred to mice that carried one or more copies of an SB transposon as a transgene concatomer to produce double transgenic males. Transposition events within the male germ line were then obtained by simply breeding the double transgenic male to a wild type female mouse. In all cases, a moderate rate of transposition was observed with each gamete from male double transgenic mice inheriting 1 or 2 de novo transposon insertions [13-15].
The moderate rate of transposition observed in these initial experiments was insufficient to make germ line transposon mutagenesis a practical approach in mice. However, a low level of transposition could be adequate for other applications. For example, a moderate transposition rate could be sufficient to induce tumors in mice if a large population of cells were mutagenized over an extended period of time. While the transposon mutagenesis rate might be low, the strong positive growth selection that drives tumor formation would allow a rare population of cells to be identified that harbor mutations in cancer genes.
The application of SB mutagenesis for cancer gene discovery required several considerations. First, prior work had shown that cellular transformation requires many independent gene mutations. Thus a tumor induced by SB mutagenesis would require multiple transposons to generate the required number of gene mutations. Second, an SB transposon used to induce tumors must be capable of mutating both tumor suppressors and proto-oncogenes, as mutations in both types of cancer genes are likely required to transform a cell. Therefore, the transposon used for this application needs to produce gain- and loss-of-function mutations. If enough copies of this type of mutagenic transposon could be mobilized by SBase in the somatic cells of mice, the resulting tumors would harbor transposon-tagged mutations that could easily be identified using a variety of molecular approaches.
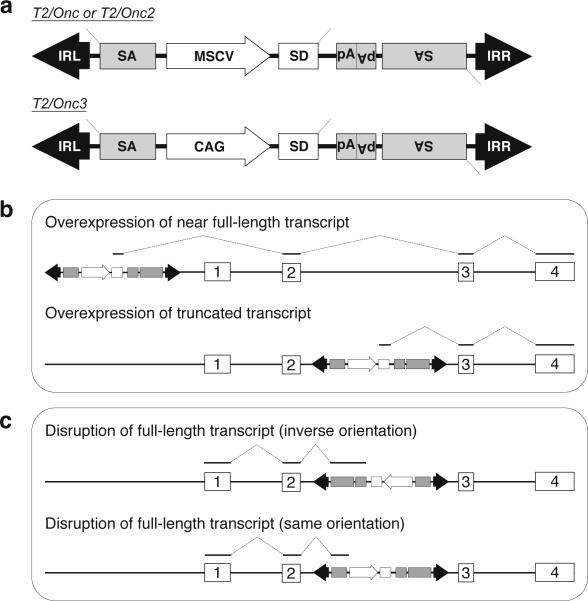
In 2005, several publications demonstrated the feasibility of this approach [22-23]. First, a mutagenic transposon was constructed that mimicked the activity of a slow transforming retrovirus (Figure 2a). This transposon, called T2/Onc, contained splice acceptors and polyadenylation sequences on both strands. These sequences allow the transposon to function as a bi-directional gene trap capable of truncating tumor suppressor gene transcripts when inserted in either the forward or reverse orientation (Figure 2c). A promoter and splice donor cassette was also included in the T2/Onc transposon. The combination of these sequences serve to initiate transcription and splice into downstream exons to drive over expression of oncogenes (Figure 2b). However, the transposon did not encode any proteins, but instead was designed to induce tumors only by mutating endogenous genes.
Figure 2.
Structure and function of mutagenic transposon vectors used in SB-induced models of cancer. (a) Two different transposon vectors have been generated to induce tumors when mobilized in the somatic cells of mice. The promoter (MSCV or CAG) together with the splice donor (SD) can cause overexpression of downstream oncogenes. The splice acceptors (SA) and polyadenylation sites (pA) included on both strands of the transposons allow it to function as a gene trap to disrupt expression of tumor suppressor genes. (b) The mutagenic transposons are capable of driving oncogene overexpression in SB-induced tumors via two main mechanisms in which the T2/Onc transposon expresses a near full-length (above) or truncated (below) transcript. These mechanisms employ the MSCV or CAG promoter along with the splice donor within the transposon. (c) Transposon-induced tumor suppressor gene disruption can be achieved through the action of the gene trap elements (e.g. splice acceptor, polyA) on the plus strand (above) or minus strand (below), depending on the orientation of the transposon relative to the mutated gene.
Work by Collier et al. produced transgenic mice harboring multiple copies of the T2/Onc transposon [22]. These mice were then bred to transgenic mice ubiquitously expressing the SBase. Unfortunately, the resulting double transgenic mice did not show signs of spontaneous tumor formation as predicted, although transposition of the T2/Onc transposon could be observed in the tissues of double transgenic mice. This suggested that the transposon mutagenesis rate was not high enough to drive cellular transformation. However, in a subsequent experiment, Collier et al. showed that transposon mutagenesis could dramatically accelerate sarcomas that developed in mice that were deficient for the p19 tumor suppressor. Analysis of the sarcomas showed that clonal transposon insertions in the Braf locus were present in nearly 80% of tumors, suggesting that transposon-induced mutations had been selected during tumor formation. This was a significant achievement since insertional mutagenesis had not been used previously to study the genetics of sarcomas.
In a separate series of experiments, Dupuy et al. further modified the SB system to improve the mutagenesis rate to more efficiently drive tumor formation [23]. First, a strain of knock-in mice (RosaSBase) was generated to express the SBase from the ubiquitously expressed ROSA26 locus. This allele provides more stable SBase expression by avoiding the epigenetic silencing experienced by some transgenes. An additional improvement was the use of an improved SBase in generating the RosaSBase allele. Transgenic mice were also produced that harbor a mutagenic transposon, called T2/Onc2, that is virtually identical to that of T2/Onc used by Collier et al. However, T2/Onc2 transgenic mice were generated that carry 150-300 copies of the T2/Onc2 transposon, in contrast to 20-30 transposon copies found in the T2/Onc transgenic mice. The combination of a more stable expression of an improved SBase along with an increased number of transposons should produce a higher transposition rate in vivo.
Double transgenic mice were generated to test the efficiency of the new RosaSBase and T2/Onc2 alleles. The result indicated that transposition rates were high enough to induce embryonic lethality among the majority of double transgenic mice [23]. However, a small number of these animals survived and were aged to determine if spontaneous tumors would form. Surprisingly, all double transgenic mice developed aggressive lymphomas within 10 weeks of age. As was seen in SB-induced sarcomas, these lymphomas harbored clonal transposon insertions, although transposon-induced mutations in the Notch1 locus were most common in lymphomas. Unlike the SB model of sarcoma, the lymphoma model described by Dupuy et al. did not require a sensitizing mutation, such as p19 deficiency, to induce tumors. This work showed that the SB system could drive tumor initiation as well as progression, under some circumstances.
While these initial publications demonstrated the feasibility of using the SB system to model cancer in mice, there were several significant limitations that needed to be overcome. First, the work of Collier et al. suggested that SB mutagenesis can be used to study a wider variety of tumor types than can be studied using retroviral insertional mutagenesis. However, the SB model described by Collier et al. was only capable of accelerating tumors in mice that were genetically predisposed to develop tumors [22]. This was not the case for the SB model described by Dupuy et al., as double transgenic mice rapidly developed spontaneous tumors. Unfortunately, the tumors were almost exclusively T-cell lymphomas, and the majority of the double transgenic died during embryonic development [23]. Therefore, the SB system required further modification to provide greater flexibility to model cancer in mice.
Controlling transposition to produce specific tumor models
Given its simplicity, the possibilities for modification of the SB system involve either altering the SBase enzyme or the structure and/or copy number of the mutagenic transposon. Several different approaches have been used, and all of these have had some success in altering the behavior of the SB system to generate more diverse cancer phenotypes in mice.
Alteration of SBase allele
The most straightforward manner in which SB mutagenesis can be controlled is to provide tissue-specific expression of the SBase enzyme. In this way, transposon mutagenesis will occur only in sites where SBase is expressed. Two different approaches could be used to achieve this goal. First, the SBase could be directly expressed from a tissue-specific promoter in transgenic or knock-in mice. This would require the production of many different mouse strains to drive SBase expression in a variety of tissue or cell types. A second approach could make use of the Cre/loxP system to generate a Cre-inducible SBase allele. This would be achieved by interrupting SBase expression from a ubiquitous promoter with a silencing cassette flanked by loxP sites. This lox-stop-lox (LsL) cassette can then be deleted by Cre recombinase, thus restoring expression of SBase. Such an SBase allele could then be activated by one of the many tissue-specific Cre transgenic mouse strains that have been developed.
The latter approach was used in a subsequent study to produce a lox-stop-lox SBase allele [24]. This was accomplished by inserting a floxed stop cassette consisting of an EGFP cDNA followed by three polyadenylation sites upstream of the SBase cDNA to produce the RosaSBase-LsL allele. In the absence of Cre recombinase, this allele expresses EGFP instead of the SBase. Thus, transposition does not occur in double transgenic mice carrying the RosaSBase-LsL allele along with a mutagenic transposon transgene, and these mice are not predisposed to form tumors. However, once Cre recombinase deletes the upstream floxed stop cassette, the downstream SBase cDNA is expressed and transposition is initiated. Thus, the RosaSBase-LsL allele can be used to perform tissue-specific transposon mutagenesis in mice in a Cre-dependent manner.
Several studies have already demonstrated the flexibility of this Cre-dependent SB system. First, Dupuy and colleagues showed that activation of transposition using a B-cell-specific Cre transgenic strain produced predominantly B-cell malignancies [24]. Starr et al. induced transposon mutagenesis using Cre recombination in the intestine to produce a model of colorectal cancer [25]. A mouse model of hepatocellular carcinoma was also produced by Keng et al. using liver-specific transposon mutagenesis [26]. These experiments provide an important demonstration of the flexibility of the SB system to model cancer in mice, as the tumors types induced using Cre-dependent transposon mutagenesis were not frequently observed in the initial SB-induced cancer models.
Modification of mutagenic transposons
The use of the RosaSBase-LsL allele shows that controlling the expression of the SBase can be used to generate tissue-specific models of cancer in mice. However, modifications to the mutagenic transposon have also been shown to alter the tumor phenotype in mice [24]. Dupuy and colleagues speculated that the tissue specificity of the promoter used in both the T2/Onc and T2/Onc2 transposons contributes to the tumor phenotype by determining the relative strength of the overexpression alleles produced in various tissues undergoing transposon mutagenesis. Since both initial transposons contain the murine stem cell virus (MSCV) promoter, the strongest overexpression alleles are most likely produced in hematopoietic cells, and this could explain the tendency for the SB system to induce lymphomas.
This hypothesis was tested by developing a new mutagenic transposon, called T2/Onc3, in which the MSCV promoter was replaced with the CMV enhancer/chicken beta-actin promoter (CAG) (Figure 2). The CAG promoter was chosen as it had been shown to be expressed at high levels in epithelial cells, but only weakly expressed in hematopoietic cell lineages. Transgenic mice harboring 10-20 copies of the T2/Onc3 transposon were then bred to the ubiquitous RosaSBase allele. Double transgenic mice did not show signs of embryonic lethality previously seen using high copy T2/Onc2 transposon transgenic strains. Moreover, mice that underwent ubiquitous T2/Onc3 mutagenesis developed a wide variety of carcinomas [24]. This result shows that direct modification of the mutagenic transposon can dramatically affect the tumor phenotype.
Another conclusion from the T2/Onc3 mutagenesis experiment is that the tumor phenotype produced in mice using SB mutagenesis is likely the result of the combined effects of the SBase expression and the properties of the mutagenic transposon. Therefore, using different combinations of existing transposon and transposase alleles could produce novel tumor phenotypes. This idea was recently tested by Collier and colleagues by generating novel combinations of SBase and transposon alleles [27]. This work was prompted by the discovery that the SBase transgenic mice initially used to study sarcomas was inefficiently expressed in most tissues. Therefore, the RosaSBase knock-in allele was combined with the low copy T2/Onc transposon allele to generate double transgenic mice. This combination of alleles drove transposon mutagenesis at a high enough rate to generate spontaneous tumors in wild type mice, but not high enough to induce embryonic lethality. Furthermore, while the double transgenic mice did develop lymphomas, a significant number developed spontaneous astrocytomas — a tumor type not commonly seen in previous SB models of cancer. This suggests that different combinations of the existing SBase and transposon alleles could modify the tumor phenotype in mice.
Currently, three different SBase alleles along with seven mutagenic transposon alleles have been described in the literature (Table 1). These studies indicate that several important factors affect the tumor phenotype produced by SB mutagenesis: 1) efficiency and pattern of SBase expression, 2) mutagenic transposon structure and 3) transposon copy number. These alleles have been used to generate a wide variety of tumor types in mice. However, the development of new SBase or transposon alleles may be required to produce some mouse models of human cancer.
Table 1.
Overview of mouse tumor models generated with the SB system. The current mutagenic transposon strains are indicated in the left column, and the three SBase strains are indicated at the top of each column. The tumor phenotype is indicated for each combination of transposon and SBase strain. For instance, the combination of T2/Onc and RosaSBase produces T-cell lymphoma and astrocytoma in mice.
| X | CAGGS-SB10 | RosaSBase | RosaSBase-LsL |
|---|---|---|---|
| T2/Onc (low copy) | sarcoma (in p19-null mice) | T-cell lymphoma, astrocytoma | gastrointestinal tumors hepatocellular carcinoma |
| References: | [26] | [31] | [29-30] |
| | |||
| T2/Onc2 (high copy) | not done | embryonic lethality, T-cell lymphoma, medulloblastoma | follicular lymphoma, diffuse large cell lymphoma |
| References: | [27] | [28] | |
| | |||
| T2/Onc3 (low copy) | not done | ~20 tumor types, mostly carcinoma | not done |
| References: | [28] | ||
Identification of driver mutations in SB-induced tumors
The goal of modifying the SB system to induce tumors in mice is to facilitate the discovery of cancer genes. A significant portion of the transposon sequence, including the inverted repeats, is divergent from the mouse genome. Primers targeting these unique sequences have been used in a variety of ligation-mediated PCR (LM-PCR) approaches to specifically amplify genomic fragments that contain a transposon insertion [28]. The DNA sequence of these PCR products can be obtained and subsequently used to map the transposon insertion to the mouse reference genome. Once the transposon insertion site is known, the gene(s) most likely affected by the transposon can be determined. A comprehensive view of the genetic events that contributed to an individual SB-induced tumor can then be obtained by mapping all transposon insertions found within the tumor.
The number of independent transposon insertion events presents a major challenge in the identification of transposon-induced mutations. In some cases, more than 20 independent clonally expanded insertion events can be detected by Southern blotting in SB-induced tumors [23]. An LM-PCR process should identify all of these events, in addition to many subclonal transposon insertion sites. Given the significant number of expected PCR products from each SB-induced tumor, traditional methods to isolate and sequence these products are not feasible. Instead, the initial studies that identified transposon insertion sites in SB-induced tumors used a shotgun cloning strategy to produce a plasmid library of the PCR products obtained from each independent tumor [22-23]. Sequencing individual clones from each library then identified the transposon-genomic DNA junctions found within each tumor.
The shotgun cloning strategy had several limitations. First, the LM-PCR procedure generated two plasmid libraries for each SB-induced tumor. Each library contained the transposon-genomic DNA junctions from either the right or left transposon inverted repeat. This was done to ensure adequate representation of the transposon insertion sites, as some transposon-genomic DNA junctions cannot be uniquely mapped due to insufficient fragment length or the presence of repetitive sequences. However, the production of the plasmid libraries was laborious. A second limitation of this approach was the significant cost of DNA sequencing, and this limited the number of independent clones that could be sequenced. As a result, only a modest number of independent transposon insertion events were identified in each tumor.
The development of next generation sequencing technologies has dramatically changed the process of identifying transposon insertion sites. Recently, a novel method was described that generates LM-PCR products that can be directly sequenced on the Genome Sequencer FLX platform (Roche/454) [24-26]. This machine is capable of simultaneously sequencing ~500 000 individual LM-PCR products, eliminating the need to produce plasmid libraries of the LM-PCR products. The significant improvement in sequence coverage also identifies hundreds of independent transposon insertion sites in each SB-induced tumor, thus producing a more comprehensive view of the transposon-induced mutations found in each sample.
While the use of next-generation DNA sequencing has greatly increased the efficiency of transposon insertion site identification, it has also complicated the interpretation of the data obtained from SB-induced tumors. As previously discussed, the identification of driver mutations in human tumors among the more abundant passenger mutations is a major challenge in the field of cancer genetics. The same challenge also exists in deciphering the genetic complexity of SB-induced tumors, given the hundreds of independent transposon insertion events that can now be readily detected in each sample.
Fortunately, transposon-induced driver mutations can be more easily detected due to the predictable activity of the SB system. Integration site bias is often a concern when performing insertional mutagenesis. Integrating viruses, such as MuLV and HIV, show a preference for insertion near genes [29]. By contrast to these vectors, several studies have shown that the SBase does not display such an integration site bias when inserting transposons into the genome [23, 30-31]. Instead, SB transposons show a distribution pattern that appears nearly random. This is a significant advantage for two reasons. First, SB insertional mutagenesis will likely generate more diverse types of mutations in a larger number of genes than will a retrovirus that displays significant insertion site bias. This provides the opportunity to identify a larger number of cancer genes. The second advantage is that the significance of any transposon-induced mutation can be assessed by simply determining if the frequency of transposon insertion within that gene occurs at a rate higher than expected, assuming a random distribution.
However, there is one type of SB transposon insertion event that is decidedly non-random, and that is the local hopping event. The SB system, like other cut-and-paste transposons, shows a preference for insertion within a region linked to the transposon donor site. This phenomenon, referred to as local hopping, has been repeatedly observed in studies in which SB transposons were mobilized in the mouse germ line [13-15]. This bias is also seen in tumors induced with the SB system [22-23]. However, the frequency and the size of the genetic interval affected by local hopping varied significantly in each study, and this variation has prevented a specific definition of local hopping for the SB system from being developed. Nevertheless, local hopping must be taken into account when determining the significance of any transposon-induced mutation in SB-induced tumors, since genes linked to the transposon transgene will acquire insertion events more often than genes that are found on different chromosomes.
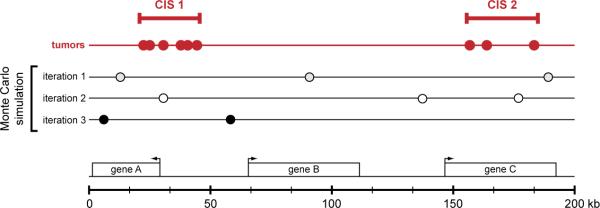
Given the predictable nature of SB-induced transposition, this process can easily be simulated using computational methods. Several approaches have been used to identify driver mutations in tumors produced by insertional mutagenesis. Monte Carlo simulation has most frequently been employed to identify driver mutations in SB-induced tumors. This approach relies on repeated random sampling to determine if the observed number of transposon insertions within any region of the genome is greater than expected [24-26]. The result of the Monte Carlo simulation establishes a series of definitions that can be used to identify common insertions sites (CISs). For insertional mutagenesis screens in mouse models of cancer, CISs are generally thought to be the driver mutations that were selected during the process of cellular transformation. The Monte Carlo simulation is used to identify CISs by determining the size of the genetic interval that contains a given number of independent insertion events (i.e. found in different tumor samples) at an expected frequency of less than 5% (Figure 3). Any region of the mouse genome that contains either a greater number of transposon insertion events or the same number of insertion events in a smaller genetic interval is defined as a CIS.
Figure 3.
Identification of common insertion sites in SB-induced tumors. High throughput PCR-based methods have recently been developed to identify a large number of transposon-induced mutations in individual SB-induced tumors. The driver mutations causally linked to transformation are identified as common insertion sites (CISs) — regions of the genome that harbor transposon insertions in multiple independent tumors. Currently, a Monte Carlo simulation is used to model random transposon insertion within the mouse genome to mimic the number of tumors and insertion events observed within each experiment. The cumulative results of thousands of interations of the Monte Carlo simulation are used to identify non-random clusters of transposon insertions in the tumor data set. These regions are then defined as CISs. The figure graphically depicts this analysis to identify CISs within a region of the genome containing three hypothetical genes (geneA-C) shown below. Each circle represents an independent transposon insertion in this 200 kilobase interval. The results of three iterations of the Monte Carlo simulation are shown, and these results are used to identify CISs in the tumor data set (shown in red).
Kernel convolution is another computational method for CIS identification that has recently been developed and modified to evaluate data from SB-induced tumors [32]. This approach places a Gaussian kernel function at each insertion site. The expected number of insertion events at any particular genomic location can then be estimated by summing all kernel functions within the data set. One advantage of the kernel convolution method is capable to evaluating integration data across multiple scales simultaneously. By contrast, the Monte Carlo method can identify CISs that contain a user-specified number of independent insertion events. The ability to evaluate multiple scale spaces (i.e. narrow or wide clusters of insertion events) allows the kernel convolution method to identify a greater number of candidate CISs in a single analysis.
It should be noted that the kernel convolution method has only recently been adapted to evaluated transposon insertion data from SB-induced tumors [33]. Additional work is needed to compare and contrast this approach with results obtained using the Monte Carlo method to determine the relative strengths and weaknesses of both methods. Furthermore, additional computational methods could also be developed to address any weaknesses shared by the existing approaches. Most likely, the interpretation of insertion site data from SB-induced tumors will involve multiple bioinformatic approaches, as with other forms of high throughput data analysis (e.g. expression arrays, CGH).
Regardless of the method used to identify CISs in SB-induced tumors, the effects of local hopping must be considered. This is important, since local hopping would otherwise lead to the identification of many false positive CISs in regions that are linked to the initial transposon transgene. Currently, the only way to adequately eliminate false positive CISs caused by local hopping is to disregard all transposon insertion events that map to the chromosome containing the transposon transgene — referred to as the “local chromosome”. While this is a conservative approach that likely leads to false negative results for some loci on the local chromosome, the number of false positive CISs eliminated by this approach is likely far greater than the number of false negative CISs. In addition, all SB cancer screens performed to date have used at least two independent transposon transgenes that map to different chromosomes, thus allowing the local chromosome in one strain to be assessed in the other.
To date, roughly 300 genes have been identified as CISs in only a few tumor types using the SB system. While the significance and relevance to human cancer has yet to be determine for most of these genes, preliminary results suggest that SB models of cancer will be useful in identifying novel cancer genes. For example, Notch1 is the most frequently mutated gene in two independent models of SB-induced T-cell lymphoma, in which transposon insertions produce gain-of-function Notch1 alleles [23, 27]. A similarly high rate of gain-of-function mutation is also seen in human T-cell acute lymphoblastic leukemia patients, though these alleles are produced by somatically acquired point mutations in the NOTCH1 gene [34]. In addition, Starr et al. recently showed that frequent disruption of the Apc locus is caused by transposon insertion in an SB-model of colorectal cancer [25]. Both spontaneous and inherited mutations in the APC locus are causally linked to colorectal cancer in humans. Examples such as these are an important proof of principle showing that SB models of cancer will be useful in identifying novel cancer genes.
Current challenges and future applications of SB in mouse cancer models
The use of the SB system to study cancer in mice is a relatively new application of transposon mutagenesis. However, the re-identification of a number of validated human cancer genes (e.g. Notch1, Apc, Braf) in SB-induced tumors validates this approach. This result also suggests that some of the novel candidate cancer genes identified in these screens will likely play a role in human cancer. Nevertheless, the broader application of SB mutagenesis in mouse cancer models still faces several challenges. The degree to which these challenges can be addressed will determine the extent to which the SB system can contribute to our understanding of cancer genetics.
As previously discussed, retroviral insertional mutagenesis has been a powerful tool that has been applied to the study of leukemia and mammary cancer in mice. The inability of retroviruses to induce tumors in other tissue types is, in part, what prompted the development of the SB system for this application. While the SB system has recently been shown to be capable of inducing a wide array of tumor types in the mouse, there are several notable exceptions. For example, cancers of the lung, mammary gland, prostate and pancreas, among others, have not yet been generated with the SB system. However, it is unclear if this failure is due to the inability of the SB system to function in these tissues, or if it is due to the lack of the appropriate tools to model these tumor types in mice. The latter is most certainly the case for mammary cancer since ubiquitous transposition of the T2/Onc3 transposon is capable of inducing spontaneous mammary tumors [24]. However, activation of the Cre-inducible SB system using mammary-specific Cre expression did not efficiently produce mammary tumors (our unpublished data). Therefore, the use of the Cre-inducible SB system may require additional optimization to produce tissue-specific tumor models. In some cases, the existing Cre transgenic strains for a given tissue may not be expressed efficiently, may not be expressed in the correct cell type within the tissue, or may not be expressed at the appropriate developmental time point to induce spontaneous tumors. In these situations, new Cre strains may need to be developed and tested to produce SB models of some specific forms of cancer.
It should also be noted that all existing SBase mouse strains do not provide the ability to shut off transposase expression once it has been established. As a consequence, transposition can occur throughout the lifetime of a mutagenized animal. However, a novel SBase allele controlled by an inducible system, such as the Tet expression system, would provide the ability to provide controlled bursts of SBase expression. This would facilitate a number of experimental approaches not currently possible using constitutive SBase expression. For example, cell lineage tracing studies could be performed to study various aspects of normal development or cancer biology using transposon tagging.
Another significant challenge involves the bioinformatic analysis of genetic data derived from SB-induced tumors. As previously discussed, several approaches have already been developed to identify CISs (i.e. driver mutations) in tumors induced by insertional mutagenesis. However, there are no well-established methods to compare insertional mutagenesis data derived from mouse tumors to data obtained from human tumors. This type of analysis is particularly challenging when performing cross-species comparisons of large data sets obtained from tumors induced by distinctly different mechanisms — insertional mutagenesis or spontaneous mutation. Nevertheless, a recent study by Mattison et al. suggests that this sort cross-species comparison will be informative [33].
Despite some challenges in generating tissue-specific models of cancer, over 20 tumor types have already been produced using the SB system. There are a number of potential applications of the SB system for these forms of cancer. For example, there is increasing interest in not only identifying driver mutations in tumors but also understanding how the combination of driver mutations within an individual tumor collectively contribute to transformation. The SB system can facilitate this type of analysis. Cooperating oncogenic mutations can be identified by determining which combinations of transposon-induced mutations in SB-induced tumors are found more often than predicted. A similar strategy has already been employed in retroviral insertional mutagenesis models of T-cell lymphoma [35]. The identification of these co-selected driver mutations could provide insight into a novel biological mechanism within the tumor that could be targeted with novel therapeutic agents.
The most obvious application of the SB system is the identification of novel cancer genes. However, SB mutagenesis could also be used to study genetic mechanisms of cancer-relevant phenotypes. One example of this would be the identification of mutations that provide resistance to chemotherapeutic agents. Such an experiment could be performed by treating mice that have developed SB-induced tumors with a drug. Tumors that proliferate in the presence of the drug could have mutations that provide resistance. Comparisons between tumors that show differential responses to drug treatment could identify candidate mutations that provide drug resistance. Another approach that could be used to study acquired drug resistance is to perform insertional mutagenesis in tumors that are initially sensitive to a specific drug and isolate resistant tumor clones that emerge in the presence of the drug. These resistant clones will have acquired insertional mutations that provide resistance. An example of this approach was recently described by Lauchle and colleagues in which retroviral insertional mutagenesis was used to identify a resistance mutation in a mouse model of leukemia [36].
Tumor metastasis is perhaps the single most significant challenge in treating human cancer. In most cases, treatment options for advanced metastatic disease are limited. Recent work has suggested that genetic selection could play a role in driving some aspects of tumor metastasis [37-38]. If this is the case, the SB system could be used to identify mutations that play a role in metastasis. Recent work by Keng and colleagues demonstrated that SB mutagenesis can drive production of metastatic hepatocellular carcinoma [26]. Interestingly, analysis of the transposon-induced mutations in both the primary and metastatic liver tumors showed that the tumors had a common origin. However, the metastatic tumors had acquired novel transposon-induced mutations that were not detected in the primary tumor. Additional work is required to determine if the metastasis-specific transposon-induced mutations directly contributed to the metastatic phenotype. Nevertheless, this result suggests that SB mutagenesis screens could be performed to determine if metastasis-specific CISs can be identified. The identification of recurrent gene mutations in metastatic tumors would support the notion that genetic selection contributes to tumor metastasis. Functional characterization of metastasis-specific gene mutations would provide significant insight into the biological mechanisms that drive metastasis, and perhaps lead to the development of novel drugs that target these processes specifically.
It should be noted that the adaptation of the SB system to study cancer in mice is a recent development. New observations are continuously made that provide more information about how to construct SB-induced models of cancer and to interpret the genetic data derived from these models. Given the early successes, the SB system will continue to evolve, as new applications and mouse strains are developed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 4.Bittner JJ. Some Possible Effects of Nursing on the Mammary Gland Tumor Incidence in Mice. Science. 1936;84:162. doi: 10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 5.Furth J, Seibold HR, Rathbone RR. Experimental studies on lymphomatosis of mice. Amer. J. Cancer. 1933;19:521–604. [Google Scholar]
- 6.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–72. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 7.Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer. 2009;9:389–99. doi: 10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- 8.Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, Jonkers J, et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39:759–69. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 9.Bazopoulou D, Tavernarakis N. The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans. Genetica. 2009;137:39–46. doi: 10.1007/s10709-009-9361-3. [DOI] [PubMed] [Google Scholar]
- 10.Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239:1121–8. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- 11.Ross-Macdonald P, Coelho PS, Roemer T, Agarwal S, Kumar A, Jansen R, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–8. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 12.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–10. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 13.Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis. 2001;30:82–8. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- 14.Fischer SE, Wienholds E, Plasterk RH. Regulated transposition of a fish transposon in the mouse germ line. Proc Natl Acad Sci U S A. 2001;98:6759–64. doi: 10.1073/pnas.121569298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horie K, Kuroiwa A, Ikawa M, Okabe M, Kondoh G, Matsuda Y, et al. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc Natl Acad Sci U S A. 2001;98:9191–6. doi: 10.1073/pnas.161071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izsvak Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem. 2002;277:34581–8. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- 17.Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–61. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 18.Baus J, Liu L, Heggestad AD, Sanz S, Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther. 2005;12:1148–56. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- 19.Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–17. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 20.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–61. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 21.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–35. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 22.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–6. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 23.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–6. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 24.Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–6. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–50. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–74. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collier LS, Adams DJ, Hackett CS, Bendzick LE, Akagi K, Davies MN, et al. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res. 2009;69:8429–37. doi: 10.1158/0008-5472.CAN-09-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Largaespada DA, Collier LS. Transposon-mediated mutagenesis in somatic cells: identification of transposon-genomic DNA junctions. Methods Mol Biol. 2008;435:95–108. doi: 10.1007/978-1-59745-232-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Q, Kong J, Stalker J, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis. 2009;47:404–8. doi: 10.1002/dvg.20508. [DOI] [PubMed] [Google Scholar]
- 31.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–94. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol. 2006;2:e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattison J, Kool J, Uren AG, de Ridder J, Wessels L, Jonkers J, et al. Novel candidate cancer genes identified by a large-scale cross-species comparative oncogenomics approach. Cancer Res. 2010;70:883–95. doi: 10.1158/0008-5472.CAN-09-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng AP, Ferrando AA, Lee W, Morris J. P. t., Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 35.Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–41. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauchle JO, Kim D, Le DT, Akagi K, Crone M, Krisman K, et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461:411–4. doi: 10.1038/nature08279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–12. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 38.Talmadge JE. Clonal selection of metastasis within the life history of a tumor. Cancer Res. 2007;67:11471–5. doi: 10.1158/0008-5472.CAN-07-2496. [DOI] [PubMed] [Google Scholar]