Abstract
Caldesmon is an actin- and myosin-binding protein found in smooth muscle that inhibits actin activation of myosin ATPase activity. The activity of caldesmon is controlled by phosphorylation and by binding to Ca2+-calmodulin. We investigated the effects of phosphorylation by p21-activated kinase 3 (PAK) and calmodulin on the 22 kDa C-terminal fragment of caldesmon (CaD22). We substituted the major PAK sites, Ser-672 and Ser-702, with either alanine or aspartic acid to mimic nonphosphorylated and constitutively phosphorylated states of caldesmon, respectively. The aspartic acid mutation of CaD22 weakened Ca2+-calmodulin binding but had no effect on inhibition of ATPase activity. Phosphorylation of the aspartic acid mutant with PAK resulted in the slow phosphorylation of Thr-627, Ser-631, Ser-635, and Ser-642. Phosphorylation at these sites weakened Ca2+-calmodulin binding further and reduced the inhibitory activity of CaD22 in the absence of Ca2+-calmodulin. Phosphorylation of these sites of the alanine mutant of CaD22 had no effect on Ca2+-calmodulin binding but did reduce inhibition of ATPase activity. Thus, the region between residues 627 and 642 may contribute to the overall regulation of caldesmon's activity.
Introduction
Regulation of myosin-based motility can be achieved by modifying either myosin or its cofactor, actin. Several actin-binding proteins inhibit actin activation of myosin ATPase activity. Troponin-tropomyosin confers Ca2+ sensitivity to striated muscle (1). Other actin-binding proteins, such as caldesmon (2), calponin (3,4), and fesselin/synaptopodin 2 (5), may inhibit actin-dependent movement in various cells.
A criterion for a biological function of these actin-binding proteins is that their inhibitory effects must be reversible. Ca2+-calmodulin reverses the effects of caldesmon (2,6) and calponin (4) on actin-activated ATPase activity. Ca2+-calmodulin also regulates the ability of synaptopodin 2 to promote actin polymerization (7). Phosphorylation by mitogen-activated protein (MAP) kinase (8) and p21-activated kinase (PAK) (9) has been reported to attenuate the inhibitory effects of caldesmon. Phosphorylation of other actin-binding proteins has been well documented (10,11).
Multiple signals, such as phosphorylation and ligand binding (i.e., Ca2+-calmodulin), may work synergistically by altering the structure of some of these actin-binding proteins. Several actin-binding proteins are known to have unfolded regions that are often targets of signaling. Unfolded regions of troponin I have been proposed to be involved in actin binding and inhibition of muscle contraction (12). Both CaD22 (13) and fesselin (14) are largely unfolded in solution. Ca2+-calmodulin binding to a COOH-terminal caldesmon fragment induces folding (13). Because the COOH-terminal region of caldesmon that regulates actomyosin ATPase activity is unfolded and is regulated by Ca2+-calmodulin and phosphorylation, we wished to explore how multiple signaling events affect its functions.
Phosphorylation by PAK was investigated here because phosphorylation of intact l-caldesmon occurs in vivo (15). The sites of phosphorylation of h-caldesmon are Ser-672 and Ser-702 (9,15). Constitutively active PAK3 produces Ca2+- independent contraction of smooth muscle (16). PAK kinase is also a major regulator of caldesmon-mediated actin dynamics in vivo (15,17).
We observed that PAK phosphorylated four additional residues within the region 627–642 of caldesmon. Phosphorylation at these sites alone had little or no effect on CaD22 binding to Ca2+-calmodulin. However, phosphorylation at these sites did enhance the effect of phosphorylation of Ser-672 and -702 (or on aspartic acid substitutions at sites 672 and 702) on weakening CaD22 binding to Ca2+-calmodulin. In addition, we found that phosphorylation at the minor sites alone partially reversed the inhibitory activity of CaD22. We made the latter observation by blocking phosphorylation at residues 672 and 702 using alanine substitutions at those positions. Because of these different effects, we observed that the effect of Ca2+-calmodulin on the activity of caldesmon is dependent on the combination of phosphorylated residues. The interplay between signals resulting in phosphorylation and Ca2+-calmodulin binding may be quite complex.
Materials and Methods
Proteins
Actin and myosin were isolated from rabbit back muscle (18,19). Myosin subfragment 1 was made by digestion of myosin with chymotrypsin (20). Recombinant Arabidopsis calmodulin (pRZ72, a gift from Dr. Ralf Zielinski, University of Illinois at Urbana-Champaign, Urbana, IL) was expressed in Escherichia coli BL21 cells and prepared as described by (21). Smooth muscle tropomyosin was isolated from turkey gizzards (22). A 22 kDa C-terminal wild-type (WT) fragment (Met-563–Pro-771) and 35 kDa C-terminal WT fragment (Met-461–Pro-771) of Gallus gallus caldesmon (NCBI Protein Data Bank BAA04539), as well as the corresponding double alanine and double aspartic acid mutants, were subcloned into a pSBET-A expression vector (gift of H. H. Steinbiss, Max Plank Institute for Plant Breeding Research, Cologne, Germany) expressed in E. coli BL21 cells and purified according to Fredricksen et al. (23). PAK (GST-mPAK3, a gift from Dr. Alan Mak, Queen's University, Kingston, Canada) was expressed in E. coli BL21 cells and prepared as described previously (16). The concentrations of the different CaD22 fragments were determined by means of a Lowry assay with bovine serum albumin as a standard. The concentrations of other proteins were determined by absorbance at 280 nm and corrected for scattering at 340 nm using the following extinction coefficients E2800.1%: actin, 1.15; smooth muscle tropomyosin, 0.22; myosin S1, 0.75; and calmodulin, 0.19. The molecular masses assumed for the key proteins were myosin S1, 120,000 Da; actin, 42,000 Da; smooth muscle tropomyosin, 68,000 Da; CaD22, 22,000 Da; CaD, CaD35, 35,000 Da; and calmodulin, 16,800 Da.
The DNA sequence of the mutants was confirmed by cycle sequencing with the ABI prism big dye terminator cycle sequencing ready reaction kit. The products were resolved on an ABI prism 377 automated sequencer (Perkin-Elmer/Applied Biosystems, Waltham, MA). The expressed mutants were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MS).
Phosphorylation of caldesmon
CaD22 (generally 1 mg/mL) was phosphorylated by constitutively active GST-mPAK3 (∼10 μg/μL). The extent of phosphorylation was determined by nonequilibrium isoelectric focusing gel electrophoresis as described by Kobayashi et al. (24). Gels contained 8 M urea, 5% acrylamide, 0.8% ampholyte pH 3.0–10.0, and 1.2% ampholyte pH 8.0–10.0 (BioRad, CA). Sample loading buffer contained 8 M urea, 10 mM EDTA, and 1% ampholyte pH 3.0–10.0. The running times were 30 min at 100 V, 40 min at 200 V, and 20 min at 500 V. The gels were soaked in fixing solution containing 10% methanol and 5% acetic acid, and then stained with Coomassie Blue R350.
The time courses of phosphorylation were determined by measuring the incorporation of 32P into caldesmon fragments at various time intervals with a filter paper assay (25). Caldesmon fragments were phosphorylated by PAK at 25°C in 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, and 1 mM 32P-ATP (4×105 cpm/nmol). Aliquots were drawn at different times after initiation of phosphorylation and placed onto Whatman grade 3 filter circles (2.3 cm diameter). Filters were dropped immediately into ice-cold 10% TCA, 8% sodium pyrophosphate (w/v), followed by three subsequent washes for 10–15 min with 10% TCA, 2% sodium pyrophosphate at room temperature. The filters were then washed in 95% ethanol and finally in acetone. The 32P activity of the filters was determined by liquid scintillation counting.
The sites of phosphorylation were identified by MS. The CaD22 protein bands were excised from SDS gels, reduced, alkylated, and digested with modified trypsin (Promega). The peptides were analyzed by liquid chromatography-tandem MS (LC-MS/MS) using capillary high-performance liquid chromatography with a 75 μm nanocolumn and a Thermo Electron LTQ quadrupole ion trap mass spectrometer. The resulting masses and spectra were searched against a database using TurboSequest (26,27) with the Proteomics Browser interface (William Lane, Harvard Microchemistry and Proteomics Analysis Facility). Extracted ion chromatograms of full MS scans were performed on the m/z values that matched the phosphorylated peptides and corresponding nonphosphorylated peptides using a mass error window of ±5 ppm. The peptide identities of integrated peaks were verified based on retention times of the MS/MS spectra. Mass spectral analysis was performed at the Wistar Proteomics Facility, Philadelphia, PA.
ATPase rate measurements
Rates were determined by measuring the time dependence of 32Pi formation from γ32P-labeled ATP at 25°C. The concentrations of actin, smooth muscle tropomyosin, and myosin S1 were 10 μM, 2.2 μM, and 0.1 μM, respectively. The final solution composition was 1 mM ATP, 3 mM MgCl2, 31 mM NaCl, 10 mM imidazole pH 7.0. The reaction volume was 1 mL. Aliquots of 0.2 mL were removed at different time points and quantified as described previously (28). Four or more time points were taken to establish each rate.
Calmodulin binding
Ca2+-calmodulin binding was determined by changes in intrinsic tryptophan fluorescence of CaD22. Fluorescence measurements were made on an Aminco Bowman II luminescence spectrometer (Thermo Electron, Madison, WI). The cell compartment was maintained at 20°C with a circulating bath in a buffer containing 10 mM MOPS pH 7.2, 100 mM NaCl, 1 mM dithiothreitol, 2 mM CaCl2. Tryptophan fluorescence was measured with excitation and emission monochomators set to 295 nm and 340 nm, respectively. Low protein concentrations were used to avoid inner filter effects. Spectra were corrected for the fluorescence contribution from calmodulin in the same buffer solution and for wavelength-dependent lamp output. Because calmodulin lacks tryptophan residues, the correction for calmodulin was <5%.
Results
To study PAK regulation of CaD22, we modified the two major phosphorylation sites of chicken gizzard CaD22: Ser-672 and Ser-702. Serine-to-alanine substitutions were made to mimic the nonphosphorylated state (CaD22AA). The phosphorylated state was mimicked by replacing both serine residues with aspartic acid residues (CaD22DD). The presence of the correct mutations was verified by sequencing both plasmids. For CaD22WT the observed mass (22,625 Da) was consistent with the calculated mass (22,630 Da). Similar agreement was obtained for the observed mass (22,690 Da) and calculated mass (22,686 Da) of CaD22DD. In both cases the observed masses were within 0.02% of the calculated ones.
The presence of additional charges in the CaD22DD mutant was verified with the use of nonequilibrium isoelectric focusing gels. The theoretical pI values for CaD22 and CaD22DD are 9.7 and 9.56, respectively. Fig. 1 shows that CaD22DD (lane 2) migrated less toward the cathode than did CaD22WT (lane 1). The migration of the AA mutant was identical to the WT (not shown). CaD22WT, CaD22AA, and CaD22DD migrated similarly on SDS gels (data not shown).
Figure 1.
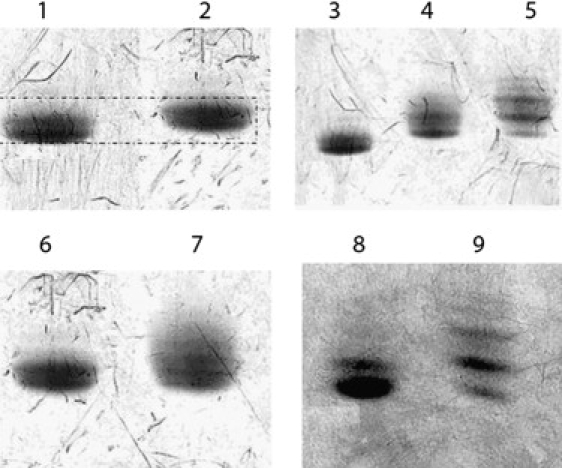
Nonequilibrium isoelectric focusing showing charge changes in CaD22 after mutations or PAK phosphorylation. Lane 1: CaD22WT; lane 2: CaD22DD; lane 3: CaD22WT; lane 4: CaD22WT PAK phosphorylated for 1.5 h; lane 5: CaD22WT PAK phosphorylated for 20 h; lane 6: CaD22DD; lane 7: CaD22DD after incubation for 20 h with PAK; lane 8: CaD22AA; lane 9: CaD22AA after incubation for 20 h with PAK. The cathode is at the bottom of each gel.
We examined the time course of PAK phosphorylation of CaD22WT by means of isoelectric focusing. Fig. 1 shows untreated CaD22WT (lane 3) and CaD22WT that was treated with PAK for 1.5 h (lane 4) and 20 h (lane 5). Untreated CaD22 focused as two closely spaced bands. The number of bands with lower isoelectric points increased with time of incubation. SDS gels showed that the CaD22WT remained intact during those periods of incubation (data not shown). These time-dependent shifts raised the possibility that more than two sites within CaD22WT became phosphorylated.
To test that possibility, we examined whether CaD22DD and CaD22AA could be phosphorylated by PAK. Fig. 1 shows CaD22DD before (lane 6) and after (lane 7) a 20 h incubation with PAK. Fig. 1 shows CaD22AA before (lane 8) and after (lane 9) a 20 h incubation with PAK. The additional acidic bands present in both cases indicate that additional phosphorylation sites were present.
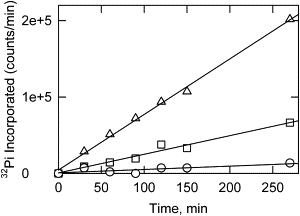
Fig. 2 shows the time courses of phosphorylation of CaD22WT and CaD22AA by PAK. CaD22WT was phosphorylated to yield 2 mol phosphate/mol CaD22 (open circles) in the first phase. Upon continued incubation, the caldesmon fragment slowly incorporated additional phosphate residues, yielding 3.3 mol phosphate/mol CaD22 after 23 h. When the study was repeated with CaD22AA having the two major PAK sites blocked, the first phosphorylation phase was absent, but the additional slow phosphorylation still occurred (solid circles).
Figure 2.
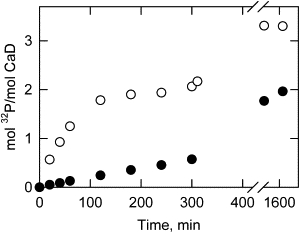
Two sets of sites on CaD22 were phosphorylated by extended treatment with PAK. The phosphorylation time courses were followed by 32Pi incorporation at 25°C in a buffer containing 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, and 1 mM 32P-ATP (4×105 cpm/nmol). Both CaD22WT (open circles) and CaDAA (solid circles) are shown. Phosphorylation at sites other than Ser-672 and Ser-702 is evident in both cases.
The effect of phosphorylation at the major and minor sites of CaD22 on Ca2+-calmodulin binding was determined with intrinsic tryptophan fluorescence measurements. Fig. 3 compares Ca2+-calmodulin binding isotherms for CaD22 that was nonphosphorylated, phosphorylated only at the major sites, and phosphorylated at both the major and minor sites. Phosphorylation at the major sites reduced the affinity of CaD22 for Ca2+-calmodulin from 0.11 μM (open circles) to 0.8 μM (dotted circles). More extensive phosphorylation (solid circles) caused a further reduction in the binding affinity, to ∼4.0 μM. An examination of the 95% confidence limits shows that the three binding curves were statistically different.
Figure 3.
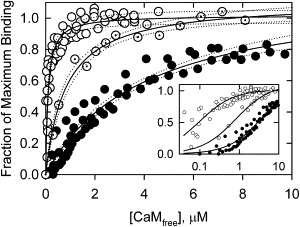
Phosphorylation at both the major and minor sites of CaD22 decreased the affinity to Ca2+-calmodulin. Binding was measured to CaD22 (open circles), CaD22 treated for 1.5 h (dotted circles), and CaD22 treated for 20 h with PAK (solid circles). The data shown are from four, one, and three data sets, respectively. Phosphorylation at the major sites (dotted circles) reduced the affinity from 0.11 μM to 0.8 μM. Additional phosphorylation to yield 3.3 Pi/caldesmon reduced the affinity to 4.0 μM. The dotted lines represent 95% confidence intervals for each curve. Inset: Plot of the fraction of maximum binding against log[CaMfree]. The reaction buffer contained 10 mM MOPS pH 7.2, 100 mM NaCl, 1 mM dithiothreitol, 2 mM CaCl2 at 20°C. CaD22 concentration was 0.6 μM.
The study shown in Fig. 3 was repeated with mutants of CaD22 lacking the major phosphorylation sites. CaD22AA lacks charges at the major sites and cannot be phosphorylated at these sites. Fig. 4 A compares unphosphorylated CaD22AA with CaD22AA that was treated with PAK for up to 20 h. Although this treatment caused phosphorylation at the minor sites, the affinity for Ca2+-calmodulin was unaffected. The affinities of both phosphorylated and nonphosphorylated CaD22AA for Ca2+-calmodulin were similar to that of CaD22WT (Fig. 4 A); that is, curves for both phosphorylated and nonphosphorylated CaD22AA fell within the 95% confidence interval for the WT curve.
Figure 4.
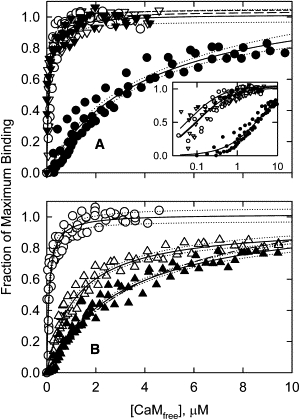
Effect of phosphorylation at the minor sites of CaD22AA (A) and CaD22DD (B) on calmodulin binding. CaD22 decreased calmodulin binding only when the major sites were negatively charged. (A) Binding to CaD22AA (open triangles, four data sets) was identical to that of WT (open circles, four data sets) and was unaffected by PAK treatment for 20 h (solid triangles, three data sets). The solid circles (from Fig. 3) show the effect of extended PAK treatment of WT caldesmon. Phosphorylation of CaD22AA increased the affinity for Ca2+-calmodulin slightly (Kd decreased from 0.1 μM to 0.04 μM). The inset shows the fraction of maximum binding versus log[CaMfree]. (B) Substituting PAK phosphorylation sites with Asp (i.e., CaD22DD; open triangles, three data sets) reduced the affinity of CaD22 to Ca2+-calmodulin to 1.15 μM compared with 0.1 μM for WT unphosphorylated (open circles, four data sets). Incubation of CaD22DD with PAK for 20 h (solid triangles, three data sets) caused a further reduction in its affinity of binding to calmodulin to 3.1 μM. The dotted lines represent 95% confidence intervals for each curve. The solid circles represent CaD22WT incubated with PAK for 20 h. Conditions are the same as in Fig. 3.
CaD22DD had negative charges at the major sites, and incubation with PAK led to phosphorylation of only the minor sites. CaD22DD bound more weakly to Ca2+-calmodulin than did CaD22WT (Fig. 4 B). Treatment of CaD22DD with PAK caused a further reduction in the binding affinity, bringing it close to that of the phosphorylated CaD22WT (Fig. 4 B). The results of Fig. 4 suggest that phosphorylation of caldesmon at the unknown sites affected Ca2+-calmodulin binding only if the two major PAK sites were already phosphorylated.
Phosphorylation of CaD22 may have altered the stoichiometry of binding to calmodulin in addition to decreasing the stability of the interaction. We determined the stoichiometry by repeating binding studies at CaD22 concentrations that exceeded the dissociation constant for the CaD22-calmodulin complex. These data are presented in Fig. S1 of the Supporting Material. As the concentration of calmodulin was increased, a plateau of binding was reached at the same ratio of calmodulin to caldesmon for all of the CaD constructs. CaD22 formed 2:1 complexes with Ca2+-calmodulin, in agreement with previously reported data (29).
Because phosphorylation of caldesmon by PAK on the minor sites affected binding to Ca2+-calmodulin, we measured the effect of Ca2+-calmodulin on the time course of phosphorylation of CaD22AA by PAK (Fig. 5). In the presence of Ca2+-calmodulin (open squares), the phosphorylation of the minor sites was significantly slower than in the absence of Ca2+-calmodulin (open triangles). PAK did not phosphorylate Ca2+-calmodulin significantly (open circles).
Figure 5.
Ca2+-calmodulin binding decreased the rate of phosphorylation of the minor sites by PAK. Phosphorylation time courses were measured as described in Fig. 2 with 0.5 μM CaD22AA and 2 μM Ca2+-calmodulin. Phosphorylation time courses of CaD22AA in the absence (open triangles) and presence (open squares) of Ca2+-calmodulin are shown. Ca2+-calmodulin was not appreciably phosphorylated by PAK (open circles).
Phosphorylation of caldesmon reduces the ability of caldesmon to inhibit actin activation of myosin S1 ATPase activity (9). We examined the contribution of the major and minor PAK phosphorylation sites on the inhibitory activity of CaD22. CaD22AA was expected to behave like CaD22WT. As shown in Fig. 6, CaD22AA had the same inhibitory activity as CaD22WT (Fig. 6, open triangles down and open circles). The mutant designed to mimic phosphorylation at sites 672 and 702 was expected to have a diminished inhibitory activity. However, CaD22DD (Fig. 6, open triangles up) had the same inhibitory activity as CaD22WT and CaD22AA. After overnight incubation with PAK, both the mutants and CaD22WT had similarly reduced inhibitory activities (Fig. 6, solid symbols). Because the minor sites were not totally phosphorylated, the total extent of reversal of activity was not determined. These results suggest that the introduction of negative charges into the minor sites (but not the major sites) attenuated the inhibitory activity of CaD22.
Figure 6.
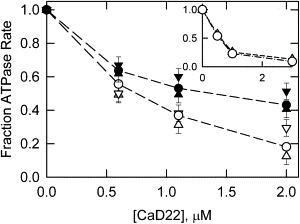
Phosphorylation of the minor sites of CaD22 reduced the inhibition of actomyosin ATPase activity. Actin-activated S1-ATPase as a function of phosphorylated and unphosphorylated CaD22WT (circles), CaD22DD (triangles up), and CaD22AA (triangles down) is shown. Open symbols show the effect of unphosphorylated CaD22, whereas solid symbols show the corresponding species after 20 h treatment with PAK. The inset shows actin-activated S1-ATPase as a function of the concentrations of CaD35WT and CaD35DD. Actin, smooth muscle tropomyosin, and myosin S1 were 10 μM, 2.2 μM, and 0.1 μM, respectively. The final solution composition was 1 mM ATP, 3 mM MgCl2, 31 mM NaCl, 10 mM imidazole pH 7.0, at 25°C.
To ensure that all our proteins functioned normally, we measured the rate of actin-activated ATPase activity as a function of CaD22WT. The CaD22 produced inhibition that was typical of caldesmon. In particular, tropomyosin markedly enhanced the inhibitory activity of CaD22. These data are shown in Fig. S2.
We also examined the effects of negative charges at sites 672 and 702 in a larger caldesmon construct, CaD35, which was previously characterized in our laboratory (23). The inset in Fig. 6 shows that CaD35DD had the same ability to inhibit actin-stimulated ATPase activity as CaD35WT.
The results presented to this point indicate that the effect of Ca2+-calmodulin on ATPase activity may depend on the level of phosphorylation of CaD22. We investigated that possibility by comparing the calmodulin dependency of ATPase activities of unphosphorylated and phosphorylated CaD22. Ca2+-calmodulin was effective in reversing CaD22WT inhibition of actomyosin ATPase (Fig. 7, open circles) but was not able to effectively reverse the inhibition by CaD22DD in which only the major PAK sites were negatively charged (triangles); that is, when the major PAK sites were negatively charged, caldesmon was inhibitory even in the presence of Ca2+-calmodulin. When CaD22 was phosphorylated at both the major and minor sites with PAK (solid circles), there was less inhibition of ATPase activity in the absence of Ca2+-calmodulin, and Ca2+-calmodulin had little effect on the degree of inhibition.
Figure 7.
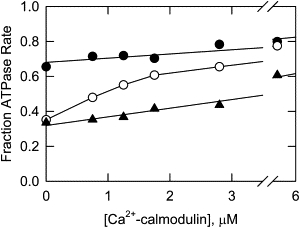
Ca2+-calmodulin has different effects on the inhibitory activities of unphosphorylated and highly phosphorylated CaD22. Shown are unphosphorylated CaD22WT (open circles), CaD22WT-PAK incubated with PAK for 20 h (solid circles), and CaD22DD (triangles). Conditions are the same as in Fig. 6 except that [CaD22] was held constant at 1.1 μM.
The minor sites of phosphorylation of CaD22 by PAK were identified by MS. The sites of phosphorylation together with the masses are given in Table 1. Phosphorylated Thr-627 and Ser-631 were more abundant than phosphorylated Ser-635 and Ser-642. The localization of these sites relative to MAP kinase sites (30), calmodulin kinase II sites (31), cdc2 kinase sites (32), and PKC sites (33) is shown in Fig. 8. Sample MS spectra of phosphorylated peptides are given in Fig. S3.
Table 1.
Mass analysis of phosphate-containing peptides
| Peptide | Expected m/z | Observedm/z | δ |
|---|---|---|---|
| RLEQYTSAVVGNK | 1464.7805 | 1464.7836 | 2.1 ppm |
| RLEQYTS(642)AVVGNK | 1544.7469 | 1544.7499 | 2.0 ppm |
| PAHTTAVVSK | 1010.5629 | 1010.5627 | 0.8 ppm |
| PAHTTAVVS(631)K | 1090.5293 | 1090.5279 | 1.3 ppm |
| IDSRLEQYTSAVVGNK | N/A | ||
| IDS(635)RLEQYTSAVVGNK | 1859.8899 | 1859.8945 | 1.0 ppm |
| SGMKPAHTTAVVSK | 1413.7519 | 1413.7548 | 0.1 ppm |
| SGMKPAHTT(627)AVVSK | 1493.7182 | 1493.7174 | 0.5 ppm |
Phosphorylated residues are in bold and residue numbers are in parentheses.
Figure 8.

Phosphorylation sites on CaD22. All phosphorylation sites are in bold. Major PAK kinase sites are in square brackets. Minor PAK sites are in parentheses. MAPK sites1, calmodulin kinase II sites2, cdc2 kinase sites3, and PKC sites4 are indicated with superscripts as shown. The tropomyosin-binding region has a single underline. The calmodulin-binding regions have a double underline.
It is not clear whether all of the minor sites, a combination of these sites, or just one site is responsible for the observed functional effects. We mutated Thr-627 to Asp in CaD22DD to mimic the phosphorylated state of that minor site and the two major sites. The sequences of the DNA and mutant were verified. The observed mass difference between the triple mutant and the WT caldesmon (68 Da) agreed with the calculated difference (70 Da).
Fig. 9 shows that the affinity of the triple mutant, CaD22DDD, for Ca2+-calmodulin was weaker than that for CaD22WT (open circles), but stronger than that of CaD22-PAK (long dashed line). The binding affinity of the triple mutant was similar to that of CaD22DD (short dashed line), which has negative charges only on the major sites.
Figure 9.
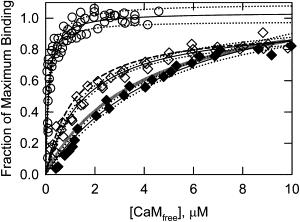
Aspartate substitution at Thr-627 in combination with aspartate substitutions at the major sites did not affect calmodulin binding. Calmodulin binding curves for WT (open circles, four data sets), triple mutant (open diamonds, three data sets), and triple mutant after 20 h PAK treatment (solid diamonds, two data sets) are shown with their 95% confidence limits (dotted lines). The binding curve for the triple mutant (Kd = 1.5 μM) was similar to that of CaD22DD (short dashed line, Kd=1.6 μM). PAK phosphorylation of the triple mutant reduced the affinity to 4.5 μM, which is similar to that observed for CaD22WT-PAK (vertical bar line). Conditions are the same as in Fig. 3.
Treatment of the triple mutant with PAK reduced the binding affinity, bringing it close to that of the phosphorylated CaD22WT. This suggests that the Thr-627 residue by itself does not affect Ca2+-calmodulin binding to caldesmon phosphorylated on the major sites.
Because phosphorylation of the minor sites plays a role in controlling inhibition of ATPase activity by caldesmon (Fig. 6), we explored the effects of the triple mutant on the ATPase activity of caldesmon. The triple mutant had the same inhibitory activity as CaD22WT and did not exhibit the reduced inhibitory activity, as the WT phosphorylated by PAK on both the major and minor residues. These data are shown in Fig. S4.
Discussion
PAK kinase phosphorylates caldesmon at Ser-672 and Ser-702 (9). Phosphorylation at these sites reduces binding to calmodulin and results in a partial reversal of the inhibitory action of caldesmon on actomyosin ATPase activity (9,15). We confirmed the effect of phosphorylation on calmodulin binding, but observed little change in ATPase inhibitory activity. Phosphorylation at Thr-627, Ser-631, Ser-635, and Ser-642 further weakened calmodulin binding and partially reversed ATPase activity. Serine residues at sites 672 and 702 were substituted with either aspartic acid (CaD22DD) or alanine (CaD22AA) to simulate the phosphorylated and nonphosphorylated states, respectively. This procedure permitted us to investigate the relationship among the major (672 and 702) and minor (627, 631, 635, and 642) sites of phosphorylation.
In the case of phosphorylated CaD22WT, the affinity for Ca2+-calmodulin decreased after further incubation with PAK. This further incubation was accompanied by phosphorylation at the minor sites. To ensure that the further reduction in affinity was not linked to an increase in phosphorylation of the major sites, we utilized CaD22DD. The affinity of CaD22DD for Ca2+-calmodulin was ∼10% of that observed with CaD22WT. Incubation of CaD22DD with PAK caused a further reduction in affinity for Ca2+-calmodulin to ∼3% of the WT affinity. The effect of phosphorylation of the minor sites on Ca2+-calmodulin binding was dependent on the prior phosphorylation of the major sites. Replacement of serine residues 672 and 702 by alanine had no effect on Ca2+-calmodulin binding compared to CaD22WT. Incubation of CaD22AA with PAK did not weaken Ca2+-calmodulin binding, although the minor sites became phosphorylated; that is, the only effect that phosphorylation of the minor sites had on Ca2+-calmodulin binding was to enhance the effect of phosphorylation at the major sites.
The effect of phosphorylation at the major PAK sites on weakening Ca2+-calmodulin binding is easy to rationalize because these sites are adjacent to the Ca2+-calmodulin binding region of caldesmon. However, the minor phosphorylation sites are ∼30 residues upstream from the closest known calmodulin-binding site (Fig. 8). We explored the effect of calmodulin binding on the time course of PAK phosphorylation of CaD22AA with the two major sites blocked. Phosphorylation of the minor sites was significantly slower in the presence of Ca2+-calmodulin than in the absence of Ca2+-calmodulin (Fig. 5). Ca2+-calmodulin binding to caldesmon impedes PAK access to the minor sites.
Our results do not indicate how the structure of caldesmon is changed by Ca2+-calmodulin binding. Because the COOH-terminal region of caldesmon is largely unstructured (13), many types of structural changes are possible. There is evidence that the COOH-terminal region of caldesmon can fold over upon itself (29) and such changes might alter the exposure of regions of caldesmon to the kinase. PAK phosphorylation does not appear to cause a gross change in the way that caldesmon binds to calmodulin, since two molecules of caldesmon bind to each molecule of calmodulin regardless of the level of phosphorylation (Fig. 5).
Foster et al. (9) found that PAK phosphorylation of intact chicken gizzard caldesmon caused a slight reduction in the affinity for actin-tropomyosin. However, they also observed a significant effect of phosphorylation on actin-tropomyosin-activated ATPase activity of myosin S1. Whereas WT caldesmon reduced the ATPase activity to 20%, caldesmon containing two phosphorylated residues produced only 45% inhibition. Eppinga et al. (15) observed an even larger effect of phosphorylation on a 39 kDa fragment of human caldesmon. They reported a complete reversal of inhibitory activity when the major PKA sites were mutated to glutamate residues (CaD39-PAKE).
We did not find a significant ATPase effect of phosphorylation at the major sites of expressed 22 and 35 kDa regions of chicken gizzard caldesmon. The phosphomimetic mutant of the major sites, CaD22DD, had the same inhibitory activity as CaD22 and CaD22AA. Furthermore, introducing negative charges at sites 672 and 702 had no effect on the inhibitory activity of the larger caldesmon construct, CaD35. Note that CaD39 and CaD35 are the same physical size despite the different notations used. Prolonged incubation of CaD22WT, CaD22DD, and CaD22AA with PAK did attenuate their inhibitory activities, and all to the same extent; that is, phosphorylation of the minor sites of our construct was necessary for reversal of inhibitory activity irrespective of the state of residues 672 and 702.
PAK phosphorylation not only attenuates the inhibitory activity of caldesmon, it also indirectly increases inhibitory activity by weakening binding to Ca2+-calmodulin. Fig. 7 shows that the net activity depends on the concentration of Ca2+-calmodulin and the number of caldesmon sites that are phosphorylated. For all caldesmon species examined, the acto-S1 ATPase rates increased with increasing calmodulin concentrations. The rates of ATP hydrolysis in the presence of CaD22DD (mimicking phosphorylation of the major sites) were lower than those of WT at all calmodulin concentrations. Caldesmon that was phosphorylated at the minor and major sites had less inhibitory activity than WT at low calmodulin concentrations, but the difference was abolished at high calmodulin concentrations.
It is interesting that a caldesmon mutant that is defective in Ca2+-calmodulin binding sites fails to be recruited to podosomes, and fails to suppress podosome formation (34). Selective control of caldesmon phosphorylation may produce similar changes in calmodulin binding and similar cellular effects.
Phosphorylation of the minor sites of caldesmon by PAK was very slow. It is possible that binding to some target molecule can alter the ability of the minor caldesmon sites to become phosphorylated. It is also possible that another kinase is able to rapidly phosphorylate the minor sites of caldesmon. To facilitate identification of possible kinases, we identified the minor PAK phosphorylation sites of caldesmon: Thr-627, Ser-631, Ser-635, and Ser-642. The location of these sites within CaD22 is shown in Fig. 8. A semiquantitative analysis of the extracted ion chromatograms showed that Thr-627 and Ser-631 carry the greatest amounts of phosphate, and Ser-642 has the least amount of phosphate. All four sites are clustered together, and are located within the tropomyosin-binding region (residues 621–672 (35)).
The location of these minor phosphorylation sites within the tropomyosin-binding region of caldesmon does not appear to have a great impact on the role of tropomyosin. The data of Fig. 6 show that phosphorylation of the minor sites of CaD22 reduced the inhibition of ATPase activity by ∼28% in the presence of tropomyosin. We observed with CaD35 that phosphorylation of these sites reduced the inhibitory activity by 18% even in the absence of tropomyosin (data not shown). We did not observe significant differences between CaD22 and CaD35, so it is unlikely that phosphorylation of the minor sites has large effects on the role of tropomyosin.
If phosphorylation of the minor PAK sites (Thr-627, Ser-631, Ser-635, and Ser-642) serves a physiological role, they are likely to be phosphorylated more rapidly than was observed here. These sites could be better substrates for other kinases, or phosphorylation may be linked to binding of caldesmon to a particular ligand. For example, it is known that Ser-635 is a substrate for calmodulin-dependent protein kinase II (31).
It also remains to be determined which of the minor sites are involved in the changes reported here. Substitution of Thr-627 with Asp did not produce the same effects observed by phosphorylation at the minor sites with PAK kinase. We conclude that phosphorylation of another minor site or a combination of minor sites is important for modulating both ATPase activity and calmodulin binding.
Acknowledgments
We thank Dr. Alan Mak for the gift of the GST-mPAK3 plasmid and for helpful comments. We thank Dr. Kaye Speicher for MS analysis.
This work was funded by grants from the National Institutes of Health (AR35216 to J.M.C.) and the Brody Brothers Foundation (M.M.S. and J.M.C.).
Supporting Material
References
- 1.Ebashi S., Kodama A., Ebashi F. Troponin. I. Preparation and physiological function. J. Biochem. 1968;64:465–477. doi: 10.1093/oxfordjournals.jbchem.a128918. [DOI] [PubMed] [Google Scholar]
- 2.Sobue K., Sellers J.R. Caldesmon, a novel regulatory protein in smooth muscle and nonmuscle actomyosin systems. J. Biol. Chem. 1991;266:12115–12118. [PubMed] [Google Scholar]
- 3.Lin Y., Ye L.H., Kohama K. Stimulatory effect of calponin on myosin ATPase activity. J. Biochem. 1993;113:643–645. doi: 10.1093/oxfordjournals.jbchem.a124096. [DOI] [PubMed] [Google Scholar]
- 4.Abe M., Takahashi K., Hiwada K. Effect of calponin on actin-activated myosin ATPase activity. J. Biochem. 1990;108:835–838. doi: 10.1093/oxfordjournals.jbchem.a123289. [DOI] [PubMed] [Google Scholar]
- 5.Schroeter M.M., Chalovich J.M. Fesselin binds to actin and myosin and inhibits actin-activated ATPase activity. J. Muscle Res. Cell Motil. 2005;26:183–189. doi: 10.1007/s10974-005-9009-6. [DOI] [PubMed] [Google Scholar]
- 6.Marston S.B., Redwood C.S. The molecular anatomy of caldesmon. Biochem. J. 1991;279:1–16. doi: 10.1042/bj2790001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroeter M.M., Chalovich J.M. Ca2+-calmodulin regulates fesselin-induced actin polymerization. Biochemistry. 2004;43:13875–13882. doi: 10.1021/bi0487490. [DOI] [PubMed] [Google Scholar]
- 8.Redwood C.S., Marston S.B., Gusev N.B. The functional effects of mutations Thr673→Asp and Ser702→Asp at the Pro-directed kinase phosphorylation sites in the C-terminus of chicken gizzard caldesmon. FEBS Lett. 1993;327:85–89. doi: 10.1016/0014-5793(93)81045-2. [DOI] [PubMed] [Google Scholar]
- 9.Foster D.B., Shen L.-H., Mak A.S. Phosphorylation of caldesmon by p21-activated kinase. Implications for the Ca(2+) sensitivity of smooth muscle contraction. J. Biol. Chem. 2000;275:1959–1965. doi: 10.1074/jbc.275.3.1959. [DOI] [PubMed] [Google Scholar]
- 10.Winder S.J., Walsh M.P. Smooth muscle calponin. Inhibition of actomyosin MgATPase and regulation by phosphorylation. J. Biol. Chem. 1990;265:10148–10155. [PubMed] [Google Scholar]
- 11.Robertson S.P., Johnson J.D., Solaro R.J. The effect of troponin I phosphorylation on the Ca2+-binding properties of the Ca2+-regulatory site of bovine cardiac troponin. J. Biol. Chem. 1982;257:260–263. [PubMed] [Google Scholar]
- 12.Blumenschein T.M., Stone D.B., Sykes B.D. Dynamics of the C-terminal region of TnI in the troponin complex in solution. Biophys. J. 2006;90:2436–2444. doi: 10.1529/biophysj.105.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Permyakov S.E., Millett I.S., Uversky V.N. Natively unfolded C-terminal domain of caldesmon remains substantially unstructured after the effective binding to calmodulin. Proteins. 2003;53:855–862. doi: 10.1002/prot.10481. [DOI] [PubMed] [Google Scholar]
- 14.Khaymina S.S., Kenney J.M., Chalovich J.M. Fesselin is a natively unfolded protein. J. Proteome Res. 2007;6:3648–3654. doi: 10.1021/pr070237v. [DOI] [PubMed] [Google Scholar]
- 15.Eppinga R.D., Li Y., Lin J.J. Requirement of reversible caldesmon phosphorylation at P21-activated kinase-responsive sites for lamellipodia extensions during cell migration. Cell Motil. Cytoskeleton. 2006;63:543–562. doi: 10.1002/cm.20144. [DOI] [PubMed] [Google Scholar]
- 16.Van Eyk J.E., Arrell D.K., Mak A.S. Different molecular mechanisms for Rho family GTPase-dependent, Ca2+-independent contraction of smooth muscle. J. Biol. Chem. 1998;273:23433–23439. doi: 10.1074/jbc.273.36.23433. [DOI] [PubMed] [Google Scholar]
- 17.Dharmawardhane S., Sanders L.C., Bokoch G.M. Localization of p21-activated kinase 1 (PAK1) to pinocytic vesicles and cortical actin structures in stimulated cells. J. Cell Biol. 1997;138:1265–1278. doi: 10.1083/jcb.138.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spudich J.A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 19.Kielley W.W., Harrington W.F. A model for the myosin molecule. Biochim. Biophys. Acta. 1960;41:401–421. doi: 10.1016/0006-3002(60)90037-8. [DOI] [PubMed] [Google Scholar]
- 20.Weeds A.G., Taylor R.S. Separation of subfragment-1 isoenzymes from rabbit skeletal muscle myosin. Nature. 1975;257:54–56. doi: 10.1038/257054a0. [DOI] [PubMed] [Google Scholar]
- 21.Pedigo S., Shea M.A. Quantitative endoproteinase GluC footprinting of cooperative Ca2+ binding to calmodulin: proteolytic susceptibility of E31 and E87 indicates interdomain interactions. Biochemistry. 1995;34:1179–1196. doi: 10.1021/bi00004a011. [DOI] [PubMed] [Google Scholar]
- 22.Bretscher A. Smooth muscle caldesmon. Rapid purification and F-actin cross-linking properties. J. Biol. Chem. 1984;259:12873–12880. [PubMed] [Google Scholar]
- 23.Fredricksen S., Cai A., Chalovich J.M. Influence of ionic strength, actin state, and caldesmon construct size on the number of actin monomers in a caldesmon binding site. Biochemistry. 2003;42:6136–6148. doi: 10.1021/bi0274017. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi T., Yang X., Solaro R.J. A non-equilibrium isoelectric focusing method to determine states of phosphorylation of cardiac troponin I: identification of Ser-23 and Ser-24 as significant sites of phosphorylation by protein kinase C. J. Mol. Cell. Cardiol. 2005;38:213–218. doi: 10.1016/j.yjmcc.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Conti M.A., Adelstein R.S. Purification and properties of myosin light chain kinases. Methods Enzymol. 1991;196:34–47. doi: 10.1016/0076-6879(91)96006-d. [DOI] [PubMed] [Google Scholar]
- 26.Chittum H.S., Lane W.S., Hatfield D.L. Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 27.Eng J.K., McCormick A.L., Yates J.R.I.I.I. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 28.Chalovich J.M., Eisenberg E. Inhibition of actomyosin ATPase activity by troponin-tropomyosin without blocking the binding of myosin to actin. J. Biol. Chem. 1982;257:2432–2437. [PMC free article] [PubMed] [Google Scholar]
- 29.Medvedeva M.V., Kolobova E.A., Gusev N.B. Mapping of contact sites in the caldesmon-calmodulin complex. Biochem. J. 1997;324:255–262. doi: 10.1042/bj3240255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adam L.P., Hathaway D.R. Identification of mitogen-activated protein kinase phosphorylation sequences in mammalian h-Caldesmon. FEBS Lett. 1993;322:56–60. doi: 10.1016/0014-5793(93)81110-l. [DOI] [PubMed] [Google Scholar]
- 31.Ikebe M., Reardon S. Phosphorylation of smooth muscle caldesmon by calmodulin-dependent protein kinase II. Identification of the phosphorylation sites. J. Biol. Chem. 1990;265:17607–17612. [PubMed] [Google Scholar]
- 32.Yamashiro S., Yamakita Y., Matsumura F. Characterization of the COOH terminus of non-muscle caldesmon mutants lacking mitosis-specific phosphorylation sites. J. Biol. Chem. 1995;270:4023–4030. doi: 10.1074/jbc.270.8.4023. [DOI] [PubMed] [Google Scholar]
- 33.Ikebe M., Hornick T. Determination of the phosphorylation sites of smooth muscle caldesmon by protein kinase C. Arch. Biochem. Biophys. 1991;288:538–542. doi: 10.1016/0003-9861(91)90232-8. [DOI] [PubMed] [Google Scholar]
- 34.Eves R., Webb B.A., Mak A.S. Caldesmon is an integral component of podosomes in smooth muscle cells. J. Cell Sci. 2006;119:1691–1702. doi: 10.1242/jcs.02881. [DOI] [PubMed] [Google Scholar]
- 35.Redwood C.S., Marston S.B. Binding and regulatory properties of expressed functional domains of chicken gizzard smooth muscle caldesmon. J. Biol. Chem. 1993;268:10969–10976. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


