Abstract
Myosin X is a molecular motor that is adapted to select bundled actin filaments over single actin filaments for processive motility. Its unique form of motility suggests that myosin X's stepping mechanism takes advantage of the arrangement of actin filaments and the additional target binding sites found within a bundle. Here we use fluorescence imaging with one-nanometer accuracy to show that myosin X takes steps of ∼18 nm along a fascin-actin bundle. This step-size is well short of the 36-nm step-size observed in myosin V and myosin VI that corresponds to the actin pseudohelical repeat distance. Myosin X is able to walk along bundles with this step-size if it straddles two actin filaments, but would be quickly forced to spiral into the constrained interior of the bundle if it were to use only a single actin filament. We also demonstrate that myosin X takes many sideways steps as it walks along a bundle, suggesting that it can switch actin filament pairs within the bundle as it walks. Sideways steps to the left or the right occur on bundles with equal frequency, suggesting a degree of lateral flexibility such that the motor's working stroke does not bias it to the left or to the right. On single actin filaments, we find a broad mixture of 10–20-nm steps, which again falls short of the 36-nm actin repeat. Moreover, the motor leans to the right as it walks along single filaments, which may require myosin X to adopt strained configurations. As a control, we also tracked myosin V stepping along actin filaments and fascin-actin bundles. We find that myosin V follows a narrower path on both structures, walking primarily along one surface of an actin filament and following a single filament within a bundle while occasionally switching to neighboring filaments. Together, these results delineate some of the structural features of the motor and the track that allow myosin X to recognize actin filament bundles.
Introduction
Myosin X is a molecular motor that localizes to filopodial tips and undergoes intrafilopodial motility (1–3). It binds to integrins (4) and is involved in filopodial formation (5,6), intrafilopodial transport (3,7), phagocytosis (8), spindle assembly (9,10), and neuronal development (11). Within the core of a filopodium is a bundle of parallel actin filaments organized by fascin (12–14). Myosin X makes long processive runs on fascin-mediated actin bundles both in vivo and in vitro (15). This motor illustrates one way in which nature has solved the problem of directing the transport of cargoes over long distances within a cell, specifically by myosin X's ability to recognize a particular arrangement of actin filaments from the rest of the actin cytoskeleton.
How the motor achieves its unique, actin architecture-specific motility is not understood. Myosin V and myosin VI are also processive myosins and have been shown to take 36-nm steps (16,17), corresponding to the pseudohelical repeat of an actin filament, making them well suited to taking multiple steps without falling off an actin filament. In comparison to myosin V, myosin X's lever arms are putatively shorter, containing only three IQ motifs for binding light chains instead of six. However, between the IQ motifs of myosin X and its coiled-coil is a highly charged single alpha-helix (SAH) that has been proposed to extend the lever arms, possibly compensating for the myosin X's shortage of IQ motifs (18). We had shown that the processivity of myosin X is enhanced on actin bundles, in that the single motors initiate runs more frequently and move for greater distances on bundled actin than they do on single actin filaments at saturating ATP. This enhancement in processivity was observed both on fascin-actin bundles and on artificial methylcellulose bundles. In contrast, myosin V showed no such enhancement (15).
Here we report the stepping behavior of myosin X under low-ATP, ADP-inhibition conditions. We tracked a dimeric head-and-neck myosin X molecule labeled at the C-terminus with a quantum-dot while walking along bundled actin and single actin filaments using fluorescence imaging with one-nanometer accuracy (FIONA) (19). We report that myosin X steps short of the 36-nm long-pitch actin repeat on single filaments and on bundled actin, taking 17.5-nm steps along bundles. We show that myosin X takes sidesteps as it moves along a bundle, allowing it to take full advantage of the many binding sites available to it within a bundle to step around any defects in the track or obstacles it may encounter within a cell. We also show that myosin X leans to the right as it takes a few short steps forward on single actin filaments, turning with the pitch of the actin helix. As a control, we tracked a similar myosin V construct walking along both bundled actin and single filaments. We did not see myosin V leaning to the right as it walks along an actin filament, and we observed that myosin V takes fewer sidesteps on bundles than myosin X, instead tracking along single actin filaments within the bundle.
Methods
Proteins
Bovine myosin X HMM-FLAG-Ctag and the affinity clamp protein with a single cysteine residue were expressed and purified as previously described (20,21). The myosin V construct with a Ctag was likewise prepared by polymerase chain reaction, using primers encoding the Ctag sequence, and cloned into the InsectDirect vector pBiEx3BS. Qdots (585 ITK; Molecular Probes, Eugene, OR) with amine-functionalized nanocrystals (cat. No. Q21511MP; Invitrogen, Carlsbad, CA) were conjugated to the affinity-clamp protein via a Sulfo-SMCC cross-linker (cat. No. 22322; Pierce, Thermo Fisher, Waltham, MA). Chicken actin was purified and polymerized as previously described (22). Fascin was purified (23), and fascin-actin bundles were assembled (15) as previously described.
Microscopy
Flow-cells were constructed with double-sided tape and No. 1.5 glass coverslips. The surface was coated with neutravidin and blocked with bovine serum albumin and then biotinylated F-actin or fascin-actin bundles labeled with ATTO-647N-phalloidin (ATTO-TEC, Siegen, Germany) were immobilized on the surface. Unbound actin was washed out of the flow-cell, and then motility buffer (25 mM imidazole pH 7.5, 25 mM KCl, 2 mM MgCl2, 100 μM dithreitol, 1% (w/v) triton X-100, 2.5 μM MgATP, 500 μM MgADP, and 0.2 nM myosin X, and 0.2 nM clamp-Qdots at a 1:1 molar ratio, neglecting losses due to protein adsorption) was perfused into the flow-cell. Images were gathered in total internal reflection fluorescence on a home-built inverted microscope using an iXon charge-coupled device (model No. DV887ECS-BV; Andor Technology, Belfast, UK) at 0.5 s exposure (see Movie S1 in the Supporting Material). The first few frames of each movie acquired an image of the actin, while the remaining frames imaged the Qdots.
Quantum dots are multivalent probes with the potential to oligomerize myosin motors, which could confound our analysis. Two lines of evidence demonstrate that our Qdots contain, at most, a single myosin X.
First, we avoid saturating the Qdots by measuring their landing rate as a function of motor concentration. Under our working conditions (0.2 nM myosin X and 0.2 nM clamp-Qdots), events on actin bundles occur at a landing rate of 13 ± 4 mm−2 s−1 (standard error, SE, from counting statistics). Increasing the amount of myosin X to 0.5 nM or 2.0 nM, while holding the clamp-Qdot concentration at 0.2 nM, resulted in observed landing rates of 30 ± 9 mm−2 s−1 and 60.0 ± 24.5 mm−2 s−1, respectively. The first-power increase in Qdot landing rate as we increase the myosin X concentration from 0.2 nM to 0.5 nM suggests that we are achieving one motor per active Qdot near our working concentration of 0.2 nM. At extreme motor concentrations, we would expect a shallower dependence on motor concentration when multiple motors adhere to each Qdot, as the concentration of Qdots would then be limiting.
Second, under our working conditions and at saturating ATP (1 mM), we observe a mean runlength of 0.5 ± 0.1 μm (n = 19, mean ± SE), similar to our previously published measurement (15) using probes with no potential to oligomerize the myosin X. However, at tenfold or higher concentrations of myosin X (>2.0 nM), observed runlengths at saturating ATP were significantly longer, due to the presence of multiple motors attached to a single clamp-Qdot. Longer runlengths are a clear signature of multiple motor interactions, because all motors must detach from the track to terminate a run (24). Thus, we avoid these high-concentration conditions and instead work at 0.2 nM myosin. Note that the mole ratio of motors to Qdots is uninformative in establishing single-motor conditions, because not all motors will bind to Qdots (or bind in an active configuration). Thus, we can ensure single molecule conditions by maintaining first-power landing rates and avoiding conditions that yield artificially high runlengths. We performed similar analysis of landing rates and runlengths to establish single-motor conditions for myosin V.
Data analysis
Coarse image position coordinates of moving Qdots were gathered using the SpotTracker plug-in (25) in ImageJ (National Institutes of Health, Bethesda, MD). These coarse coordinates were used as a guide for fitting the raw image (9000 photons per frame, mean count from 105 frames) to a two-dimensional Gaussian by maximum likelihood estimation (26) in MATLAB (The MathWorks, Natick, MA). The center of the two-dimensional Gaussian fits were used as the refined image position coordinates and were filtered to avoid low intensities from the Qdot blinking (minimum 1000 counts) and to avoid the occurrence of another spot within close proximity (minimum distance 600 nm, approximately two spot radii). For each movie, a function for the recorded stage drift was calculated and subtracted from the raw data of each run (see Fig. S1 in the Supporting Material). We tested our method of tracking points and measuring step-sizes by way of recording data while stepping the piezo stage 30 nm or 10 nm (Fig. S2).
We defined a coordinate system for an observer looking down at the motor as it moves along actin with the coverslip underneath (Fig. 1 a). The actin image establishes the position of the x axis (see Fig. S3); using the actin image, we rotate our observed motor trajectories so that all runs move toward the +x direction as follows. Coordinates of the endpoints of segments describing the position of the actin tracks were picked using the NeuronJ plug-in (27) in ImageJ (National Institutes of Health). Additional points along the actin tracks were interpolated at half-pixel spacing along the line segments. A straight line fit to the actin track point nearest the centroid of each run and the five nearest actin track points in each direction was used to define a rotation matrix to rotate our coordinates in the laboratory frame onto the actin filament frame. The predominant direction of a run establishes the positive x direction. The y axis for each run was perpendicular to that run's x axis, describing the relative distance from the x axis in each frame with the positive y direction to the left from the perspective of a forward-moving motor. The total distance moved by a motor in an event was short (0.16 ± 0.02 μm on bundles, 0.17 ± 0.02 μm on single filaments, mean ± SE; n = 29 and 32 motors, respectively) relative to the length of the actin tracks (5.9 ± 0.4 μm for bundles, 2.6 ± 0.2 μm for single filaments, mean ± SE; n = 99 and 133 tracks, respectively). The total distance moved under these nucleotide conditions is limited largely by the length of the movie, not the motor's inherent properties, so these total runlength distances are not comparable to our previously published runlength measurement (15). Because our observed runs are much shorter than the persistence length of the tracks (>10 μm), we expect that the tracks are largely straight on these length scales.
Figure 1.
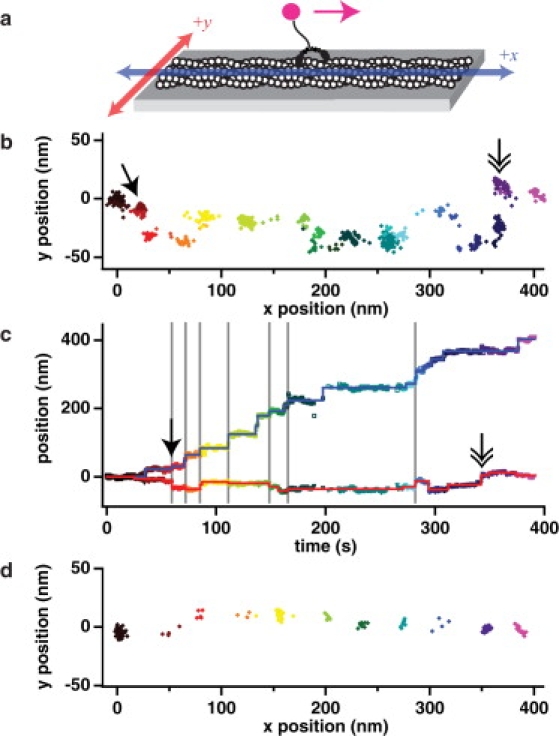
Myosin X takes sidesteps as it walks along a fascin-actin bundle. (a) Myosin X HMM labeled with a quantum dot (magenta) at its tail was imaged walking along surface-attached tetramethylrhodamine-phalloidin-labeled fascin-actin bundles (or single filaments). The actin image and the predominant direction of motion of the quantum dot defined the position (blue) and positive direction (magenta arrow) of the x axis, with the y axis perpendicular (red). Our coordinate system is defined for an observer looking down on the top of the actin, with the coverslip underneath. We verified the arrangement of our coordinate system by translating the microscope stage in known directions in the laboratory frame while tracking objects observed on the camera. (b) Example position trace of one motor walking along a bundle. The motor is moving predominantly from left to right while taking some side steps, up and down. We rotate all measured trajectories to show the motor moving from the origin along the +x direction. This coordinate rotation is derived from the actin filament image as described in the Methods. (c) Plot of the x position (open squares) and y position (open triangles) versus time of the motor shown in panel b (see Fig. S5 for more examples). The step-fitting algorithm (28) was used to pick the steps independently in x (blue trace) and in y (red trace). Each position is given a unique color (from black to magenta) in panels b and c. Some x steps occur concurrently with a y step (gray vertical lines, solid arrow) resulting in a single step oblique to both the x and y axes. Multiple, successive y steps with no corresponding x step are also observed (double-headed arrow). At ∼200 s and again at ∼275 s, the quantum dot blinked to low intensity, and these frames were filtered out as described in text (see Methods). (d) Myosin V walks straight along an actin filament. An example FIONA position-trace of myosin V, labeled at the tail as for myosin X, walking along a single actin filament is shown. Over these short lengths, we do not expect to see significant evidence of the slow spiraling with >1 μm pitch observed by Ali et al. (30). Note the lack of direct sidesteps. Myosin V walked with a step-size of 37.2 ± 0.9 nm (SE, n = 259).
Steps were picked from plots of runlength, x position, or y position versus time using the step-fitting algorithm described in Kerssemakers et al. (28). From the independently picked x and y step traces, oblique steps were identified as the occurrence of a simultaneous x step and y step where the difference in time between the steps was three frames or fewer.
Results and Discussion
Myosin X takes sidesteps as it walks along a fascin-actin bundle
Fluorescence imaging with 1-nm accuracy (FIONA) (19) allows us to resolve steps of 10 nm or less (see Fig. S3). By tracking the center of Qdot-labeled myosin X in two dimensions as it steps along fascin-actin bundles, stepwise motion is observed (see Movie S1). Our myosin X construct consists of amino-acid residues 1–920 of native bovine myosin X, followed by a GCN4 sequence to assist in dimerization, and C-terminal tags for purification and labeling. Similar constructs, with and without artificial dimerization, did not exhibit functional defects in previous in vivo studies (see Conclusions) (1,3,5,6). For labeling, we used a clamp-tag system, where we encoded a short eight-residue epitope tag at the C-terminus of the motor (20,21). This tag is recognized by a Qdot-labeled affinity clamp with high affinity. The position of our clamp-tag at the C-terminus of the heavy chain means that we track the approximate center-of-mass of the motor while it moves.
To resolve single steps of the motor using FIONA,
-
1.
The motor must be moving slowly enough that the dwell time for each position it occupies along actin is longer than the image acquisition frame rate, and
-
2.
The frame rate must be sufficiently long to collect enough photons in each frame to obtain a good two-dimensional Gaussian maximum likelihood estimate to the image data.
One accessible way to slow the motor's stepping rate is by using ADP inhibition to slow its catalytic cycle. Under the experimental nucleotide conditions of 2.5 μM ATP and 500 μM ADP, we observed myosin X moving at a velocity of 1.2 ± 0.2 nm s−1 along bundles (mean ± SE, n = 29 motors), equal to 15–30 0.5 s frames per step for a step-size of 20–40 nm and uniform stepping rate. We chose to use these conditions because they satisfied the necessary criteria for resolving single steps and enforce a single ADP-bound waiting-state.
The actin image serves to define an x axis (see Fig. S3) that describes the larger component of the total runlength, with the motor moving primarily in the positive x direction (Fig. 1 a). Perpendicular to the x axis, a y axis describes motion to the left or right of the positive x direction. Examining the plot of x and y position coordinates reveals mostly forward steps in the positive x direction (Fig. 1 b; see Fig. S4 for more examples). Some steps occur straight along the y axis, including multiple such steps in a row (Fig. 1 b, double-headed arrow), suggesting that the motor is spanning more than one filament within the bundle and can adjust to contacting other nearby parallel filaments. Some steps occur at an oblique angle to the axes (Fig. 1 b, solid-headed arrow), demonstrating that the motor can move forward and to the side simultaneously. Note that we somewhat underestimate the number of sideways steps, because some of our myosin X molecules may walk along the side of a filament or bundle, where sidesteps would project along the unresolved z axis.
Plots of the x position and the y position over time show that the motion in each direction is stepwise (Fig. 1 c; see also Fig. S5). The x position trace contributes largely to the overall runlength, while the y position trace illustrates the clear side-to-side behavior occurring over the course of the much longer run. The step-fitting algorithm (28) was used to pick the locations of steps in both x and y independently of one another. The occurrence of x and y steps simultaneously illustrates that steps that are oblique to both axes are detectable using this method, although oblique steps with a small component in one direction will likely be detected only in one dimension. Sideways stepping has been previously reported in kinesins stepping onto adjacent protofilaments within a microtubule (29), but this behavior has not been investigated in myosins beyond single motors switching from one actin filament to a crossing actin filament (30,31).
The observed stepping pattern of myosin X on an actin bundle contrasts sharply with that of myosin V on a single filament or on a bundle (Fig. 1 d; see also Fig. S6). Myosin V walks almost straight along the x axis in a stepwise manner with very little motion along the y axis. We interpret these results to mean that myosin V steps primarily along one face (azimuth) of an actin filament. Only occasionally will myosin V step longer or shorter than the 36-nm repeat distance and thus rotate around the filament, or step to a nearby filament within a bundle.
Myosin X steps short of the 36-nm pseudohelical repeat along bundles and single filaments
Steps can also be picked from a plot of the runlength (distance from the starting point of the event) versus time (Fig. 2 a). The plot of runlength versus time is remarkably similar to the plot of x position versus time (as shown in Fig. 1 c) because motion in the x direction, or along the length of the bundle, is the primary contributor to runlength. The distribution of step-sizes measured from runlength traces for myosin X stepping on bundles yield a histogram with Gaussian peaks at −12.8. ± 2.5 nm and 17.5 ± 1.9 nm (n = 239 steps measured from 29 motors; Fig. 2 b; errors derived from curve fits to 200 bootstrap sampled histograms). The motor predominantly takes forward steps (83%), but backward steps are common (17%). This forward step-size is roughly half the 36-nm pseudohelical repeat distance of actin.
Figure 2.
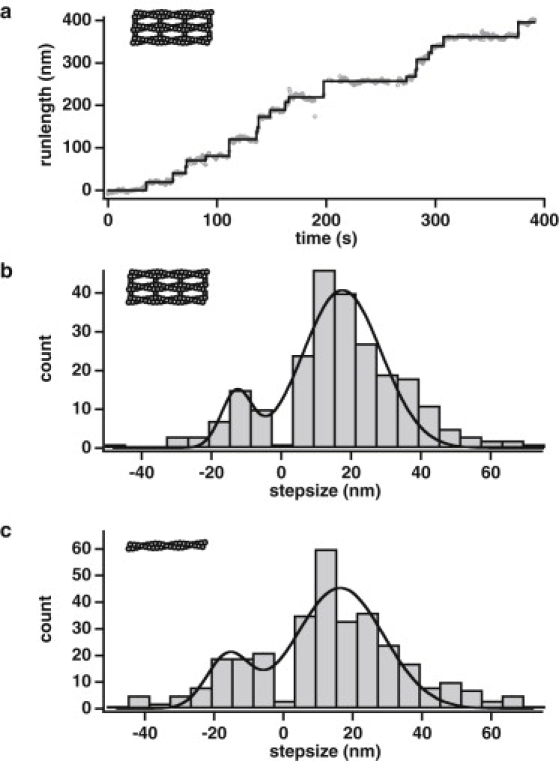
Myosin X steps short of the 36-nm pseudohelical repeat of actin taking 17.5-nm steps along fascin-actin bundles. (a) Plot of runlength over time (gray circles) for the same example event as in Fig. 1b. The runlength trace is remarkably similar to the x trace in Fig. 1c, because the x direction is the primary contributor to runlength. Steps (black trace) were picked from runlength traces using the step-fitting algorithm (28). (b) Step-size histogram of myosin X on fascin-actin bundles (n = 239 steps). The histogram was fit to the sum of two Gaussians (black trace), centered at −12.8 ± 2.5 nm (σ = 4.4 ± 1.5 nm) and 17.5 ± 1.9 nm (σ = 11.1 ± 2.0 nm). (c) Step-size histogram of myosin X on single actin filaments (n = 327 steps; picked from plots of runlength over time traces. See Fig. S7). The histogram was fit to the sum of two Gaussians (black trace), centered at −16.0 ± 2.0 nm (σ = 6.1 ± 1.3 nm) and 16.4 ± 1.8 nm (σ = 12.6 ± 2.0 nm). Errors were obtained by curve fits to 200 bootstrap-sampled histograms with the same bin parameters.
We also tracked the motor stepping along single actin filaments under the same conditions. We find that myosin X takes more frequent processive runs under ADP-inhibition compared to saturating ATP conditions (15). This behavior is not surprising, because ADP-inhibition would increase the residence time of the bound head on an actin filament, allowing the free head additional time to find a binding site along the actin filament. We find that myosin X also moves primarily in the forward direction with some side-to-side motion on single filaments (see Fig. S4 for examples).
Step-sizes for myosin X on single actin filaments are also well short of 36 nm (n = 327 steps measured from 32 motors; Fig. 2 c and Fig. S7). On single actin filaments, the step-size distribution has peaks at −16.0 ± 2.0 nm and 16.4 ± 1.8 nm (errors derived from curve fits to 200 bootstrap sampled histograms). Backward steps occur at a slightly higher frequency on single actin than on bundles (26% backward steps and 74% forward steps). Due to the short step-size observed on single filaments, we cannot exclude the possibility of a fraction of missed events that are shorter than 10 nm because step-sizes near zero are always difficult to detect. However, we note that our dwell-time distributions are consistent with a single exponential without an apparent lag-phase (see below), suggesting that missed events are not a major contaminant in our datasets. Given the spacing of myosin binding-sites along the actin filament, we would expect that any processive, missed steps would be ∼5.5 nm.
We find it remarkable that myosin X disfavors taking 36-nm steps on both actin architectures. Myosin V and myosin VI are processive on single actin filaments and can take 36-nm steps, allowing them to walk along one face of the actin helix (16,17). In fact, on both bundles and on single filaments we measure a step-size for myosin V that closely matches this distance (37.1 ± 0.9 nm, mean ± SE, n = 244 steps from 12 motors on single filaments; 35.7 ± 1.3 nm, mean ± SE, n = 252 steps from 10 motors on bundles). Therefore, the step-size distribution for myosin X both on bundles and single filaments indicate that the myosin X follows a different path than these other motors as it walks along actin.
Myosin X spirals as it walks along single actin filaments
When looking at a gallery of x-versus-y position plots for events on single actin filaments (Fig. 3; see Fig. S4), we see that many local clusters of points appear to have a negative slope as the trace moves in the positive x direction. Although the overall trace is straight along the x axis, the local negative slope would correspond to the motor leaning to the right as it walks forward. In contrast, the slope of local clusters of points in x versus y position plots for events on bundled actin does not appear to have any visible correlation within each local cluster. Additionally, local clusters of points that likely containing several motor steps for events on single actin filaments are located at intervals along the x axis of 40 ± 23 nm, measured center-to-center distance of the clusters (mean ± SD, picked manually, n = 111 clusters). This is very near the length of the actin pseudohelix. Our interpretation of this cluster spacing is that short steps along the pseudohelix proceed until steric interactions with the coverslip force a long step to the other actin protofilament, to allow the processive run to continue. The clusters of accessible sites along actin repeat every 36 nm. For events on bundles, the local clusters occur at shorter intervals along the x axis, 27 ± 17 nm center-to-center (mean ± SD, picked manually, n = 146 clusters). The difference in these cluster spacing measurements is statistically significant (p < 10−6 by t-test).
Figure 3.
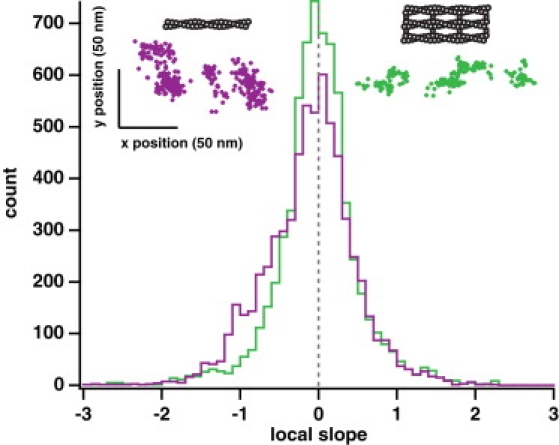
Myosin X takes a spiraling path along single actin filaments. (Upper left, violet), example position trace of an event on a single actin filament. (Upper right, green) Example position trace of an event on a fascin-actin bundle. Each event is moving predominantly in the forward direction (from left to right, as plotted; see Fig. S7 for more examples). The position traces on single actin filaments show local trends of movement forward and to the right (upper left to lower right, as plotted). Local clusters of points within position traces on bundled actin are uncorrelated. Large axes, histograms of the local slope calculated from position traces of events on fascin-actin bundles (green, n = 6675 slopes) and on single actin filaments (violet, n = 6466 slopes). Each slope was calculated using a sliding window of 25 consecutive points in a position trace. Along fascin-actin bundles, the motor shows no asymmetry, with the distribution of slopes centered at 0 (gray dashed line). There is no statistically significant difference between the positive slopes and the absolute value of the negative slopes (p > 0.9 by Wilcoxon rank-sum test). On single actin filaments, the distribution of slopes is asymmetric ∼0. The difference between the positive slopes and the absolute value of the negative slopes is statistically significant (p < 10−34 by Wilcoxon rank-sum test). This shoulder of negative slopes in the distribution demonstrates that the motor is moving to the right locally as it moves forward over short distances, suggesting it turns with the actin helix over the course of a few small steps. The difference between the overall bundle and single filament local slope distributions is statistically significant (p < 10−20 by Wilcoxon rank-sum test). These conclusions do not change when long runs are omitted (>200 points) or the sliding window size is changed (10 or 50 points). The spread in the y position data (∼50 nm) is approximately twice the sum of the actin filament radius (4 nm), the radius of the quantum dot (∼10 nm), and the distance from the binding domain to the C-terminal end of the coiled-coil (∼10 nm or more), and this spread agrees with a recent measurement for the twirling radius of a similarly sized myosin constructs on single actin filaments (41).
A histogram of the distribution of local slope, taken from a sliding window of 25 points, highlights this difference (Fig. 3). This negative slope and the intervals of clusters of points at 40 nm strongly suggests that the motor is turning with the right-handed helical pitch of the actin filament as it takes a few short steps along a single actin filament. If the motor largely follows the actin protofilament (long-pitch helix) on single actin filaments, it must dramatically reorient at ∼36-nm intervals to continue processive stepping. The motor likely steps across to the other protofilament of actin when available binding sites on the same protofilament are occluded by the coverslip, accounting for the long steps between clusters. We had previously reported that myosin X twists single actin filaments, seen in the formation of plectonemes in gliding filament assays when the pointed end of an actin filament becomes stuck (15), a result explained by the motor walking along the helical pitch of a filament. In contrast, the lack of any trend in the local slope on bundled actin suggests that the motor is walking along largely in the forward direction with little or no bias to the left or the right. The lack of a rightward bias on bundles suggests that myosin X is not walking along a single filament within the bundle for the majority of its steps. Instead, it uses a neighboring filament, which presents additional binding sites that do not require the lean to the right. As we will show in our model (below), bundles can accommodate the short steps of myosin X, because they present binding sites at an appropriate azimuth for repeated 18-nm steps.
Myosin X steps with little left-right bias along fascin-actin bundles
From the independently picked x and y stepping traces, we identified concurrent x and y steps, or steps that are oblique to the x and y axes (Fig. 1, a and b; see Fig. S5). After identifying oblique steps, a scatter plot of the x and y step-sizes for all transitions demonstrates the lack of left-right bias in myosin X's stepping pattern on bundled actin (Fig. 4 a). The bias for forward steps over backward steps is clear from the step-size distributions of steps parallel to the x axis (Fig. 4 c) and the x components of oblique steps (Fig. 4 d). Interestingly, myosin X is more likely to take a perpendicular step than it is to take an oblique step. The remarkable balance in right and left steps is also apparent in the step-size distributions of steps parallel to the y axis (Fig. 4 b) and the y components of oblique steps (Fig. 4 e), demonstrating that myosin X has considerable lateral flexibility yet little lateral bias in where it steps. The frequency of sideways steps (36% perpendicular and 22% oblique) is much higher than has been observed in kinesin (13%) (29), and also contrasts with our observations here for myosin V on bundles or on single filaments (<20% total for both actin structures; see Fig. S8 and Fig. S9). This suggests that myosin X and kinesin both can make use of sideways steps to navigate around obstacles on their tracks. However, myosin X may take a greater number of sideways steps as a side effect of it being able to span multiple filaments within a bundle and its necessity to do so during processive runs. Interestingly, the majority of all steps detected for myosin V on both bundles and single filaments are directly along the x axis, demonstrating that myosin V's stepping behavior is not greatly perturbed when a filament is a part of a fascin-actin bundle and that most steps it takes within a bundle are along a single filament.
Figure 4.
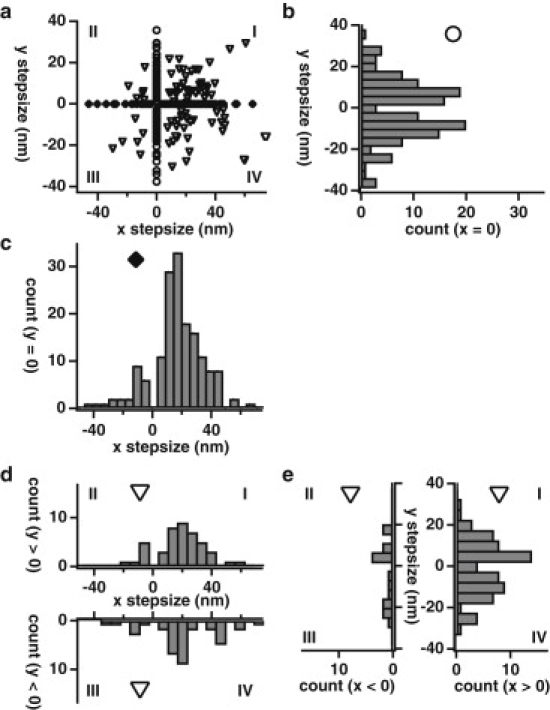
Myosin X meanders symmetrically along fascin-actin bundles. (a) Scatter plot of the x and y components for all steps on fascin-actin bundles (n = 379). Steps occur either along the x axis (solid diamonds, n = 161), along the y axis (open circles, n = 135), or in one of the four Cartesian quadrants (open triangles, n = 83). Quadrants are noted with Roman numerals. (b) Histogram of y step-sizes with no corresponding x step-size (n = 135; 68 positive y step-sizes, 67 negative y step-sizes). (c) Histogram of x step-sizes with no corresponding y step-size (n = 161; 137 positive x step-sizes, 24 negative x step-sizes). (d) (Top) Histogram of x step-sizes of concurrent steps with a positive y step-size (n = 45; Quadrants I and II, n = 37 and n = 8, respectively). (Bottom) Histogram of x step-sizes of concurrent steps with a negative y step-size (n = 38; Quadrants III and IV, n = 7 and n = 31, respectively). (e) (Left) Histogram of y step-sizes of concurrent steps with a negative x step-size (n = 15; Quadrants II and III, n = 8 and n = 7, respectively). (Right) Histogram of y step-sizes of concurrent steps with a positive x step-size (n = 68; Quadrants I and IV, n = 37 and n = 31, respectively). Note that the motor takes steps to the left or right (up or down, as plotted) at nearly equal frequencies.
Myosin X steps in all directions from a common intermediate
Single-molecule stepping traces also reveal information about the kinetic events that lead to a motor step. Under our nucleotide conditions where the stepping process is deliberately slowed, we find that the dwell times before steps, irrespective of stepping direction, are drawn from a single exponential distribution (Fig. 5), with a mean dwell time of 9.2 ± 0.5 s (mean ± SE, n = 363 steps). Similar plots for steps in each direction, along the axes or obliquely, show that each type of dwell is drawn from the same exponential distribution. None of these distributions, when compared pairwise with the others, show significant statistical differences by a Wilcoxon rank-sum test (p ≥ 0.08). The simplest explanation of this result is that all stepping directions emerge from a common kinetic intermediate (32). In such a scheme, the kinetic partitioning ensures that our observed dwell-time distributions are identical. However, the partition probabilities do vary in each direction; these probabilities and the overall stepping rate (6.2 ± 0.4 min−1) may be used to obtain the stepping rate in each direction (see Fig. S10) (33). These rates show the motor has a strong kinetic preference for forward steps (straight or oblique) and straight left or straight right steps. By comparison, straight backward steps occur less frequently, and oblique backward steps occur even less frequently. These individual rates are estimates, because the presence of missed events would alter the partition probabilities. We expect that we detect all steps along the x axis; however, some of the sidesteps or oblique steps may be difficult to observe if they occur along the side of the bundle, where components of the motion perpendicular to the bundle could occur along the unresolved z axis.
Figure 5.
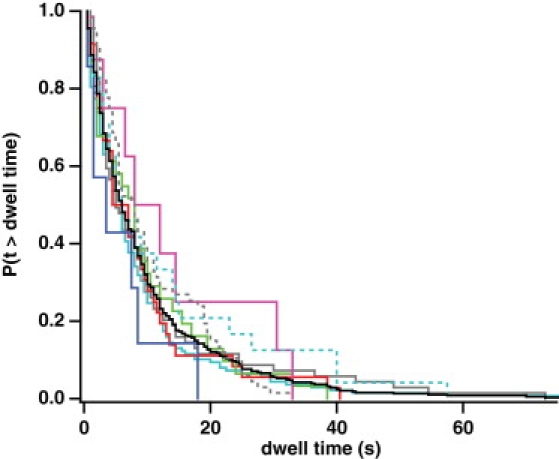
Myosin X steps in all directions from a common intermediate. On a fascin-actin bundle, the dwell time of myosin X before a step in any direction is part of the same single-exponential distribution. (Black solid trace) Survivor function of dwell times preceding steps in any direction (n = 363 dwells, 9.2 ± 0.5 s, mean ± SE). (Cyan solid trace) Positive x steps (along the x axis; n = 122 dwells, 8.5 ± 0.8 s). (Cyan dashed trace) Negative x steps (n = 22 dwells, 11.2 ± 2.6 s). (Gray solid trace) Positive y steps (along the y axis; n = 59 dwells, 8.1 ± 1.4 s). (Gray dashed trace) Negative y steps (n = 58 dwells, 10.6 ± 1.4 s). (Red trace) Quadrant I steps (n = 45 dwells, 7.2 ± 1.1 s). (Blue trace) Quadrant II steps (n = 9 dwells, 10.9 ± 3.9 s). (Magenta trace) Quadrant III steps (n = 8 dwells, 7.1 ± 2.1 s). (Green trace) Quadrant IV steps (n = 40 dwells, 9.7 ± 1.8 s). We detected no significant statistical differences among these nine distributions by a Wilcoxon rank-sum test at the α = 0.5 level.
Conclusions
Our goal in this study was to determine how myosin X uses the additional filaments in fascin-actin bundles to its advantage in processive movement. The bovine myosin X HMM-like construct (lacking the native cargo-binding domains) used in this report is essentially identical to the one used in our previous study (15), containing the head, the neck (3 IQ motifs), the entire SAH domain (amino acids 807–861), and the native predicted coiled-coil sequence (amino acids 862–920). Others have shown that dimerization is required for proper localization and filopodial formation activity (1,3,5,6). A forced dimer containing the head, neck, and only the first 34 residues of the SAH domain (75 residues of native sequence shorter than our construct) shows similar localization and motility in vivo as longer, nonforced dimer constructs (3). A drug-induced dimer construct of the myosin X head, neck, and SAH domain localizes to filopodial tips in the presence of the dimer-inducing drug, but not in the absence of it (6). Moreover, overexpression of the putative coiled-coil sequence acts as a dominant negative against myosin X's filopodial formation activity by disrupting the functional dimer (5). These results suggest that a dimer of the head, neck, and SAH domain of myosin X are required for its motor function, but the exact nature of dimerization beyond the SAH domain is unimportant for localization and in vivo function.
The previously reported enhancement in myosin X's processivity on bundled actin filaments (15), our finding of ∼18-nm steps on bundles (which would force rapid spiraling while walking on a single filament), and our observation of more than one sideways steps all strongly suggest that the motor often binds to more than one filament at a time within a bundle. This adds a level of complexity to a hand-over-hand stepping mechanism where there could be a difference between left and right steps based on the configuration of binding (Fig. 6). If the rear head is to the left of the motor's center, a straight forward step makes the newly rear head to the right of the motor's center. Such configurations would view the array of available actin binding sites as approximate mirror images of one another. This could lead to a given two-headed binding state having a higher probability of going right or left on the next step, but it would be balanced out over the course of many steps. Although we have examined our traces for alternating left-right asymmetries, we could not find clear signatures of such behavior, likely due to the presence of a small number of missed events.
Figure 6.
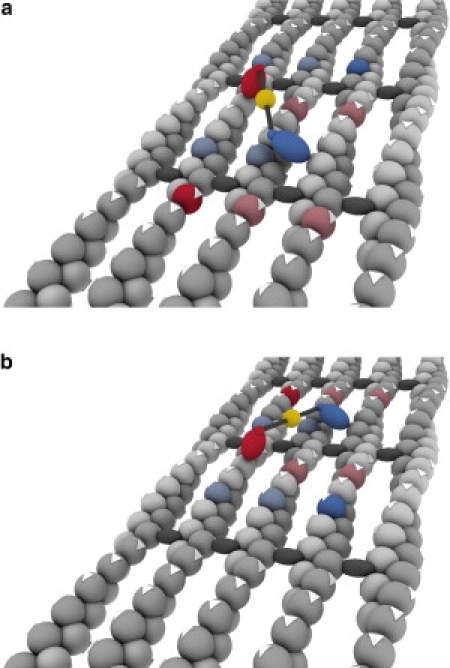
Model of potential stepping pattern of myosin X on a fascin-actin bundle. Myosin X walks toward the barbed end of actin (away from the observer). (a) A possible configuration for myosin X to bind to actin in a two-headed fashion, with the rear head (blue) to the right of the motor's center, the forward head (red) to the left of the motor's center, and the quantum dot (gold) at the approximate center-of-mass. If the rear head detaches and lands on the bright blue actin subunit 36 nm away from the head's starting position, the motor takes a straight and forward step of 18 nm. However, the rear head can also reach a number of other binding sites indicated by, but not limited to, the light-blue actin subunits, resulting in oblique forward steps or sideways steps to the left. The forward head may also detach instead of the rear head. If it lands on the bright-red actin subunit, the motor takes a straight backward step, but it could land on one of the light-red actin subunits, resulting in an oblique backward step or a sideways step to the right. (b) After the motor steps from its starting position in a to the solid-blue subunit in a, it could adopt a binding configuration as shown in b. Here, the pattern of positions of reachable subunits is the mirror image of that shown in a because the rear head is now to the left of the motor's center.
We imagine that myosin X operates similar to other myosin motors, with a working stroke biasing movement in the forward (positive x) direction, followed by a diffusional search for the next binding site. Such a diffusional search has been directly observed in myosin V (34,35). This ability to take sideways steps in either direction makes myosin X well-adapted to negotiating a path along the length of the fascin-actin bundle in a filopodium while anchored to the plasma membrane, avoiding potential obstacles attached to either the actin bundle or the membrane. When allowed to freely rotate around an actin filament while stepping, myosin V showed a slight bias for a left-handed spiral and myosin VI showed a bias for a right-handed spiral (36,37), both with a pitch >1 μm. In contrast, myosin X shows no preference for consistent right-leaning or left-leaning steps on bundles. If it did, we would expect to see runs on bundles frequently terminating to either the motor's left or right as it spirals into the coverslip. Instead, the motor appears to take a random walk perpendicular to the bundle. This observation suggests that the structural features responsible for bundle selection do not force the motor to wander in a particular direction around the bundle. We can rule out models where the working stroke of the motor forces steps to the left with each step (or, alternatively, steps to the right). Indeed, the observation of an equal fraction of steps to the left or right suggests a degree of lateral flexibility of the motor.
We speculate that the SAH domains may contribute to this lateral flexibility. These extended helical domains can contribute to the working strokes of myosin motors at loads <1 pN (38,39). At the same time, they are sufficiently flexible (compared to the 10-times-more-rigid IQ domains) to adopt multiple conformations, including 90° overall bends in the SAH domain under thermal forces (39). The SAH domains of myosin X appear to play a role in orienting the myosin X head such that the motor is selective for processive runs on bundled actin. To test this, we have separately observed the loss of selectivity in chimeras of myosin X and myosin V and in a myosin X mutant with a swivel introduced between the SAH and native coiled-coil (40). We further speculate that the SAH domains serve here as semiflexible springs that must be splayed-open for both motor domains to bind to actin. Binding along a bundle with an 18-nm head spacing might result in less distortion of the SAH domains compared to binding along a single actin filament, which would drive bundle selection. The precise arrangement of the two motor domains on a bundle, as well as the orientation of the lever arms in the prestroke and poststroke states, would be useful information to constrain this model; these details will be pursued in future work.
Our findings contrast with the recent work from Sun et al. (41), who report a step-size for myosin X of 34 nm. Although their native sequence of myosin X is 19-amino-acids longer than ours, their myosin X construct contains the myosin V coiled-coil sequence to assist in dimerization. Unlike our GCN4 sequence, this myosin V coiled-coil sequence is placed out of register with the heptad repeat of the native myosin X predicted coiled-coil. This could alter the structural contribution of the myosin X SAH domains to the motor's stepping, representing a potentially interesting mode of regulation where the motor can be switched from a state where its motility is unselective for bundles to a state where it is selective for bundles. Future work into the regulation of the myosin X motor domain will provide insight into these observed differences.
Myosin X, with its short step-size and ability to take some sideways steps while moving toward the barbed-end of actin, is well adapted to making long processive runs on fascin-actin bundles, allowing it to fulfill its roles in intrafilopodial transport. It also exemplifies the diversity among the myosin superfamily, where a single mechanochemical cycle has diverged both chemically and structurally to serve a wide variety of biological functions.
Acknowledgments
The authors thank Stanislav Nagy and Alicja Janik for help with protein purification; Stanislav Nagy, Crista Brawley, Melanie Norstrom, David Courson, and Alicja Janik for helpful discussions; and Shohei Koide and Jin Huang for the affinity clamp construct. The authors especially thank Kim Mortensen, L. Stirling Churchman, Jim Spudich, and Henrik Flyvbjerg for sharing preprints and code of their algorithm for maximum likelihood estimation of a two-dimensional Gaussian.
R.S.R. is supported by a grant from the National Institutes of Health (No. GM 078450).
Supporting Material
References
- 1.Berg J.S., Cheney R.E. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 2.Berg J.S., Derfler B.H., Cheney R.E. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J. Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 3.Kerber M.L., Jacobs D.T., Cheney R.E. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr. Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H., Berg J.S., Strömblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 5.Bohil A.B., Robertson B.W., Cheney R.E. Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl. Acad. Sci. USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokuo H., Mabuchi K., Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J. Cell Biol. 2007;179:229–238. doi: 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tokuo H., Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 2004;319:214–220. doi: 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- 8.Cox D., Berg J.S., Greenberg S. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat. Cell Biol. 2002;4:469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- 9.Weber K.L., Sokac A.M., Bement W.M. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- 10.Woolner S., O'Brien L.L., Bement W.M. Myosin-10 and actin filaments are essential for mitotic spindle function. J. Cell Biol. 2008;182:77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sousa A.D., Berg J.S., Cheney R.E. Myo10 in brain: developmental regulation, identification of a headless isoform and dynamics in neurons. J. Cell Sci. 2006;119:184–194. doi: 10.1242/jcs.02726. [DOI] [PubMed] [Google Scholar]
- 12.Adams J.C. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Vignjevic D., Kojima S., Borisy G.G. Role of fascin in filopodial protrusion. J. Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medalia O., Beck M., Gerisch G. Organization of actin networks in intact filopodia. Curr. Biol. 2007;17:79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Nagy S., Ricca B.L., Rock R.S. A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta A.D., Rock R.S., Cheney R.E. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 17.Rock R.S., Rice S.E., Sweeney H.L. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight P.J., Thirumurugan K., Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 19.Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 20.Huang J., Koide A., Koide S. Design of protein function leaps by directed domain interface evolution. Proc. Natl. Acad. Sci. USA. 2008;105:6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J., Nagy S.S., Koide S. A peptide tag system for facile purification and single-molecule immobilization. Biochemistry. 2009;48:11834–11836. doi: 10.1021/bi901756n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardee J.D., Spudich J.A. Purification of muscle actin. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- 23.Vignjevic D., Peloquin J., Borisy G.G. In vitro assembly of filopodia-like bundles. Methods Enzymol. 2006;406:727–739. doi: 10.1016/S0076-6879(06)06057-5. [DOI] [PubMed] [Google Scholar]
- 24.Block S.M., Goldstein L.S., Schnapp B.J. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 25.Sage D., Neumann F.R., Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- 26.Mortensen K.I., Churchman L.S., Flyvbjerg H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Methods. 2010;7:377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijering E., Jacob M., Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 28.Kerssemakers J.W., Munteanu E.L., Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 29.Yildiz A., Tomishige M., Vale R.D. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali M.Y., Krementsova E.B., Warshaw D.M. Myosin Va maneuvers through actin intersections and diffuses along microtubules. Proc. Natl. Acad. Sci. USA. 2007;104:4332–4336. doi: 10.1073/pnas.0611471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross J.L., Ali M.Y., Warshaw D.M. Cargo transport: molecular motors navigate a complex cytoskeleton. Curr. Opin. Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolomeisky A.B., Stukalin E.B., Popov A.A. Understanding mechanochemical coupling in kinesins using first-passage-time processes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;71:031902. doi: 10.1103/PhysRevE.71.031902. [DOI] [PubMed] [Google Scholar]
- 33.Norstrom M.F., Smithback P.A., Rock R.S. Unconventional processive mechanics of non-muscle myosin IIB. J. Biol. Chem. 2010;285:26326–26334. doi: 10.1074/jbc.M110.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn A.R., Spudich J.A. Dynamics of the unbound head during myosin V processive translocation. Nat. Struct. Mol. Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 35.Uemura S., Higuchi H., Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat. Struct. Mol. Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- 36.Ali M.Y., Homma K., Ikebe M. Unconstrained steps of myosin VI appear longest among known molecular motors. Biophys. J. 2004;86:3804–3810. doi: 10.1529/biophysj.103.037416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali M.Y., Uemura S., Ishiwata S. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat. Struct. Biol. 2002;9:464–467. doi: 10.1038/nsb803. [DOI] [PubMed] [Google Scholar]
- 38.Baboolal T.G., Sakamoto T., Peckham M. The SAH domain extends the functional length of the myosin lever. Proc. Natl. Acad. Sci. USA. 2009;106:22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivaramakrishnan S., Sung J., Spudich J.A. Combining single-molecule optical trapping and small-angle x-ray scattering measurements to compute the persistence length of a protein ER/K alpha-helix. Biophys. J. 2009;97:2993–2999. doi: 10.1016/j.bpj.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagy S., Rock R.S. A structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J. Biol. Chem. 2010;285:26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Y., Sato O., Goldman Y.E. Single-molecule stepping and structural dynamics of myosin X. Nat. Struct. Mol. Biol. 2010;17:485–491. doi: 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


