Abstract
A complete set of genetic tools is still being developed for the micro-alga Chlamydomonas reinhardtii. Yet even with this incomplete set, this photosynthetic single-celled plant has demonstrated significant promise as a platform for recombinant protein expression. In recent years, techniques have been developed that allow for robust expression of genes from both the nuclear and plastid genome. With these advances, many research groups have examined the pliability of this and other micro-algae as biological machines capable of producing recombinant peptides and proteins. This review describes recent successes in recombinant protein production in Chlamydomonas, including production of complex mammalian therapeutic proteins and monoclonal antibodies at levels sufficient for production at economic parity with existing production platforms. These advances have also shed light on the details of algal protein production at the molecular level, and provide insight into the next steps for optimizing micro-algae as a useful platform for the production of therapeutic and industrially relevant recombinant proteins.
Keywords: Chlamydomonas, Micro-algae, Recombinant protein expression, Therapeutic protein production
Introduction
As genetically accessible photosynthetic organisms, algae are now recognized for their potential as a platform for recombinant protein expression where large scale and reduced material costs are important. The most-commonly used eukaryotic model alga, Chlamydomonas reinhardtii, has recently been shown to be able to fill this role, and this review will primarily discuss the technical and biological advances made in recombinant protein production in this alga. Algae have now come of age as a platform for recombinant protein expression.
All three genomes (chloroplast, mitochondrial, and nuclear) can be transformed in Chlamydomonas, and each has distinct transcriptional, translational, and post-translational properties that make them distinct. Each of these genomes has been fully sequenced, providing a wealth of information and a strong foundation for targeted manipulation (Maul et al. 2002; Popescu and Lee 2007; Merchant et al. 2007). Recent efforts have primarily focused on understanding and improving gene transformation, mRNA transcript accumulation, and protein accumulation of nuclear and chloroplast recombinant genes.
Chlamydomonas is an excellent system for reasons beyond its genetic and metabolic malleability. This alga has a rapid doubling time (about 10 h), it is easily scaled in homogenous culture as an aqueous microbe, it can be grown either photoautotrophically or with acetate as a reduced carbon source, and it has a controllable and rapid sexual cycle (about 2 weeks) with stable and viable haploids.
Advantages of algal protein production
Protein production in plants provides a number of advantages not found in other production platforms. First, a major advantage that all plant protein production systems have over cell culture systems (including bacteria, yeast, and mammalian cell culture) is the potential for significant reduction in cost. It is estimated that protein production in transgenic plants can be as much as four orders of magnitude less expensive than production in mammalian cell culture, on a per gram of unpurified protein basis (Dove 2002). Secondly, plant-produced proteins are not susceptible to viral or prion contamination that can harm humans, as is always a concern with animal cell culture (Chebolu and Daniell 2009). Third, as eukaryotes, algae and other plants possess the chaperones and cellular machinery required to fold complex human proteins that bacteria and yeast may not be able to process properly (Franklin and Mayfield 2004). Finally, many species of green algae are considered GRAS (generally regarded as safe) (Rosenberg et al. 2008), meaning that if the protein can be expressed in a bioavailable form, purification steps could potentially be eliminated altogether.
Algae possess a number of advantages over transgenic plant systems for the production of recombinant proteins. They can be grown in contained bioreactors, reducing the risk of contamination of the production system by airborne contaminants, and also protecting the environment from any potential flow of transgenes into the surrounding ecosystem. Growth in containment also greatly reduces the potential for loss of the crop due to predation or pathogen attack. Algae progress from initial transformation to large-scale protein production in a matter of weeks, compared to timescales on the order of months or years in higher plants such as corn or tobacco (Franklin and Mayfield 2004). As micro-algae are all a single cell type, there should also be less variation in recombinant protein accumulation, making downstream processing more uniform.
Production of recombinant proteins in chloroplasts also possesses several unique attributes. At present transgenic proteins can accumulate to much higher levels in the chloroplast than when expressed from the nuclear genome, mainly because plastids lack gene silencing mechanisms and other mechanisms that reduce recombinant protein production from nuclear encoded genes (Bock 2007). Chloroplasts can be transformed with multiple genes in a single event, due to the availability of multiple insertion sites, as well as an ability to process polycistronic transcripts, allowing an entire gene cassette to be regulated by a single promoter (Rymarquis et al. 2006; Bock 2007). Additionally, proteins produced within the chloroplast are not glycosylated (Franklin and Mayfield 2005), which can prove useful in many applications such as producing antibodies that are similar to native antibodies in their ability to recognize their antigen, but whose lack of glycosylation prevents them from recruiting killer cells (Tran et al. 2009). In fact, it is estimated that over two-thirds of the therapeutic human monoclonal antibodies in the testing pipeline do not require glycosylation for therapeutic function (Dove 2002).
Genetic tools and techniques
Transformation techniques
The plastid genome can be reliably transformed through homologous recombination using bombardment by DNA-coated gold or tungsten particles (Koop et al. 2007). Nuclear transformation in algae can also be achieved by biolistic bombardment, but the preferred methods are electroporation or agitation with glass beads using a cell-wall defective strain (Eichler-Stahlberg et al. 2009; Leon and Fernandez 2007). New transformation techniques using the Cre/lox recombination system have been demonstrated to recombine in the nuclear genome of Chlamydomonas (Heitzer and Zschoernig 2007). Robust in vivo recombinant reporters, including GFPs (Fuhrmann et al. 1999; Franklin et al. 2002) and luciferases (Mayfield and Schultz 2004; Shao and Bock 2008), have been developed for tracking both nuclear and chloroplast gene expression. Techniques that have been employed previously on higher plants, such as transformation by Agrobacterium tumefaciens, have also been demonstrated to work with Chlamydomonas (Kumar et al. 2004). However, nuclear transformants still generally fail to accumulate recombinant proteins to the levels observed in plastids, most likely due to nuclear silencing mechanisms (see Nuclear Gene Silencing below).
Codon optimization
As with other expression systems, codon optimization has played a large role in the success of recombinant protein expression in both the chloroplast and nuclear genomes of Chlamydomonas (Heitzer et al. 2007). The nuclear genome and the plastid genome have highly divergent codon usage, with the chloroplast preferring an A or T in the wobble position while the nuclear genome prefers a G or C (Nakamura et al. 2000). Using GFP, early work showed that codon optimization to reflect the genome bias could increase transgene protein accumulation 5-fold in the nucleus (Fuhrmann et al. 1999) and up to 80-fold in the chloroplast (Franklin et al. 2002). Today, recombinant genes are universally codon optimized for improved protein expression in almost every system (Xia 2007; Puigbo et al. 2007, 2008).
Chloroplast gene regulation
Promoter and regulatory mRNA untranslated region (UTR) choices are both crucial factors in transgene expression levels. In chloroplasts, the most successful promoter to date in algae is the psbA promoter in combination with the psbA UTRs (Manuell et al. 2007; Surzycki et al. 2009). However, there are two caveats with using the psbA regulatory elements. First, they appear to be highly auto-attenuated; if any of the psbA gene product (D1 protein) is present, it will strongly decrease expression of any recombinant coding sequence under the control of its 5′-UTR. Secondly, since D1 is essential for the function of photosystem II, psbA knockouts are nonphotosynthetic. This would clearly negate the benefits of using a photosynthetic organism for protein production. There is evidence that reintroducing an attenuated psbA gene at a new locus elsewhere in the plastid genome can restore photosynthesis while only mildly reducing recombinant protein production (Manuell et al. 2007).
Other UTRs in use for transgene expression include those from the endogenous atpA, rbcL, and psbD genes (Fletcher et al. 2007; Hallmann 2007). These have been used with varying levels of success, though as with the psbA promoter, it is unclear why certain regulatory elements engender high expression levels with some genes but not others (Marin-Navarro et al. 2007). While endogenous promoters have been primarily used, other exogenously induced expression systems have been explored in the chloroplast. It has been demonstrated that inducible systems, such as the lac operon system from E. coli, can be engineered into the Chlamydomonas chloroplast (Kato et al. 2007), and more recently a riboswitch was shown to work to regulate translation in Chlamydomonas chloroplasts using a small molecule for induction (Croft et al. 2007), similar to their use in many other expression platforms (Suess 2005; Winkler and Breaker 2005).
While regulation of gene expression occurs at both the transcriptional and translational level in the nucleus, it appears that most regulation is post-transcriptional in the plastid (Marin-Navarro et al. 2007). However, many of the activators and suppressors of mRNA splicing and processing in the chloroplast are indeed encoded by nuclear genes (Boudreau et al. 2000; Somanchi et al. 2005; Raynaud et al. 2007; Schwarz et al. 2007; Loiselay et al. 2008). A multi-component copper-induced system has been designed as a switch for chloroplast protein expression. This system utilizes the nuclear-encoded Nac2 chloroplast protein necessary for stable accumulation of psbD RNA by acting on its 5′-regulatory region. By transforming a copper induced cytochrome c6 promoter fused to the Nac2 coding sequence into a Nac2 deficient strain, proteins encoded with the psbD regulatory region only accumulate in the presence of copper (Surzycki et al. 2007). Additionally, nuclear gene products are required for splicing of group I introns in the chloroplast 23S rRNA and psbA genes, and can potentially be used to regulate plastid gene expression (Li et al. 2002).
Post-transcriptional control in the chloroplast is mediated by both cis- and trans-acting-elements. The 5′-UTR of chimeric chloroplast mRNAs was shown to significantly impact recombinant protein production, while 3′-UTRs had little if any effect (Barnes et al. 2005). Cis-acting elements in 3′-UTRs include UG-repeats that are used in circadian protein regulation (Kiaulehn et al. 2007) and inverted repeat sequences that contribute to mRNA processing (Goldschmidt-Clermont et al. 2008). Recent work has identified a complex comprising an RNA stabilizing factor and a translational activator that appears to be specific to the D2 protein of photosystem II (Schwarz et al. 2007). Post-transcriptional control mechanisms can also have synergistic or antagonistic effects. Kasai et al. (2003) used the GUS reporter controlled by various endogenous 3′- and 5′-UTRs to determine that there is an inverse correlation between protein accumulation and transcript stability, suggesting a feedback mechanism. Together all of these data suggest that regulation of mRNA translation has the greatest impact on recombinant protein accumulation, and is clearly an area where additional research seems likely to identify mechanisms for increased recombinant protein accumulation. For a comprehensive review of chloroplast translation regulation, see Marin-Navarro et al. 2007.
Nuclear gene regulation
In the nucleus, the most commonly used promoters are those from the HSP70A, psaD, and rbcS2 genes (Schroda et al. 2002; Fischer and Rochaix 2001; Berthold et al. 2002). Chimeric promoters have also demonstrated high levels of transcription and expression (Schroda et al. 2000; Fischer and Rochaix 2001; Wu et al. 2008). Additionally, placing endogenous intronic sequences in transgenes has been shown to enhance expression regardless of orientation or position. The first intron from the endogenous rbcS2 gene has shown particular efficacy in increasing mRNA and protein accumulation and is now commonly used to enhance recombinant gene expression (Lumbreras et al. 1998).
In the nucleus, gene expression can be induced by a number of factors, including heat-shock or metal addition (Wu et al. 2008; Ferrante et al. 2008). The Nit1 promoter suppresses transcription in the presence of ammonia, but induces transcription when cells are grown in nitrate- or nitrite-containing media (Ohresser et al. 1997). More recently, iron-deficiency response elements (FeREs) in nuclear gene promoters have been characterized in Chlamydomonas (Fei and Deng 2007). Signaling cascades triggered by photo-oxidative stress in the chloroplast can also activate transcription of specific nuclear genes, indicating that gene regulation in Chlamydomonas is not necessarily localized to the site of signal production (Fischer et al. 2007). More recently, signaling molecules such as Mg-protoporphyrin and heme produced in the plastid have been shown to activate transcription of nuclear genes such as HSP70A through an interaction with the plastid response element (PRE) (von Gromoff et al. 2008). While these inducible systems provide great insight into gene regulatory strategies, there is still significant work to be done before robust production of recombinant proteins can be routinely achieved from nuclear genes in algae.
Nuclear gene silencing
Transgene silencing is a significant obstacle for recombinant protein expression in Chlamydomonas nuclear transformants, but recent work is helping to overcome this problem (Casas-Mollano et al. 2007, 2008a, b). Neupert et al. (2009) have developed strains with impaired transgene silencing by using UV mutagenesis and selection on media that permits higher antibiotic tolerance proportional to higher expression of the transgene product, to select strains with improved protein accumulation. However, their most impressive yields of exogenous protein accumulation are only 0.2% of total soluble protein (TSP), as compared to nearly 10% TSP obtained in plastids (Manuell et al. 2007). It has been postulated that gene silencing may be difficult to eliminate because it may have evolved as a protective measure against intracellular pathogens or viruses (Rosenberg et al. 2008; Neupert et al. 2009). It appears that combating gene silencing will be a major hurdle before recombinant proteins can be expressed at economically viable levels from nuclear transgenes in Chlamydomonas.
Current successes in algal protein production
Plastid transformation in higher plants has identified a few recombinant proteins that accumulate to very high levels, reported up to as high as 70% of total protein for some antibiotic proteins in tobacco leaves (Oey et al. 2009), but in general recombinant protein expression is highly variable. For an extensive review of vaccine production in plants, see Davoodi-Semiromi et al. (2009). However, very recent work in Chlamydomonas has demonstrated that fully bioactive proteins can indeed be produced to appreciable levels in green algae. A recent technique has been exploited to improve protein accumulation and stability by expression of cleavable fusions to highly expressed endogenous proteins (Muto et al. 2009), or to highly expressed recombinant proteins in the chloroplast (Rasala et al. 2010).
The first demonstration of mammalian protein expression in the chloroplast was of a large single-chain antibody (HSV8-lsc) directed against glycoprotein D of the herpes simplex virus (Mayfield et al. 2003). The protein was soluble, suggesting that it was correctly folded, and electrophoresis indicated the formation of the dimer by disulfide bond formation. This work was followed up by the expression of a single chain fragment variable antibody that accumulated to 0.54% TSP (Mayfield and Franklin 2005). More recently a full-length human IgG1 monoclonal antibody, directed against anthrax protective antigen 83 (83K7C), was expressed in the chloroplast of Chlamydomonas (Tran et al. 2009). Unlike the previously expressed lsc antibody, this antibody was assembled in the chloroplast from separately expressed light chain and heavy chain proteins, and it could be purified at 100 μg per 1 g dry algal biomass, and was found to have binding activity identical to that of the same antibody expressed in a traditional mammalian cell culture system (Tran et al. 2009).
A host of non-antibody recombinant proteins have been expressed in the chloroplast for therapeutic purposes. The human metallothionein-2 gene product, which is considered to have anti-radiation function, was expressed and demonstrated to improve the survivorship of transgenic algae compared to wild type algae (Zhang et al. 2006). The human tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) protein, known to induce apoptosis in virus-infected and tumor cells, accumulated in the chloroplast at 0.43–0.67% TSP based on densitometric analysis of Western blot (Yang et al. 2006). The expression of human glutamic acid decarboxylase (hGAD65) was also achieved in the chloroplast of Chlamydomonas (Wang et al. 2008). This protein is an important autoantigenic marker in type I diabetes, and the algae-derived protein remained immunologically active, accumulating at 0.25–0.3% TSP. Human erythropoietin (Epo), used in the treatment of anemia, was fused to an export sequence and expressed from the nuclear genome (Eichler-Stahlberg et al. 2009). Protein was detected in and isolated from the culture medium, although with uncharacterized post-translational modification closely matching the mass of endogenous human Epo. Also, biologically active bovine mammary-associated serum amyloid (M-SAA) was expressed in the chloroplast (Manuell et al. 2007). Notably, the accumulation of this soluble protein was above 5% TSP as quantified by Western blot, with levels of expression determined to be more than twice that amount using ELISA quantification.
A fusion protein containing the foot-and-mouth disease virus VP1 gene and the cholera toxin B subunit (CTBVP1) was produced in the chloroplast as well (Sun et al. 2003). The protein was reported to accumulate to 3% total protein by ELISA quantification, and retained GM1-ganglioside binding activity and antigenicity. Another viral protein, classical swine fever virus E2 structural protein, was expressed in the chloroplast (He et al. 2007). ELISA quantification indicated the accumulation of the E2 protein to 1.5–2% TSP, and retained immunological activity. Similarly, along with a list of other recombinant proteins, the white spot syndrome virus protein 28 (VP28) was expressed in the chloroplast (Surzycki et al. 2009). VP28 was reported to accumulate to a striking 10.5% TSP, although no data was presented to show how this level of expression was determined. Recently, the D2 fibronectin-binding domain from a Staphylococcus aureus protein was fused to the B subunit of cholera toxin, and expressed in the chloroplast (Dreesen et al. 2010). The transgenic algae were fed to mice, and induced resistance against lethal doses of S. aureus, presumably by eliciting a systemic antigenic response to the S. aureus peptide. This is the first demonstration of the functional possibility of orally delivered vaccines from algal production.
More recently, Rasala et al. (2010) attempted the expression of a set of seven recombinant proteins in the chloroplast and were met with very good success (four out of the seven genes expressed at economically viable levels). This work demonstrates that recombinant protein expression in algal chloroplasts is on par with any other expression platform, and shows that expression of complex mammalian proteins is as likely to be achieved in algae as it is in any eukaryotic system. These data are summarized in Table 1.
Table 1.
Recent successes in therapeutic protein production in algae
| Gene expressed | Function | Expression level achieved | Application | Source |
|---|---|---|---|---|
| HSV8-lsc | First mammalian protein expressed, antibody | Detectable | Pharmaceutical | Mayfield et al. (2003) |
| CTB-VP1 | Cholera toxin B subunit fused to foot and mouth disease VP1 | 3% TSP | Vaccine | Sun et al. (2003) |
| HSV8-scFv | Classic single-chain antibody | 0.5% TSP | Pharmaceutical | Mayfield et al. (2005) |
| hMT-2 | Human metallothionine-2 | Detectable | Pharmaceutical, UV-protection | Zhang et al. (2006) |
| hTRAIL | Human tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) | ~0.67% TSP | Pharmaceutical | Yang et al. (2006) |
| M-SAA | Bovine mammary-associated serum amyloid | ~5% TSP | Therapeutics, oral delivery | Manuell et al. (2007) |
| CSFV-E2 | Swine fever virus E2 viral protein | ~2% TSP | Vaccine | He et al. (2007) |
| hGAD65 | Diabetes-associated anutoantigen human glutamic acid decarboxylase 65 | ~0.3% TSP | Diagnostics and therapeutics | Wang et al. (2008) |
| ARS2-crEpo-his6 | Human erythropoietin fused to ARS2 export sequence w/6xhis tag | 100 μg/l culture | Pharmaceutical, protein export | Eichler-Stahlberg et al. (2009) |
| 83K7C | Full-length IgG1 human monoclonal antibody against anthrax protective antigen 83 | 0.01% dry algal biomass | Therapeutics | Tran et al. (2009) |
| IgG1 | Murine and human antibodies (LC and HC) | Detectable | Therapeutics | Tran et al. (2009) |
| VP28 | White spot syndrome virus protein 28 | ~10.5% TSP | Vaccine | Surzycki et al. (2009) |
| CTB-D2 | D2 fibronectin-binding domain of Staphylococcus aureus fused with the cholera toxin B subunit | 0.7% TSP | Oral vaccine | Dreesen et al. (2010) |
| 10NF3, 14FN3 | Domains 10 and 14 of human fibronectin, potential antibody mimics | 14FN3: 3% TSP 10FN3: detectable | Therapeutics | Rasala et al. (2010) |
| M-SAA-Interferon β1 | Multiple sclerosis treatment fused to M-SAA | Detectable | Therapeutics | Rasala et al. (2010) |
| Proinsulin | Blood sugar level-regulating hormone, type I diabetes treatment | Detectable | Therapeutics | Rasala et al. (2010) |
| VEGF | Human vascular endothelial growth factor isoform 121 | 2% TSP | Therapeutics | Rasala et al. (2010) |
| HMGB1 | High mobility group protein B1 | 2.5% TSP | Therapeutics | Rasala et al. (2010) |
Potential future applications
Oral vaccines
Traditional vaccines are normally produced from an attenuated or killed form of the pathogenic organism itself. An alternative approach is to produce a pathogen antigen as a recombinant protein, and this is now used for some select vaccines like the hepatitis A vaccine (Powdrill and Johnston 1991). As algae contain very sophisticated protein folding machinery that bacteria and other prokaryotes lack, algae can be used to produce complex eukaryotic proteins that cannot be easily produced on large scale in bacterial culture without costly denaturation and refolding steps. Algae are also ideal for producing vaccines against pathogens which exhibit little or no glycosylation, such as those from the parasites plasmodium (Gowda and Davidson 1999).
Algae are also especially suited for the production of oral vaccines on a large scale due to their GRAS status. Once the process of oral delivery of a vaccine protein is refined, algae could produce inexpensive oral vaccines, making vaccination an accessible form of disease management for a whole host of third-world diseases. Furthermore, the oral delivery method and the option to store doses at ambient temperature would allow vaccines to be transported and administered to remote populations without the need for expensive refrigeration or highly trained medical personnel (Chebolu and Daniell 2009).
Evidence for the feasibility of plastid-produced vaccines has been provided by several groups who have produced antigens that elicit similar immune responses as the actual pathogen when injected in standard vaccine adjuvants (Tregoning et al. 2003; Koya et al. 2005; Molina and Shoenfeld 2005; Chebolu and Daniell 2009). Furthermore, fusions of antigens with the cholera toxin B subunit show promise for eliciting immune response from mucosal delivery alone, as the cholera toxin B subunit allows a fused protein to penetrate the intestinal lining (Sun et al. 2003; Harakuni et al. 2005).
Discovery of novel bioactive molecules
Screening of diverse algal species for novel bioactive compounds has become a field of intense interest lately, as algae are a rich source of secondary metabolites that often have industrial or nutritional implications. A wide range of functional molecules produced by algae—everything from antioxidants to pigments used in laboratory analytical techniques—have been isolated (Plaza et al. 2009). High-throughput screening methods comprising pressurized liquid extraction; functional characterization of antioxidant or antimicrobial activity; and chemical characterization by HPLC and GC-MS have been optimized to streamline the screening process (Plaza et al. 2010).
Protein secretion
Figure 1 illustrates the basic structure of Chlamydomonas reinhardtii, as well as the cellular locations of recombinant protein accumulation depending on the gene construct and transformation method. Plastid transformation results in accumulation of the transgene product in the single large chloroplast. Nuclear gene products accumulate in the cytosol by default, but nuclear transformation with appropriate signal sequences allows for targeting to the endoplasmic reticulum and Golgi for export (Griesbeck et al. 2006) or for localization to the cell membrane, which may be sufficient for antigenic recognition in the case of an oral vaccine (Eichler-Stahlberg et al. 2009).
Fig. 1.
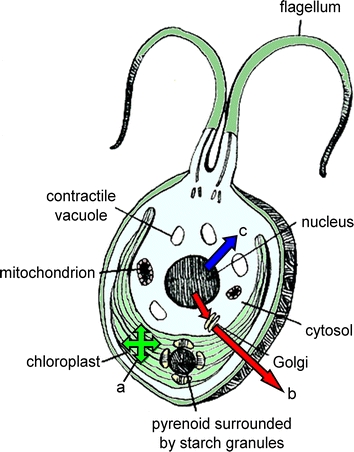
Chlamydomonas reinhardtii as a versatile recombinant protein production platform. Protein expressed from the chloroplast genome is accumulated inside the single large chloroplast (a). The reducing environment of the chloroplast allows for proper folding of heavily disulfide bonded proteins, which is not easily accomplished in bacterial production platforms. Protein expressed from the nuclear genome accumulates in the cytosol (b), unless it is given an export signal sequence. In this case, it is sent to the endoplasmic reticulum for translocation and processing and then moves to the Golgi apparatus for packaging and export to the extracellular media (c)
Other algal transformation efforts
Genetic manipulation of algae is no longer limited to Chlamydomonas reinhardtii. Recent successes in generating transgenic algae are growing in number. Exogenous genes have been expressed in the unicellular charophyte alga, Closterium peracerosum–strigosum–littorale complex (Abe et al. 2008). The nuclear genome of volvocine alga Gonium pectorale has been stably transformed (Lerche and Hallmann 2009), as has the chlorophyceae Haematococcus pluvialis (Kathiresan and Sarada 2009), by co-cultivation with Agrobacterium (Kathiresan et al. 2009). Some successes have been made by transient transformation of marine chlorarachniophyte Lotharella amoebiformis (Hirakawa et al. 2008), chlorophyta alga Ulva pertusa (Kakinuma et al. 2009), and red alga Cyanidioschyzon merolae (Ohnuma et al. 2008). Methods have also been improved for previously transformed alga, such as Dunaliella salina (Feng et al. 2009), and cyanobacterial genetics have also been extensively explored, but will not be discussed in this review.
Future challenges
A significant obstacle for algal protein expression systems is the lack of production systems optimized for large-scale growth and harvesting of algae under photoautotrophic conditions. The main limitations in photobioreactor size arise from inhibited gas exchange and light penetration in large cultures, especially at the high cell densities required to keep costs low (Ugwu et al. 2008). An alternative approach, which has been discussed extensively by Chen and Chen (2006), is to grow algae heterotrophically in conventional fermentation bioreactors. This is certainly an economically viable option for high-value products such as therapeutics or enzymes, and indeed is currently used in much of the micro-algae products industry (Chen and Chen 2006) with yields reported as high as 83 g dry weight per liter of culture for some species (de Swaaf et al. 2003). Maximum yields for Chlamydomonas reinhardtii are around 1.5 g dry biomass per liter when grown in continuous flow on acetate media (Chen and Johns 1996).
The scale and cost requirements for algal biofuels will likely necessitate photoautotrophic open cultivation systems. These systems are being improved, as are processes for efficiently harvesting the algae. For example, new methods have been optimized for harvesting algae using microbes capable of flocculating 90% of the algal mass with no deleterious effect on algal viability (Lee et al. 2009). More efficient methods and systems for large-scale growth are critical if algal-derived biofuels are to become a reality. If these issues can be resolved, algae represent a far superior source of biofuel than terrestrial plants. It is estimated algae can produce up to ten times as much oil per acre than any current terrestrial crop (Cooney et al. 2009). For a detailed recent review on the potential of algal biofuels, see Mata et al. 2010.
Conclusion
Several decades of work in Chlamydomonas has elucidated a better understanding of the transcriptional and translational machinery and regulation of the cell, ultimately generating improved methods for transgenic expression of recombinant proteins in algae. However, only in the past few years have these advances been used to successfully express large, fully bioactive, therapeutically relevant proteins at a level sufficient for economically viable large-scale production. Extensive research on optimal transformation constructs and gene optimization have greatly increased yields of recombinant protein, though further work is still needed to address nuclear gene silencing, plastid auto-attenuation, and to optimize reactor design for large-scale use. Algae have proven their utility and tractability as a production system for therapeutic or industrial proteins and peptides, and algae now seem poised to become the “green” alternative to current mammalian, yeast, or bacterial recombinant protein production systems.
Acknowledgements
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases to SPM (AI059614). ES is supported by a National Science Foundation pre-doctoral fellowship.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abe J, Hiwatashi Y, Ito M, et al. Expression of exogenous genes under the control of endogenous HSP70 and CAB promoters in the Closterium peracerosum-strigosum-littorale complex. Plant Cell Physiol. 2008;49:625–632. doi: 10.1093/pcp/pcn039. [DOI] [PubMed] [Google Scholar]
- Barnes D, Franklin S, Schultz J, et al. Contribution of 5′- and 3′-untranslated regions of plastid mRNAs to the expression of Chlamydomonas reinhardtii chloroplast genes. Mol Genet Genomics. 2005;274:625–636. doi: 10.1007/s00438-005-0055-y. [DOI] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W. An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153:401–412. doi: 10.1078/14344610260450136. [DOI] [PubMed] [Google Scholar]
- Bock R. Plastid biotechnology: prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr Opin Biotechnol. 2007;18:100–106. doi: 10.1016/j.copbio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Boudreau E, Nickelsen J, Lemaire SD, et al. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. Eur Mol Biol Organ J. 2000;19:3366–3376. doi: 10.1093/emboj/19.13.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano JA, van Dijk K, Eisenhart J, et al. SET3p monomethylates histone H3 on lysine 9 and is required for the silencing of tandemly repeated transgenes in Chlamydomonas. Nucleic Acids Res. 2007;35:939–950. doi: 10.1093/nar/gkl1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano JA, Jeong B-r, Xu J, et al. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc Natl Acad Sci. 2008;105:6486–6491. doi: 10.1073/pnas.0711310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano JA, Rohr J, Kim E-J, et al. Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics. 2008;179:69–81. doi: 10.1534/genetics.107.086546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebolu S, Daniell H (2009) Chloroplast-derived vaccine antigens and biopharmaceuticals: expression, folding, assembly and functionality. Plant-Produced Microbial Vaccines, pp 33–54 [DOI] [PMC free article] [PubMed]
- Chen G-Q, Chen F. Growing phototrophic cells without light. Biotechnol Lett. 2006;28:607–616. doi: 10.1007/s10529-006-0025-4. [DOI] [PubMed] [Google Scholar]
- Chen F, Johns MR. Heterotrophic growth of Chlamydomonas reinhardtii on acetate in chemostat culture. Process Biochem. 1996;31:601–604. doi: 10.1016/S0032-9592(96)00006-4. [DOI] [Google Scholar]
- Cooney M, Young G, Nagle N. Extraction of bio-oils from microalgae. Sep Purif Rev. 2009;38:291–325. doi: 10.1080/15422110903327919. [DOI] [Google Scholar]
- Croft MT, Moulin M, Webb ME, et al. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi-Semiromi A, Samson N, Daniell H. The green vaccine: a global strategy to combat infectious and autoimmune diseases. Hum Vaccin. 2009;5:488–493. doi: 10.4161/hv.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Swaaf ME, Pronk JT, Sijtsma L. Fed-batch cultivation of the docosahexacaoic-acid-producing marine alga Crypthecodinium colunii on ethanol. Appl Microbiol Biotechnol. 2003;61:40–43. doi: 10.1007/s00253-002-1118-1. [DOI] [PubMed] [Google Scholar]
- Dove A. Uncorking the biomanufacturing bottleneck. Nat Biotechnol. 2002;20:777–779. doi: 10.1038/nbt0802-777. [DOI] [PubMed] [Google Scholar]
- Dreesen IAJ, Charpin-El Hamri G, Fussenegger M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J Biotechnol. 2010;145:273–280. doi: 10.1016/j.jbiotec.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Eichler-Stahlberg A, Weisheit W, Ruecker O, et al. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta. 2009;229:873–883. doi: 10.1007/s00425-008-0879-x. [DOI] [PubMed] [Google Scholar]
- Fei XW, Deng XD. A novel Fe deficiency-responsive element (FeRE) regulates the expression of atx1 in Chlamydomonas reinharditii. Plant Cell Physiol. 2007;48:1496–1503. doi: 10.1093/pcp/pcm110. [DOI] [PubMed] [Google Scholar]
- Feng S, Xue L, Liu H, et al. Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol Biol Rep. 2009;36:1433–1439. doi: 10.1007/s11033-008-9333-1. [DOI] [PubMed] [Google Scholar]
- Ferrante P, Catalanotti C, Bonente G, et al. An optimized, chemically regulated gene expression system for Chlamydomonas. PLoS ONE. 2008;3(9):e3200. doi: 10.1371/journal.pone.0003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Rochaix JD. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genomics. 2001;265:888–894. doi: 10.1007/s004380100485. [DOI] [PubMed] [Google Scholar]
- Fischer BB, Krieger-Liszkay A, Hideg E, et al. Role of singlet oxygen in chloroplast to nucleus retrograde signaling in Chlamydomonas reinhardtii. FEBS Lett. 2007;581:5555–5560. doi: 10.1016/j.febslet.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Fletcher SP, Muto M, Mayfield SP. Optimization of recombinant protein expression in the chloroplasts of green algae. In: Leon R, Galvan A, Fernandez E, editors. Advances in experimental medicine and biology, vol 616: transgenic microalgae as green cell factories. New York, NY: Landes Bioscience and Springer Science Business Media LLC; 2007. pp. 90–98. [DOI] [PubMed] [Google Scholar]
- Franklin SE, Mayfield SP. Prospects for molecular farming in the green alga Chlamydomonas reinhardtii. Curr Opin Plant Biol. 2004;7:159–165. doi: 10.1016/j.pbi.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Franklin SE, Mayfield SP. Recent developments in the production of human therapeutic proteins in eukaryotic algae. Expert Opin Biol Ther. 2005;5:225–235. doi: 10.1517/14712598.5.2.225. [DOI] [PubMed] [Google Scholar]
- Franklin S, Ngo B, Efuet E, et al. Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast. Plant J. 2002;30:733–744. doi: 10.1046/j.1365-313X.2002.01319.x. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Hegemann P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313X.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M, Rahire M, Rochaix JD. Redundant cis-acting determinants of 3′ processing and RNA stability in the chloroplast rbcL mRNA of Chlamydomonas. Plant J. 2008;53:566–577. doi: 10.1111/j.1365-313X.2007.03365.x. [DOI] [PubMed] [Google Scholar]
- Gowda DC, Davidson EA. Protein glycosylation in the malaria parasite. Parasitol Today. 1999;15:147–152. doi: 10.1016/S0169-4758(99)01412-X. [DOI] [PubMed] [Google Scholar]
- Griesbeck C, Kobl I, Heitzer M. Chlamydomonas reinhardtii. Mol Biotechnol. 2006;34:213–223. doi: 10.1385/MB:34:2:213. [DOI] [PubMed] [Google Scholar]
- Hallmann A. Algal transgenics and biotechnology. Transgenic Plant J. 2007;1:81–98. [Google Scholar]
- Harakuni T, Sugawa H, Komesu A, et al. Heteropentameric cholera toxin B subunit chimeric molecules genetically fused to a vaccine antigen induce systemic and mucosal immune responses: a potential new strategy to target recombinant vaccine antigens to mucosal immune systems. Infect Immun. 2005;73:5654–5665. doi: 10.1128/IAI.73.9.5654-5665.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D-M, Qian K-X, Shen G-F, et al. Recombination and expression of classical swine fever virus (CSFV) structural protein E2 gene in Chlamydomonas reinhardtii chloroplasts. Colloids Surf B Biointerfaces. 2007;55:26–30. doi: 10.1016/j.colsurfb.2006.10.042. [DOI] [PubMed] [Google Scholar]
- Heitzer M, Zschoernig B. Construction of modular tandem expression vectors for the green alga Chlamydomonas reinhardtii using the Cre/lox-system. Biotechniques. 2007;43:324–328. doi: 10.2144/000112556. [DOI] [PubMed] [Google Scholar]
- Heitzer M, Eckert A, Fuhrmann M et al (2007) Influence of codon bias on the expression of foreign genes in microalgae. Transgenic Microalgae as Green Cell Factories, pp 46–53 [DOI] [PubMed]
- Hirakawa Y, Kofuji R, Ishida K-i. Transient transformation of a chlorarachniophyte alga, Lotharella amoebiformis (Chlorarachniophyceae), with uidA and egfp reporter genes. J Phycol. 2008;44:814–820. doi: 10.1111/j.1529-8817.2008.00513.x. [DOI] [PubMed] [Google Scholar]
- Kakinuma M, Ikeda M, Coury D, et al. Isolation and characterization of the rbcS genes from a sterile mutant of Ulva pertusa (Ulvales, Chlorophyta) and transient gene expression using the rbcS gene promoter. Fish Sci. 2009;75:1015–1028. doi: 10.1007/s12562-009-0101-5. [DOI] [Google Scholar]
- Kasai S, Yoshimura S, Ishikura K, et al. Effect of coding regions on chloroplast gene expression in Chlamydomonas reinhardtii. J Biosci Bioeng. 2003;95:276–282. doi: 10.1016/s1389-1723(03)80029-4. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Sarada R. Towards genetic improvement of commercially important microalga Haematococcus pluvialis for biotech applications. J Appl Phycol. 2009;21:553–558. doi: 10.1007/s10811-009-9414-0. [DOI] [Google Scholar]
- Kathiresan S, Chandrashekar A, Ravishankar GA, et al. Agrobacterium-mediated transformation in the green alga Haematococcus pluvialis (Chlorophyceae, Volvocales) J Phycol. 2009;45:642–649. doi: 10.1111/j.1529-8817.2009.00688.x. [DOI] [PubMed] [Google Scholar]
- Kato K, Marui T, Kasai S, et al. Artificial control of transgene expression in Chlamydomonas reinhardtii chloroplast using the lac regulation system from Escherichia coli. J Biosci Bioeng. 2007;104:207–213. doi: 10.1263/jbb.104.207. [DOI] [PubMed] [Google Scholar]
- Kiaulehn S, Voytsekh O, Fuhrmann M, et al. The presence of UG-repeat sequences in the 3′-UTRs of reporter luciferase mRNAs mediates circadian expression and can determine acrophase in Chlamydomonas reinhardtii. J Biol Rhythms. 2007;22:275–277. doi: 10.1177/0748730407301053. [DOI] [PubMed] [Google Scholar]
- Koop H-U, Herz S, Golds T. The genetic transformation of plastids. In: Bock Ralph., editor. Topics in current genetics, vol 19: cell and molecular biology of plastids. Berlin/Heidelberg: Springer; 2007. pp. 457–510. [Google Scholar]
- Koya V, Moayeri M, Leppla SH, et al. Plant-based vaccine: mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect Immunol. 2005;73:8266–8274. doi: 10.1128/IAI.73.12.8266-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Misquitta RW, Reddy VS, et al. Genetic transformation of the green alga Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004;166:731–738. doi: 10.1016/j.plantsci.2003.11.012. [DOI] [Google Scholar]
- Lee AK, Lewis DM, Ashman PJ. Microbial flocculation, a potentially low-cost harvesting technique for marine microalgae for the production of biodiesel. J Appl Phycol. 2009;21:559–567. doi: 10.1007/s10811-008-9391-8. [DOI] [Google Scholar]
- Leon R, Fernandez E. Nuclear transformation of eukaryotic microalgae—historical overview, achievements and problems. In: Leon R, Fernandez E, editors. Transgenic microalgae as green cell factories. New York: Landes Bioscience and Springer Science Business Media LLC; 2007. pp. 1–11. [DOI] [PubMed] [Google Scholar]
- Lerche K, Hallmann A. Stable nuclear transformation of Gonium pectorale. BMC Biotechnol. 2009;9:64. doi: 10.1186/1472-6750-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Holloway SP, Lee J, et al. Nuclear genes that promote splicing of group I introns in the chloroplast 23S rRNA and psbA genes in Chlamydomonas reinhardtii. Plant J. 2002;32:467–480. doi: 10.1046/j.1365-313X.2002.01437.x. [DOI] [PubMed] [Google Scholar]
- Loiselay C, Gumpel NJ, Girard-Bascou J, et al. Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol Cell Biol. 2008;28:5529–5542. doi: 10.1128/MCB.02056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Stevens DR, Purton S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998;14:441–447. doi: 10.1046/j.1365-313X.1998.00145.x. [DOI] [Google Scholar]
- Manuell AL, Beligni MV, Elder JH, et al. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant Biotechnol J. 2007;5:402–412. doi: 10.1111/j.1467-7652.2007.00249.x. [DOI] [PubMed] [Google Scholar]
- Marin-Navarro J, Manuell AL, Wu J, et al. Chloroplast translation regulation. Photosynth Res. 2007;94:359–374. doi: 10.1007/s11120-007-9183-z. [DOI] [PubMed] [Google Scholar]
- Mata TM, Martins AA, Caetano NS. Microalgae for biodiesel production and other applications: a review. Renew Sustain Energy Rev. 2010;14:217–232. doi: 10.1016/j.rser.2009.07.020. [DOI] [Google Scholar]
- Maul JE, Lilly JW, Cui L, et al. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell. 2002;14:2659–2679. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Franklin SE. Expression of human antibodies in eukaryotic micro-algae. Vaccine. 2005;23:1828–1832. doi: 10.1016/j.vaccine.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Mayfield S, Schultz J. Development of a luciferase reporter gene, luxCt, for Chlamydomonas reinhardtii chloroplast. Plant J. 2004;37:449–458. doi: 10.1046/j.1365-313X.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- Mayfield SP, Franklin SE, Lerner RA. Expression and assembly of a fully active antibody in algae. Proc Natl Acad Sci. 2003;100:438–442. doi: 10.1073/pnas.0237108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina V, Shoenfeld Y. Infection, vaccines and other environmental triggers of autoimmunity. Autoimmunity. 2005;38:235–245. doi: 10.1080/08916930500050277. [DOI] [PubMed] [Google Scholar]
- Muto M, Henry Ryan E, Mayfield Stephen P. Accumulation and processing of a recombinant protein designed as a cleavable fusion to the endogenous Rubisco LSU protein in Chlamydomonas chloroplast. BMC Biotechnol. 2009;9:26. doi: 10.1186/1472-6750-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international dna sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009;57:1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, et al. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 2009;57:436–445. doi: 10.1111/j.1365-313X.2008.03702.x. [DOI] [PubMed] [Google Scholar]
- Ohnuma M, Yokoyama T, Inouye T, et al. Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2008;49:117–120. doi: 10.1093/pcp/pcm157. [DOI] [PubMed] [Google Scholar]
- Ohresser M, Matagne RF, Loppes R. Expression of the arylsulphatase reporter gene under the control of the nit1 promoter in Chlamydomonas reinhardtii. Curr Genet. 1997;31:264–271. doi: 10.1007/s002940050204. [DOI] [PubMed] [Google Scholar]
- Plaza M, Herrero M, Cifuentes A, et al. Innovative natural functional ingredients from microalgae. J Agric Food Chem. 2009;57:7159–7170. doi: 10.1021/jf901070g. [DOI] [PubMed] [Google Scholar]
- Plaza M, Santoyo S, Jaime L, et al. Screening for bioactive compounds from algae. J Pharm Biomed Anal. 2010;51:450–455. doi: 10.1016/j.jpba.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Popescu CE, Lee RW. Mitochondrial genome sequence evolution in Chlamydomonas. Genetics. 2007;175:819–826. doi: 10.1534/genetics.106.063156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powdrill TF, Johnston JM. Immunologic priming with recombinant hepatitis A virus capsid proteins produced in Escherichia coli. J Virol. 1991;65:2686–2690. doi: 10.1128/jvi.65.5.2686-2690.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Guzman E, Romeu A, et al. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Bravo I, Garcia-Vallve S. E-CAI: a novel server to estimate an expected value of codon adaptation index (eCAI) BMC Bioinformatics. 2008;9:65. doi: 10.1186/1471-2105-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Muto M, Lee PA, et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol J. 2010;8:1–15. doi: 10.1111/j.1467-7652.2009.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C, Loiselay C, Wostrikoff K, et al. Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc Natl Acad Sci. 2007;104:9093–9098. doi: 10.1073/pnas.0703162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg JN, Oyler GA, Wilkinson L, et al. A green light for engineered algae: redirecting metabolism to fuel a biotechnology revolution. Curr Opin Biotechnol. 2008;19:430–436. doi: 10.1016/j.copbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Rymarquis LA, Higgs DC, Stern DB. Nuclear suppressors define three factors that participate in both 5′ and 3′ end processing of mRNAs in Chlamydomonas chloroplasts. Plant J. 2006;46:448–461. doi: 10.1111/j.1365-313X.2006.02711.x. [DOI] [PubMed] [Google Scholar]
- Schroda M, Blcker D, et al. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Schroda M, Beck CF, Vallon O. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant J. 2002;31:445–455. doi: 10.1046/j.1365-313X.2002.01371.x. [DOI] [PubMed] [Google Scholar]
- Schwarz C, Elles I, Kortmann J, et al. Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell. 2007;19:3627–3639. doi: 10.1105/tpc.107.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N, Bock R. A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr Genet. 2008;53:381–388. doi: 10.1007/s00294-008-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanchi A, Barnes D, Mayfield SP. A nuclear gene of Chlamydomonas reinhardtii, Tba1, encodes a putative oxidoreductase required for translation of the chloroplast psbA mRNA. Plant J. 2005;42:341–352. doi: 10.1111/j.1365-313X.2005.02378.x. [DOI] [PubMed] [Google Scholar]
- Suess B. Engineered riboswitches control gene expression by small molecules. Biochem Soc Trans. 2005;33:474–476. doi: 10.1042/BST0330474. [DOI] [PubMed] [Google Scholar]
- Sun M, Qian KX, Su N, et al. Foot-and-mouth disease virus VP1 protein fused with cholera toxin B subunit expressed in Chlamydomonas reinhardtii chloroplast. Biotechnol Lett. 2003;25:1087–1092. doi: 10.1023/A:1024140114505. [DOI] [PubMed] [Google Scholar]
- Surzycki R, Cournac L, Peltier G, et al. Potential for hydrogen production with inducible chloroplast gene expression in Chlamydomonas. Proc Natl Acad Sci. 2007;104:17548–17553. doi: 10.1073/pnas.0704205104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surzycki R, Greenham K, Kitayama K, et al. Factors effecting expression of vaccines in microalgae. Biologicals. 2009;37:133–138. doi: 10.1016/j.biologicals.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Tran M, Zhou B, Pettersson PL, et al. Synthesis and assembly of a full-length human monoclonal antibody in algal chloroplasts. Biotechnol Bioeng. 2009;104:663–673. doi: 10.1002/bit.22446. [DOI] [PubMed] [Google Scholar]
- Tregoning JS, Nixon P, Kuroda H, et al. Expression of tetanus toxin Fragment C in tobacco chloroplasts. Nucleic Acids Res. 2003;31:1174–1179. doi: 10.1093/nar/gkg221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugwu CU, Aoyagi H, Uchiyama H. Photobioreactors for mass cultivation of algae. Bioresour Technol. 2008;99:4021–4028. doi: 10.1016/j.biortech.2007.01.046. [DOI] [PubMed] [Google Scholar]
- von Gromoff ED, Alawady A, Meinecke L, et al. Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell. 2008;20:552–567. doi: 10.1105/tpc.107.054650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Brandsma M, Tremblay R, et al. A novel expression platform for the production of diabetes-associated autoantigen human glutamic acid decarboxylase (hGAD65) BMC Biotechnol. 2008;8:87. doi: 10.1186/1472-6750-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Wu JX, Hu ZL, Wang CG, et al. Efficient expression of green fluorescent protein (GFP) mediated by a chimeric promoter in Chlamydomonas reinhardtii. Chin J Oceanol Limnol. 2008;26:242–247. doi: 10.1007/s00343-008-0242-x. [DOI] [Google Scholar]
- Xia X. An improved implementation of codon adaptation index. Evol Bioinform Online. 2007;3:53–58. [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Li y, Chen F, et al. Expression of human soluble TRAIL in Chlamydomonas reinhardtii chloroplast. Chin Sci Bull. 2006;51:1703–1709. doi: 10.1007/s11434-006-2041-0. [DOI] [Google Scholar]
- Zhang Y-K, Shen G-F, Ru B-G. Survival of human metallothionein-2 transplastomic Chlamydomonas reinhardtii to ultraviolet B exposure. Acta Biochim Biophys Sin. 2006;38:187–193. doi: 10.1111/j.1745-7270.2006.00148.x. [DOI] [PubMed] [Google Scholar]


