Abstract
The use of immunofluorescence (IF) and fluorescence in situ hybridisation (FISH) underpins much of our understanding of how chromatin is organised in the nucleus. However, there has only recently been an appreciation that these types of study need to move away from cells grown in culture and towards an investigation of nuclear organisation in cells in situ in their normal tissue architecture. Such analyses, however, especially of archival clinical samples, often requires use of formalin-fixed paraffin wax-embedded tissue sections which need addition steps of processing prior to IF or FISH. Here we quantify the changes in nuclear and chromatin structure that may be caused by these additional processing steps. Treatments, especially the microwaving to reverse fixation, do significantly alter nuclear architecture and chromatin texture, and these must be considered when inferring the original organisation of the nucleus from data collected from wax-embedded tissue sections.
Keywords: chromatin, fluorescence in situ hybridization, heterochromatin, nuclear organisation, tissue sections
Introduction
Whilst historically electron microscopy (De Boni 1988) and a variety of bright-field staining (Barr and Bertram 1949) light microscopy techniques were important in laying the groundwork for an investigation of chromatin structure and nuclear organisation, it has been the more recent approaches of immunofluorescence (IF) and fluorescence in situ hybridization (FISH) that have been key to our increasing understanding of mammalian genome function and dysfunction in the nucleus. With a few exceptions (Solovei et al. 2009), this has been mainly done on nuclei prepared from cultured cell lines (Bolzer et al. 2005; Cremer et al. 2003; Boyle et al. 2001; Chambeyron and Bickmore 2004; Wiblin et al. 2005), on primary cells prepared from the haematopoietic system of the animal (Kim et al. 2004; Osborne et al. 2007; Brown et al. 2001, 2006, 1997) or on primary cells isolated from disaggregated tissues (Parada et al. 2004). These approaches have used both 2D and 3D FISH, and whereas gross features and general trends of nuclear organisation are preserved between these two methods, 3D FISH does better preserve chromatin architecture at the sub-micron-size scale (Croft et al. 1999; Morey et al. 2007; Eskeland et al. 2010). However, it still remains unclear to what extent the nuclear and chromatin organisation of cultured cell lines relates to the cells of origin from which these lines were established, or whether chromatin structure is altered as the cells habituate to survival and proliferation ex vivo. Similarly, the isolated myeloid and lymphoid cells of the haematopoietic system that have been studied are not typical of cells, e.g. epithelial cells, that are embedded within complex tissue architectures where cell–cell adhesion, cell polarity and cell–cell signalling may well impact on nuclear organisation, and this context is lost when such tissues are disaggregated. Therefore, there is a need to extend our understanding of nuclear organisation to cells in situ in a variety of tissues.
Both IF and FISH require that nuclei be fixed and permeabilised, then DNA must also be denatured prior to FISH. By definition, these processes must alter chromatin structure at some level. Several studies have investigated the extent of this for cells grown in culture. Imaging of green fluorescent protein-tagged centromeres in the nuclei of living cells and then analysis of the relative position and morphology of these exact same centromeres after fixation in 4% paraformaldehyde (pFa) and also after 3D DNA-FISH with probes for centromeric DNA sequences have shown that, at the level of resolution afforded by light microscopy, any perturbations of nuclear organisation during such fixation and 3D FISH are small (Mahy et al. 2002; Kim et al. 2007). Similarly, electron microscopy analysis of nuclei during the process of FISH suggests that large-scale (~1 MB) chromatin organisation is preserved, but that nuclear ultrastructure is disrupted (Solovei et al. 2002).
Texture analysis can be used to quantitatively assess the variation in chromatin structures within the nucleus, by providing a measure of the unevenness, complexity and coarseness of chromatin staining derived from the distribution of grey levels in the pixel intensities. This approach has important applications since it has been used to try and distinguish normal from cancerous cells and to assess the grade of various cancers (Gil et al. 2002). Various textural features have been used to evaluate chromatin staining, such as “granularity and heterogeneity” (Young et al. 1986), “entropy” (Jorgensen et al. 1996; Yogesan et al. 1996), “textons” (Deligdisch et al. 1993), “autocorrelation” (Deligdisch et al. 1999), “fractals” (Einstein et al. 1998) and, more recently, contour complexity (Kiyuna et al. 2008).
Although there has been limited FISH analysis of fresh-frozen tumour specimens (Lukasova et al. 2004), archival samples of human tissue are most commonly available as formalin-fixed, paraffin wax-embedded (FFPE) blocks, which can then be cut into sections for analysis. Both the strong fixation and the presence of the paraffin wax present additional challenges and complexities in preparing such samples for analysis by high-resolution light microscopy. Here we quantify changes in gross features of chromatin and nuclear organisation that occur in different cell types from two different human tissues with epithelial architecture, the mammary and thyroid glands, during the different steps of processing of tissue sections in preparation for FISH. We find that these treatments, though known to be effective in our hands, at producing good FISH signal (Chambeyron et al. 2005; Dundas et al. 2001; Wiech et al. 2005, 2009; Newsome et al. 2003; Solovei et al. 2009), significantly alter nuclear size, the distribution of chromatin at the nuclear periphery and the texture of chromatin. Moreover, the nature of these changes depends on the tissue architecture.
Materials and methods
Sample processing
Paraffin-embedded formalin-fixed sections of thyroid and breast tissue were cut from tissue blocks at 4-6μm thickness and mounted on slides. Sections on slides were heated at 60°C for 30 min and washed four times in xylene each for 10 min to remove the paraffin wax, followed by rehydration through an ethanol (EtOH) series, before washing and storing in dH2O prior to 4,6-diaminidino-2-phenylidole (DAPI) staining. Slides stopped at this stage were designated as x (xylene only). Other sections, cut from the same tissue block at the same time as those designated as x, were processed through xylene as above and then microwaved for 27 min (thyroid) and 25 min (breast), respectively, in 0.1 M citrate buffer (pH 6.0), cooled, then rinsed and stored in dH2O prior to DAPI staining (xm). Further sections from the same tissue block were processed up to the xm stage and were then washed in 0.005% pepsin/0.01 M HCl at 37°C for 12 min, rinsed in PBS, then 2× sodium saline citrate (SSC) and dehydrated through an EtOH series and air-dried prior to DAPI staining (xmp). Finally, for slides being processed as for FISH, sections processed up to the xmp stage were then incubated at 75°C for 5 min in 2× SSC, denatured for 3 min at 75°C in 70% formamide/2× SSC, plunged into ice-cold 70% EtOH for 3 min, dehydrated through an EtOH series and air-dried (xmpf).
Image acquisition
For visualisation of chromatin after each appropriate step, slides were mounted in Vectashield (Vector) containing 1 μg/ml DAPI and visualised using a Zeiss Axioskop fluorescence microscope with Plan apochromat objectives (NA 1.4), a 50-W Hg source (Carl Zeiss, Welwyn Garden City, UK) and Chroma #83000 triple band pass filter set (http://www.chroma.com/products/catalog/80000_Series/83000v2, Chroma Technology Corp., Rockingham, VT, USA) with the excitation filters installed in a motorised filter wheel (Ludl Electronic Products, Hawthorne, NY, USA). A piezoelectrically driven objective mount (PIFOC model P-721, Physik Instrumente GmbH & Co, Karlsruhe, Germany) was used to control movement in the z-dimension. 3D image stacks were captured at 0.25 μm intervals through the z-axis with a Hamamatsu Orca AG CCD camera (Hamamatsu Photonics (UK) Ltd, Welwyn Garden City, UK). Hardware control, image capture and analysis were performed using in-house scripts written for IPLab Spectrum (Scanalytics Corp, Fairfax, VA, USA). Images were captured at a binning of 1 pixel = 0.067 μm.
Image analysis
3D images were deconvolved using the no neighbour (single image) or nearest neighbour (three images) algorithm of Hazebuster (Scanalytics Corp, Fairfax, VA, USA). To quantify the distribution of peripheral compact chromatin, we assessed the proportion of DAPI staining present in each of five shells, of approximately the same area as defined by the segmented DAPI image, eroded from the edge of the nucleus into the nuclear centre in single mid-plane deconvolved images of nuclei (Boyle et al. 2001).
We used contour length analysis to quantify chromatin texture in single mid-plane deconvolved images of the nuclei. We “extracted” the segmented nuclear area from a given image—i.e. set pixel values outside the nucleus to 0, leaving pixel values inside the nucleus unchanged. Because, de facto, contour length must increase as nuclei become larger (Kiyuna et al. 2008), the contour length at each threshold value (L i) was normalized to the nuclear contour length at intensity threshold of 0 (L *), to produce a value for the contour complexity (cc) given by the equation:
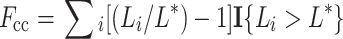 |
where:
- Li:
contour length at threshold i
- L*:
contour length of original nucleus
- IC:
1 if condition C holds, 0 otherwise
Thus, the chromatin contour length at 0 intensity thresholding = 1 and at maximum intensity thresholding (256) = 0.
The significance of differences in measurements between samples was assessed in the Minitab statistical software package, using the non-parametric Mann–Whitney U test.
Results
Pre-treatments for FISH can distort nuclear size and shape
Typical approaches that have been taken to prepare paraffin-embedded tissue blocks for use in FISH include removal (clearing) of the wax using the organic solvent xylene and heating, e.g. by microwaving, to retrieve epitopes masked by fixation and to increase DNA probe access (Chambeyron et al. 2005; Dundas et al. 2001; Wiech et al. 2005, 2009; Newsome et al. 2003; Solovei et al. 2009; http://www.ihcworld.com/epitope_retrieval.htm). Whilst this is adequate in some instances for freshly embedded tissue, archival specimens also require treatment with proteases prior to denaturation of DNA (Wiech et al. 2005, 2009).
We investigated other methods for antigen retrieval that do not involve heat, e.g. HCl (http://www.ihcworld.com/_protocols/epitope_retrieval/hcl.htm) or protease treatments, but these did not give reliable FISH signals in comparison to microwaving.
To determine how nuclear morphology might be affected by these processing steps, we analysed the pattern of DNA counterstaining with DAPI in 3D image stacks from human thyroid or mammary gland sections that had been treated just with xylene (x), xylene followed by microwaving (xm), xylene–microwaving and pepsin treatment (xmp) or finally fully processed sections that had then been denatured as if subject to FISH (xmpf; Figs. 1a and 2a). After image deconvolution, the dimensions of the DAPI-stained nuclei were taken from an image in the mid-plane of the z-axis.
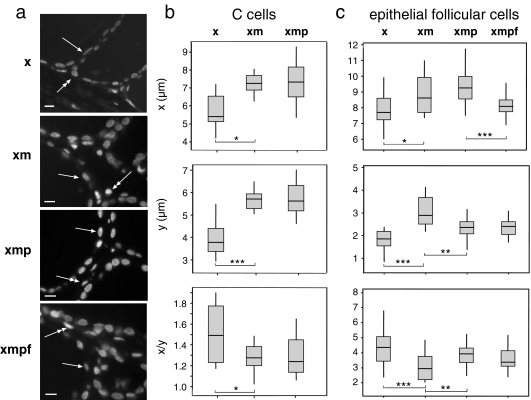
Fig. 1.
Alterations of nuclear size and shape during treatment of thyroid sections. a DAPI-stained 4-μm thyroid sections treated with xylene only (x), xylene + microwaving (xm), xylene + microwaving + protease (xmp) and as for FISH (xmpf), showing the positions of epithelial follicular cells (arrow) and parafollicular C cells (double-headed arrow). Bars = 10 μm; b box plots of nuclear diameter (micrometres) in x- or y-dimensions and nuclear shape (x/y) for C cells in sections treated with xylene only (x), xylene + microwaving (xm) or xylene + microwaving + protease (xmp). The boxed areas show the 25–75 percentiles and the medians are indicated by horizontal lines through these boxed areas. Statistically significant changes in nuclear size or shape between different treatments are indicated by asterisks (*p < 0.05 and >0.01; **p ≤ 0.01 and >0.001; ***p < 0.001). n = 10–20; c as in b, but for epithelial cells and including analysis of these cells as for FISH (xmpf). n = 22
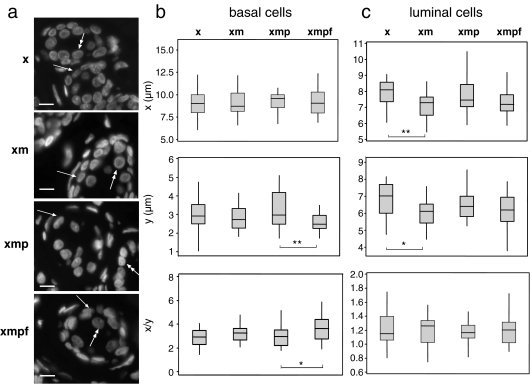
Fig. 2.
Alterations of nuclear size and shape during treatment of breast sections. a DAPI-stained 6-μm breast sections treated with xylene only (x), xylene + microwaving (xm), xylene + microwaving + protease (xmp) and after FISH (xmpf), showing the position of basal (arrow) and luminal (double-headed arrow) epithelial cells. Bars = 10 μm; b box plots of nuclear diameter (micrometres) in x- or y-dimensions and nuclear shape (x/y) for basal epithelial cells in sections treated with xylene only (x), xylene + microwaving (xm), xylene + microwaving + protease (xmp) and as for fish (xmpf). The boxed areas show the 25–75 percentiles and the medians are indicated by horizontal lines through these boxed areas. Statistically significant changes in nuclear size or shape between different treatments are indicated by asterisks (*p < 0.05 and >0.01; **p ≤ 0.01 and >0.001; ***p < 0.001). n = 28; c as in b, but for luminal epithelial cells. n = 28
The thyroid gland has a simple tissue architecture composed of the epithelial follicular cells that secrete thyroid hormone, which from a single-layered cuboidal epithelium, and the parafollicular C cells that synthesis calcitonin. The diameters of the perpendicular axes of the equatorial nuclear profiles were recorded, with the x-axis being the axis parallel to the basement membrane for the epithelial follicular cells of the thyroid (arrowed in Fig. 1a), or the longer axis for parafollicular C cells (double-headed arrow in Fig. 1a). The y was the axis perpendicular to the basement membrane and was usually the short axis. The shapes of the nuclei were determined from the x/y ratio. The significance of changes in these values between treatments was assessed with a non-parametric Mann–Whitney U test.
For C cells, microwaving after xylene treatment resulted in significantly increased nuclear size in the x and especially the y-dimensions compared to xylene treatment alone, resulting in an overall rounding of nuclear shape (decreased x/y ratio; Fig. 1b). No further significant change in size or shape was seen with subsequent protease treatments. There were insufficient numbers of C cells in the sections to analyse by FISH. For the follicular epithelial cells of the thyroid, microwaving also significantly increases nuclear size along the x- and y-axes (Fig. 1c) accompanied by a significantly more rounded nuclear shape compared to the epithelial nuclei in the xylene-only treated sections. Subsequent protease treatments then decreased the y-axis dimensions compared to those seen in xylene-microwaved samples, resulting in a return to a more elongated nuclear shape. Finally, the FISH process decreased the length of epithelial follicular cells along the x, but not the y-axis.
Changes in nuclear morphology induced by treatments were less extensive and quite different in the more complex stratified epithelium of the mammary gland compared to the thyroid. For the basal (myoepithelial) cell layer (arrowed in Fig. 2a), there was no significant change in size along the x- or y-axes during the processing steps from xylene to xmp (Fig. 2b). We consider this likely reflects the constraints imposed on these cells by their attachment to the basement membrane on one side and their position underlying another epithelial layer—the luminal cells—on the other side. The processing as for FISH did significantly decrease the width (y) of these cell nuclei, but not their length (x). For the luminal epithelial cell themselves (double-headed arrow in Fig. 2a), microwaving resulted in a small, but significant, decrease in nuclear size along both the x- and y-axes (Fig. 2c).
Pre-treatments for FISH change chromatin texture
Analysis of changes in chromatin architecture, at the level of both light and electron microscopy, has indicated that the steps of FISH, especially heat denaturation, alter chromatin ultrastructure, though larger-scale structures (at the level of ~1-Mb sub-chromosomal domains) are not detectably perturbed (Solovei et al. 2002). It has been noted that the DAPI counterstaining of nuclei appears more blurry after FISH also indicating an alteration of chromatin structure at the local level (Kim et al. 2007). Any changes in chromatin structure that may occur during the pre-FISH processing steps of paraffin wax-embedded have not been studied.
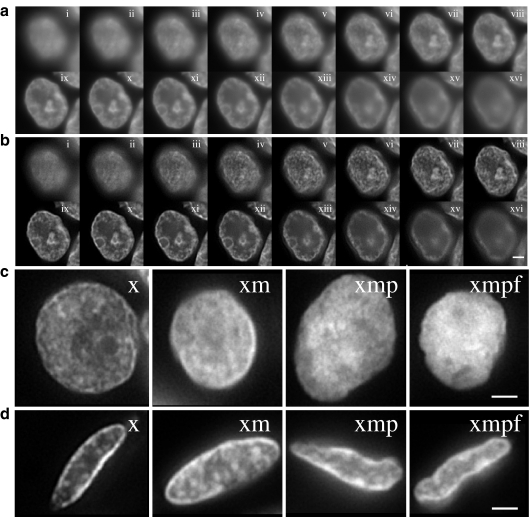
Images, taken at 0.25 μm steps through the z-axis of DAPI-stained nuclei (Fig. 3a), were subject to deconvolution to minimize the contribution of out-of-focus information to the subsequent analyses (Fig. 3b). Examination of these deconvolved DAPI-stained images during tissue processing stages revealed subjective changes in chromatin texture (Fig. 3c, d). In particular, there was a loss of crispness in the staining induced by microwave treatments. For luminal epithelial cells of the mammary gland, the peripheral heterochromatin also became less apparent during processing (Fig. 3c). This layer of compact chromatin around the nuclear edge was more prominent in the thyroid epithelial cells and, although less distinct, was still visible even after FISH pre-treatments (Fig. 3d).
Fig. 3.
Visualisation of chromatin texture during tissue processing. a Grey-scale images of DAPI-stained nucleus from the luminal epithelium of normal mammary gland after xylene treatment to remove wax. Images are taken (i to xvi) at 0.25 μm intervals through the z-axis of the nucleus. Scale bar = 2 μm; b as in a but after image deconvolution to remove out-of-focus information; c single plane deconvolved images of DAPI-stained nuclei from luminal cells of the mammary gland epithelium after treatments with; xylene (x), xylene + microwaving (xm), xylene + microwaving + protease (xmp) and as for fish (xmpf). Scale bar = 2 μm; d as in c but with nuclei from the epithelium of the thyroid gland
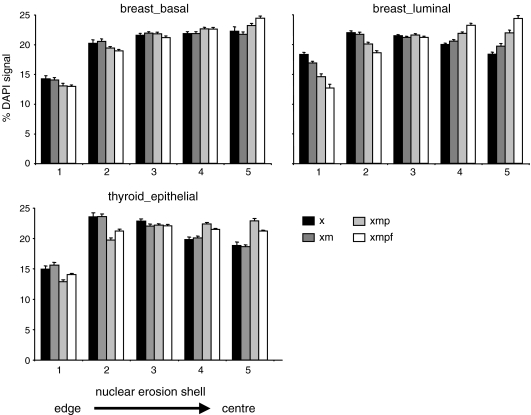
To quantify the loss of peripheral compact chromatin during tissue processing, we assessed the proportion of DAPI staining present in each of five shells eroded from the edge (shell 1) into the centre (shell 5) of single mid-plane images of nuclei from basal and luminal cells of the breast epithelium and from thyroid epithelial cells (Fig. 4; Boyle et al. 2001). This showed that the relative proportion of DAPI-stained chromatin at the nuclear periphery of mammary gland epithelial cells declined steadily as samples were processed through wax removal (x), epitope retrieval (xm), protease (xmp) and FISH (xmpf) steps, especially for breast luminal cells. There was also a detectable decline in the more prominent peripheral chromatin layer of thyroid epithelial cells and a subsequent redistribution of DAPI-stained chromatin away from the nuclear periphery and towards the centre, but this did not occur until the protease treatment step.
Fig. 4.
Changes in the distribution of chromatin at the nuclear periphery during tissue processing. Histograms showing the mean (± SEM) percent of DAPI staining in each of five shells of equal area, eroded from the edge (shell 1) to the centre (shell 5) of the nucleus for; top row—basal (left) and luminal (right) breast epithelial cells, bottom row—thyroid epithelial cells, in tissue sections treated with xylene only (x, black bars), xylene + microwave (xm, dark grey bars), xylene + microwave + pepsin (xmp, light grey bars) and treated as for FISH (xmpf, white bars). n > 21
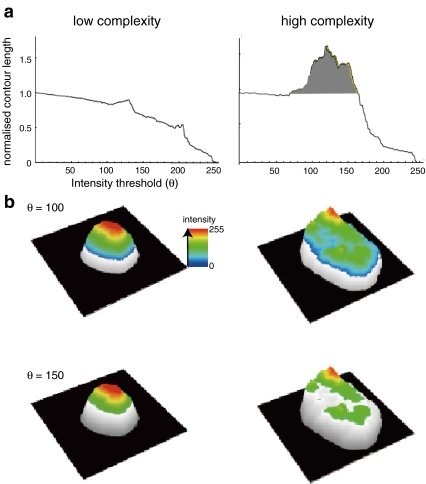
We have described that contour complexity analysis can be used to provide a quantitative measure of chromatin texture that is more sensitive than other features used previously, such as the fractal dimension (Kiyuna et al. 2008). In this analysis, the segmented DAPI-counterstained grey-scale image of a nucleus is analysed at a range of intensity thresholds, and the contour length of the resulting segmented images is measured. The chromatin contour length at 0 intensity thresholding = 1 and at maximum intensity thresholding (256) = 0 (Fig. 5a). If DAPI staining is uniform across the nucleus, then the contour length decreases with increasing threshold intensity, but if the staining is more irregular and complex in texture, then the contour length increases within particular windows of intensity thresholds and the normalized contour length can even be >1 (Fig. 5a, b).
Fig. 5.
Contour length analysis as a measure of chromatin texture. a Graphs showing the relationship between the normalized chromatin contour length (contour length/nuclear contour) and increasing intensity threshold (θ) for examples of (left) rather homogeneous chromatin staining/low complexity and for (right) irregular chromatin texture/high complexity; b three-dimensional visualisation of the data in a for intensity thresholds set at 100 and 150. As in a, example images on the left are for low complexity chromatin texture and on the right for high complexity. Adapted from Kiyuna et al. 2008
We used this method of analysis to quantify any changes in chromatin texture that accompany the various processing steps of FFPE sections from normal thyroid and mammary epithelia. Single mid-plane deconvolved images of the nuclei were subject to contour length analysis. The contour length at each threshold value was then normalized to the nuclear contour length at intensity threshold of 0, to produce a value for the cc (Kiyuna et al. 2008). Cc was also measured in the corresponding images before deconvolution to make sure that any changes in cc value during the steps of tissue processing were not just a reflection of the deconvolution process itself. As expected, given the removal of out-of-focus signals, deconvolution did increase the cc of each image, but this was seen at each tissue processing step and so did not influences conclusion drawn by comparing images at different tissue processing steps.
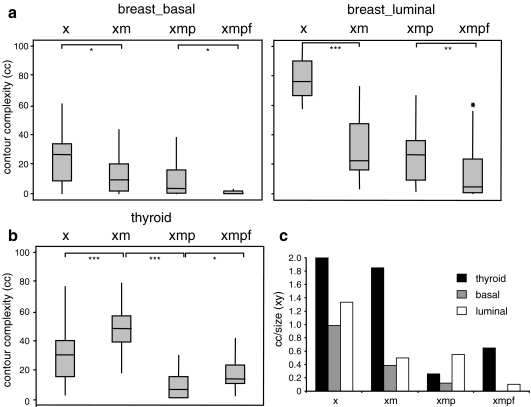
For mammary epithelial cells, there was drastic loss of contour complexity in chromatin texture during microwave treatment and then a further decrease during pre-treatments for FISH (Fig. 6a). Thyroid epithelial cells behaved differently in that contour complexity apparently increased during microwaving, before decreasing during the subsequent protease processing step (Fig. 6b). We consider this likely due to the large increase in nuclear size of these cells during these processing steps (Fig. 1c) that is not seen for mammary epithelial cells. This could provide extra spatial resolution in which to detect changes in chromatin texture. To assess this, we divided the measured contour complexity of cells at each step of pre-treatment by a simple measure of nuclear size—xy (nuclear diameter along the x-axis multiplied by diameter along the y). This clearly showed (Fig. 6c) that once changes in nuclear size are accounted for, the contour complexity of chromatin is indeed reduced in all cells as they proceed from the xylene step to the microwave step (xm) and then to treatment with proteases (xmp).
Fig. 6.
Chromatin contour complexity changes during tissue processing. Box plots showing the contour complexity for basal and luminal mammary epithelial cells (a) and thyroid follicular epithelial cells; b in sections treated with xylene only (x), xylene + microwaving (xm), xylene + microwaving + protease (xmp) and as for fish (xmpf). The boxed areas show the 25–75 percentiles and the medians are indicated by horizontal lines through these boxed areas. Statistically significant changes in nuclear size or shape between different treatments are indicated by asterisks (*p < 0.05 and >0.01; **p ≤ 0.01 and >0.001; ***p < 0.001). n ≥ 21; c histogram showing contour complexity (cc) normalized to nuclear size (xy) at each of the pre-treatment steps for epithelial cells of the thyroid (black bars) and basal (grey bars) or luminal (white bars) cells of the mammary epithelium
Discussion
We have used DAPI staining to investigate how the treatments required to obtain good FISH signals within the nuclei of cells in FFPE tissue sections, themselves might affect nuclear and chromatin organisation in terms of overall nuclear size/shape, chromatin organisation in the nucleus and chromatin texture. The most damaging pre-treatment generally appears to be the use of microwaving to reverse the formalin fixation. Whereas pFa fixation of cells grown ex vivo in culture uses short (typically 10 min) time periods of fixation, which are easily reversed during FISH to enable probe penetration and DNA denaturation, the embedding of tissues typically involves fixation in formalin for much longer time periods, typically overnight or up to 24 h.
In the simple epithelium of the thyroid gland, microwaving results in increased nuclear size along the x but especially the y-axis of both the epithelial and C cells, the cells apparently being free to expand in size into the space of the large lumen of the gland (Fig. 1). The consequences of microwave pre-treatment are completely different in the more complex stratified epithelium of the mammary gland. There is no detectable change in nuclear size in the basal myoepithelial cells (Fig. 2), which are in contact with the laminin-rich basement membrane to one side, the layer of cuboidal luminal epithelial cells to the other side and with each other laterally. Thus, the tissue architecture may constrain their expansion. The luminal cells of the mammary gland face the lumen of the duct and are not in contact with the basement membrane, but do make lateral cell–cell homotypic interactions. These cells also show no increased nuclear size upon microwaving in contrast to the thyroid epithelium cells. Indeed, they show a small but significant decrease in nuclear size. However, the nuclei of luminal mammary epithelium cells are already larger in volume than those of the thyroid and appear tightly packed together, and this may affect their response.
Pre-treatments also seem to lead to a gradual loss of peripheral heterochromatin as judged visually (Fig. 3) and by assessing the proportion of DAPI staining in the most peripheral shell of the nucleus (Fig. 4). This is especially pronounced in luminal cells. Thus, the radial organisation of chromatin in thyroid epithelium seems more robust to microwave treatment than that in mammary gland epithelium. Finally, chromatin texture as measured by contour complexity is degraded during pre-treatments, particularly microwave treatments of mammary epithelial cells (Fig. 6a). The apparent increase in contour complexity of thyroid epithelial cells during this step (Fig. 6b) we show is due to the increase of nuclear size of these cells induced by microwave treatment (Fig. 1c). Once normalized to nuclear size, the contour complexity of chromatin is indeed reduced in all cells as they proceed from the xylene step to the microwave step (xm) and then to treatment with proteases (xmp; Fig. 6c).
Investigation in cultured cells has been very productive in illuminating how nuclear organisation can control gene expression, and in some cases, features of altered nuclear organisation uncovered in ex vivo systems seem to faithfully reflect changes occurring in cells in vivo in the developing organism (Chambeyron et al. 2005). However, some dramatic cases of tissue-specific altered nuclear organisation have only been uncovered by studying cells in their correct tissue context and at specific points in development (Solovei et al. 2009). Indeed, tissue architecture itself probably plays a direct role in controlling chromatin structure and nuclear organisation and so affects gene expression and the differentiation state of epithelial cells (Chandramouly et al. 2007; Jolivet et al. 2005; Le et al. 2007; Kim et al. 2005).
Even with the advent of more molecular biomarkers, histological and cytological analyses remain linchpins of cancer diagnosis, with changes in nuclear size, shape and chromatin texture being used to diagnose tumour type, stage and grade (Zink et al. 2004). If a better understanding of the molecular basis of cancer could be married with a high-resolution cellular approach to examination of the cancer phenotype, then this might lead to better diagnostic criteria. However, this will likely involve the quantitative analysis of FISH images of biopsies, and hence, an understanding of how nuclear parameters in these clinical specimens are altered by the pre-treatments for FISH is crucial to the interpretation of such data. Our study shows that, whilst FISH will undoubtedly continue to be useful for examining nuclear organisation in cells in situ, the interpretation of the results of such experiments must be considered alongside how the different tissue processing steps involved may alter gross features of nuclear architecture and chromatin organisation and that different cell types might respond differently due to constraints imposed upon them by their tissue architecture. Whilst in this study we have used DAPI to counterstain DNA, it may well be worth investigating whether other methods of DNA detection reveal similar or different levels of chromatin changes during sample processing steps. Although the study of nuclear organisation in FFPE sections by FISH has not so far revealed any major discrepancies with data collected from studies in cells grown ex vivo in tissue culture (Chambeyron et al. 2005), this may become more of a problem as analyses move to higher levels of resolution with the advent of new sub-diffraction methods of light microscopy (Schermelleh et al. 2008).
Acknowledgements
This work was supported by Breakthrough Breast Cancer and by the Medical Research Council, UK. We thank Prof. Jonathan Rees (Dermatology University of Edinburgh) for samples of normal human breast tissue and Dr. Anne-Marie McNicol and Prof. Barry A. Gusterson (Division of Cancer Sciences and Molecular Pathology, Faculty of Medicine, University of Glasgow) for thyroid samples.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- 2D
two-dimensional
- 3D
three-dimensional
- cc
contour complexity
- dH2O
deionised water
- EtOH
ethanol
- FFPE
formalin-fixed paraffin-embedded
- FISH
fluorescence in situ hybridisation
- GFP
green fluorescent protein
- IF
immunofluorescence
- pFa
paraformaldehyde
- SSC
saline sodium citrate buffer
References
- Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Amoils S, Horn JM, Buckle VJ, Higgs DR, Merkenschlager M, Fisher AG. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat Cell Biol. 2001;3:602–606. doi: 10.1038/35078577. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/S0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouly G, Abad PC, Knowles DW, Lelievre SA. The control of tissue architecture over nuclear organization is crucial for epithelial cell fate. J Cell Sci. 2007;120:1596–1606. doi: 10.1242/jcs.03439. [DOI] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De BU. Chromatin and nuclear envelope of freeze-fractured, neuronal interphase nuclei, resolved by scanning electron microscopy. Biol Cell. 1988;63:1–8. doi: 10.1016/0248-4900(88)90102-5. [DOI] [PubMed] [Google Scholar]
- Deligdisch L, Miranda C, Barba J, Gil J. Ovarian dysplasia: nuclear texture analysis. Cancer. 1993;72:3253–3257. doi: 10.1002/1097-0142(19931201)72:11<3253::AID-CNCR2820721120>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Deligdisch L, Gil J, Kerner H, Wu HS, Beck D, Gershoni-Baruch R. Ovarian dysplasia in prophylactic oophorectomy specimens: cytogenetic and morphometric correlations. Cancer. 1999;86:1544–1550. doi: 10.1002/(SICI)1097-0142(19991015)86:8<1544::AID-CNCR22>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Dundas SR, Boyle S, Bellamy CO, Hawkins W, Garden OJ, Ross JA, Bickmore W. Dual Y-chromosome painting and immunofluorescence staining of archival human liver transplant biopsies. J Histochem Cytochem. 2001;49:1321–1322. doi: 10.1177/002215540104901015. [DOI] [PubMed] [Google Scholar]
- Einstein AJ, Wu HS, Sanchez M, Gil J. Fractal characterization of chromatin appearance for diagnosis in breast cytology. J Pathol. 1998;185:366–381. doi: 10.1002/(SICI)1096-9896(199808)185:4<366::AID-PATH122>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan YH, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Wu H, Wang BY. Image analysis and morphometry in the diagnosis of breast cancer. Microsc Res Tech. 2002;59:109–118. doi: 10.1002/jemt.10182. [DOI] [PubMed] [Google Scholar]
- Jolivet G, Pantano T, Houdebine LM. Regulation by the extracellular matrix (ECM) of prolactin-induced alpha s1-casein gene expression in rabbit primary mammary cells: role of STAT5, C/EBP, and chromatin structure. J Cell Biochem. 2005;95:313–327. doi: 10.1002/jcb.20397. [DOI] [PubMed] [Google Scholar]
- Jorgensen T, Yogesan K, Tveter KJ, Skjorten F, Danielsen HE. Nuclear texture analysis: a new prognostic tool in metastatic prostate cancer. Cytometry. 1996;24:277–283. doi: 10.1002/(SICI)1097-0320(19960701)24:3<277::AID-CYTO11>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Kim IH, Nagel J, Otten S, Knerr B, Eils R, Rohr K, Dietzel S. Quantitative comparison of DNA detection by GFP-lac repressor tagging, fluorescence in situ hybridization and immunostaining. BMC Biotechnol. 2007;7:92. doi: 10.1186/1472-6750-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, McQueen PG, Lichtman MK, Shevach EM, Parada LA, Misteli T. Spatial genome organization during T-cell differentiation. Cytogenet Genome Res. 2004;105:292–301. doi: 10.1159/000078201. [DOI] [PubMed] [Google Scholar]
- Kim YB, Yu J, Lee SY, Lee MS, Ko SG, Ye SK, Jong HS, Kim TY, Bang YJ, Lee JW. Cell adhesion status-dependent histone acetylation is regulated through intracellular contractility-related signaling activities. J Biol Chem. 2005;280:28357–28364. doi: 10.1074/jbc.M412608200. [DOI] [PubMed] [Google Scholar]
- Kiyuna T, A Saito, E Kerr, W Bickmore 2008 Characterization of chromatin texture by contour complexity for cancer cell classification. In: Proceedings of the 8th IEEE international conference on bioinformatics and bioengineering
- Le BJ, Xu R, Lee SY, Nelson CM, Rizki A, Alcaraz J, Bissell MJ. Cell shape regulates global histone acetylation in human mammary epithelial cells. Exp Cell Res. 2007;313:3066–3075. doi: 10.1016/j.yexcr.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasova E, Kozubek S, Falk M, Kozubek M, Zaloudik J, Vagunda V, Pavlovsky Z. Topography of genetic loci in the nuclei of cells of colorectal carcinoma and adjacent tissue of colonic epithelium. Chromosoma. 2004;112:221–230. doi: 10.1007/s00412-003-0263-3. [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- Newsome PN, Johannessen I, Boyle S, Dalakas E, McAulay KA, Samuel K, Rae F, Forrester L, Turner ML, Hayes PC, Harrison DJ, Bickmore WA, Plevris JN. Human cord blood-derived cells can differentiate into hepatocytes in the mouse liver with no evidence of cellular fusion. Gastroenterology. 2003;124:1891–1900. doi: 10.1016/S0016-5085(03)00401-3. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, Leonhardt H, Sedat JW. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D, Cremer C, Fakan S, Cremer T. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH) Exp Cell Res. 2002;276:10–23. doi: 10.1006/excr.2002.5513. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci. 2005;118:3861–3868. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- Wiech T, Stein S, Lachenmaier V, Schmitt E, Schwarz-Finsterle J, Wiech E, Hildenbrand G, Werner M, Hausmann M. Spatial allelic imbalance of BCL2 genes and chromosome 18 territories in nonneoplastic and neoplastic cervical squamous epithelium. Eur Biophys J. 2009;38:793–806. doi: 10.1007/s00249-009-0474-5. [DOI] [PubMed] [Google Scholar]
- Wiech T, Timme S, Riede F, Stein S, Schuricke M, Cremer C, Werner M, Hausmann M, Walch A. Human archival tissues provide a valuable source for the analysis of spatial genome organization. Histochem Cell Biol. 2005;123:229–238. doi: 10.1007/s00418-005-0768-3. [DOI] [PubMed] [Google Scholar]
- Yogesan K, Jorgensen T, Albregtsen F, Tveter KJ, Danielsen HE. Entropy-based texture analysis of chromatin structure in advanced prostate cancer. Cytometry. 1996;24:268–276. doi: 10.1002/(SICI)1097-0320(19960701)24:3<268::AID-CYTO10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Young IT, Verbeek PW, Mayall BH. Characterization of chromatin distribution in cell nuclei. Cytometry. 1986;7:467–474. doi: 10.1002/cyto.990070513. [DOI] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]