Abstract
Proteasomes are multisubunit proteases that initiate degradation of many Ags presented by MHC class I molecules. Vertebrates express alternate forms of each of the three catalytic proteasome subunits: standard subunits, and immunosubunits, which are constitutively expressed by APCs and are induced in other cell types by exposure to cytokines. The assembly of mixed proteasomes containing standard subunits and immunosubunits is regulated in a tissue specific manner. In this study, we report that the presence of mixed proteasomes in immune cells in LMP2−/− mice compromises multiple components that contribute to the generation of antiviral Ab responses, including splenic B cell numbers, survival and function of adoptively transferred B cells, Th cell function, and dendritic cell secretion of IL-6, TNF-α, IL-1β, and type I IFNs. These defects did not result from compromised overall protein degradation, rather they were associated with altered NF-κB activity. These findings demonstrate an important role for immunoproteasomes in immune cell function beyond their contribution to Ag processing.
Proteasomes are abundant complex proteases that initiate degradation of full-length proteins and play a critical role in the generation of many MHC class I-associated peptides (1). Ag presentation is a nonessential proteasome function added during vertebrate evolution. Proteasomes are essential components of lower eukaryotes and archaeobacteria that function to initiate degradation of full-length proteins that are defective or deemed disposable by cellular control mechanisms. Although proteasomes typically generate peptides of 3–30 residues that are rapidly disposed of by endopeptidases and exopeptidases (2), there are several well-defined examples of proteasomes regulating protein function by liberating biologically active fragments. Protesomal processing regulates NF-κB, Spt23p, and Mga2p transcription factors (3, 4).
Proteasomes consist of a four-ring structure with dyad symmetry of the form αββα (5). Fourteen distinct subunits assemble to generate seven subunit α and β rings. Only three of the 14 subunits (β1, β2, β5; also referred to as Δ, Z, and X), are known to possess proteolytic activities. In vertebrates, each catalytic subunit is encoded by two genes. One set of genes is constitutively expressed by most cell types. The other set, termed immunosubunits (β1i, β2i, β5i; nominally LMP2, MECL-1, LMP7 in mice), are coordinately expressed in dendritic cells (DCs) and monocyte-derived cells, where they assemble to form immunoproteasomes (6, 7). Immunoproteasomes are also expressed by most cell types after exposure to interferons, where they supplement and eventually replace standard proteasomes. Immunoproteasomes are upregulated by viruses that induce high levels of interferons, such as hepatitis B virus (8, 9) and lymphocytic choriomeningitis virus (10).
Immunoproteasome assembly is intricately coordinated; the generation of “mixed” proteasomes with standard subunits and immunosubunits is under tissue-specific control (11–14). The codependent incorporation of LMP2 and MECL1 suggests that mixed proteasomes with Δ/MEC1 or LMP2/Z may be detrimental in certain cell types. Standard and immunoproteasomes generate an overlapping repertoire with differences in cleavage at both N-and COOH-termini (15), and there are numerous examples of defined peptides whose generation is enhanced by immunoproteasomes (16). Based on these findings, the favored hypothesis is that immunoproteasomes principally function to optimally generate peptides for immunosurveillance.
There are, however, a few findings that undermine this hypothesis. A number of defined determinants are presented more efficiently following knockout of LMP2 or LMP7 (16). T cells lacking one or more of the immunosubunits demonstrate altered proliferation upon activation by cognate Ags (17) or mitogens (18) and differ in homeostatic proliferation (19). Based on these findings, it has been proposed that major functions of immunoproteasomes are unrelated to Ag processing (20, 21). To explore potential alternative functions of immunoproteasomes, we characterized antiviral innate and humoral immune responses in immunoproteasome-deficient mice.
Materials and Methods
Mice and Viruses
C57BL/6 mice were purchased from Taconic (Germantown, NY) and CD45.1 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). LMP2−/− were originally obtained from Luc Van Kaer (Vanderbilt Medical Center, Nashville, TN), who backcrossed them 10 times, and then bred at Taconic. LMP2−/− MECL1−/− and LMP7/MECL1−/− mice were bred at the Cincinnati Children’s Hospital Medical Center. Mice were maintained under specific pathogen-free conditions and all procedures involving mice were approved by the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases. Influenza A virus (IAV; A/Puerto Rico/8/34) was propagated in 10-d-old embryonated chicken eggs. For all vaccinations, the virus was first inactivated by treating with paraformaldehyde (1:864 dilution) for 2 d at 4°C, and then ~100 hemagglutination units of inactivated IAV was injected i.p. At several time points after vaccination, mice were retro-orbitally bled, and RBCs were removed using serum-gel tubes (Sarstedt, Newton, NC). Serum was inactivated by incubating at 56°C for 30 min.
Hemagglutination inhibition titrations
Hemagglutination inhibition (HAI) titrations were performed in 96-well round polystyrene plates (Corning, Corning, NY). Serial 2-fold dilutions of sera in 100 ul PBS were mixed with four agglutinating doses of IAV in 100 μl of PBS. Next, 25 μl of a 2% (v/v) human RBC solution was added, and the reaction was allowed to proceed for 60 min at room temperature. The plates were held perpendicularly to the ground, and HAI was observed as the agglutinated RBCs formed streaks in each well. HAI titers are expressed as the reciprocal of the highest Ab dilution that was able to inhibit four agglutinating doses of virus in a total volume of 225 μl.
IAV ELISAs
Flat bottom microtiter plates (Thermo Fisher Scientific, Waltham, MA) were coated with ~75 hemagglutination units of IAV per well in a 0.1 M sodium carbonate coating solution (BD Biosciences, San Jose, CA) overnight at 4°C and then blocked with PBS containing 10% FBS for 2 h at room temperature. Mouse sera was then added and incubated for 2 h at room temperature. Next, rabbit (IgG) anti-mouse secondary Abs of different isotypes (Calbiochem, San Diego, CA) were added and incubated for 1 h at room temperature, and then HRP conjugated goat anti-rabbit IgG (Calbiochem) was added and incubated for 1 h at room temperature. Finally, tetramethylbenzidine substrate (Kirkegaard & Perry Laboratories, Gaithersburg, MD) was added, and then 1 M H3PO4 was quickly added to stop the reaction. OD was read at 450 nm. In between each step of the anti-IAV ELISAs, the plates were washed with PBS-Tween (0.05%).
Cell phenotyping
Spleens and bone marrow were isolated, single cell suspensions were created, and ACK lysing buffer (BioWhittaker, Walkersville, MD) was added to lyse RBCs. The cells were then passed through a 70-μm nylon cell strainer (BD Falcon, San Jose, CA) and washed twice with balanced salt solution containing 0.1% BSA. Cells were incubated with different combinations of fluorescently labeled Abs (eBiosciences, San Diego, CA and BD Pharmingen, San Jose, CA) and samples were analyzed using an LSR II machine (BD Biosciences).
Lymphocyte Transfer Studies
Splenocytes were isolated as described above, and CD4+ T cells and B cells were purified by negative selection using commercially available kits (Miltenyi Biotec, Auburn, CA). For some experiments, APCs (CD11c+ and CD11b+) were removed from splenocytes preparations after staining the cells with anti-CD11c FITC and anti-CD11b FITC and then using a commercially available kit (Miltenyi Biotec) that selects FITC-negative cells. Mice were irradiated with ~800 rad; 6 h later, purified cell populations that were 83–98% pure were injected i.v. through the tail vein; 1.5 × 107 cells were injected for APC-depletion studies, and 8 × 106 B cells and 3 × 106 CD4+ T cells were injected for fractionated splenocytes studies. For all studies, equal numbers of C57BL/6 and LMP2−/− cells were injected. Two hours later, mice were injected i.p. with ~100 hemagglutination units of inactivated IAV, and anti-IAV Abs were determined at several time points as described above. For experiments involving CD45.1 mice, we did not inject IAV, but instead isolated the spleens from these irradiated mice 7 d after transferring cells. For these experiments, single cell suspensions were prepared as described above and cells were stained using Abs against CD45.1 and CD45.2 (eBiosciences) and analyzed on an LSR II flow cytometer (BD Biosciences).
Lymphocyte proliferation assays
Proliferation assays were performed as described previously (18). CD4+ T cells and B cells were purified by negative selection as described above and plated in RPMI 1640 with 10% FCS in flat bottom polystyrene plates (Corning). For B cell assays, 1 × 105 cells per well were plated; for CD4+ T cell assays, 2 × 105 cells per well were plated. Various amounts of Con A or LPS were added to CD4+ T cells or B cells, respectively. The cells were pulsed with [3H]thymidine 2 d later and [3H] incorporation was determined 1 d after pulsing.
DC experiments
Bone marrow-derived DCs were cultured for 10 d in RPMI 1640 containing 20 ng/ml GM-CSF (PeproTech, Rocky Hill, NJ) and then reseeded at 2.5 × 105 cells per well in round-bottom polystyrene plates (Corning). IAV was added to the cells, and supernatant was collected 18 h later. Cytokine levels in supernatants were measured by Luminex technology (Bio-Rad, Hercules, CA) and by an IFN-α ELISA that was performed using a previously described protocol (22).
Immunoblots
B cells were purified by negative or positive selection using magnetic beads. For some experiments, cells were treated with LPS and then removed from culture and resuspended in lysis buffer (20 mM Tris pH 7.4, 150 mM mNaCl, 2mM sodium pyrophosphate, 25 mM β-glycerol phosphate, 5 mM NEM, 1% NP-40) and incubated on ice for 30 min. Nuclei and insoluble material were pelleted by centrifugation, and the resulting supernatants were mixed with SDS-PAGE buffer (Quality Biologicals, Gaithersburg, MD) and run on a 12% SDS-PAGE gel followed by blotting onto nitro-cellulose. For polyubiquitin and immunoproteasome subunit staining, total cell lysates were prepared. Blots were probed with rabbit polyclonal anti-IκBα (Santa Cruz Biotechnology, Santa Cruz, CA) and mouse monoclonal anti-actin (Abcam, Cambridge, MA) Abs or with mouse monoclonal anti-polyubiquitin (clone FK2; BIOMOL, Plymouth Meeting, PA) Ab followed by infrared dye coupled secondary Abs. For immunoproteasome subunit expression studies, blots were probed with rabbit anti-LMP2 (Abcam), rabbit anti-LMP7 (Abcam), or rabbit anti-MECL1 (BIOMOL), as well as a GAPDH loading control Ab (Abcam). Blots were analyzed using an Odyssey imaging system (LI-COR, Lincoln, NE) or a STORM phosphorimager (GE Healthcare Bio-Sciences, Uppsala, Sweden).
Proteasome purification
Cells were homogenized in ice cold homogenization buffer (50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 25 mM KCl, 0.2 M sucrose, 0.5% NP-40, 100 μg/ml CHX, EDTA-free protease inhibitors [Roche, Basel, Switzerland], 10 U/Ml RNase Out [Invitrogen, Carlsbad, CA], diethyl pyrocarbonate water), and the lysate was centrifuged at 20,000 × g for 10 min at 4°C. The cleared lysate was layered on a 15–50% sucrose gradient in the same buffer. After centrifugation at 35,000 rpm (Beckman, SW41.Ti) for 2.5 h at 4°C, the gradient was fractionated. Each fraction was pelleted to concentrate proteins and analyzed by immunoblotting as described above.
Results
Mixed proteasomes inhibit B cell development
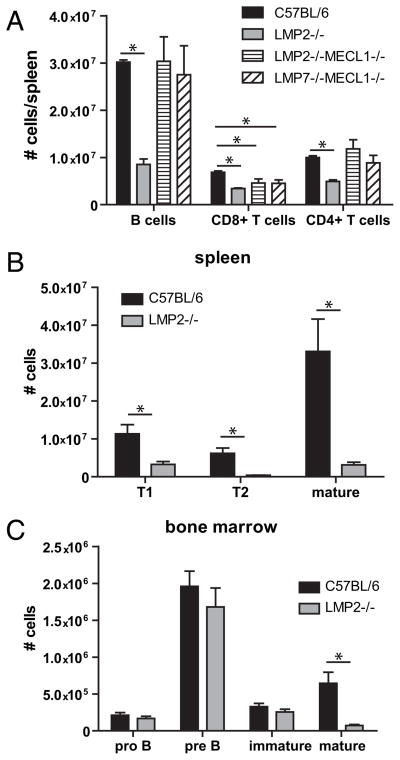
Immunoproteasomes are constitutively expressed in professional APCs, such as DCs (23), and other cell types, such as T cells (24). Immunoblotting revealed that B cells purified from the spleen express immunoproteasomes (Fig. 1). To determine whether immunoproteasome expression is required for maintaining homeostatic levels of resting B cells, we enumerated B cells in spleens isolated from immunoproteasome-deficient and wild type (WT; C57BL/6) mice. Consistent with previous studies (25, 18), immunoproteasome-deficient mice had fewer CD8+ T cells compared with WT mice (Fig. 2A), an observation originally attributed to aberrant thymic selection. Although we found that most strains of immunoproteasome-deficient mice posses normal numbers of B cells, LMP2−/− mice had dramatically reduced numbers of splenic B cells compared with WT mice (Fig. 2A). Further analysis revealed that transitional type 1 (T1) and T2 B cells were underrepresented in the spleens of LMP2−/− mice (Fig. 2B), yet normal numbers of pro B, pre B, and immature B cells were present in the bone marrow of these mice (Fig. 2C). The defect in mature B cells in LMP2−/− mice is not simply caused by the absence of LMP2, because LMP2−/− MEC1−/− mice have normal numbers of B cells (Fig. 2A). Rather, this effect is likely due to the expression of the mixed proteasomes (11, 12). Immunoblotting of sucrose gradient fractions from LMP2−/− B cell lysates revealed that these cells express Δ/MECL1/LMP7 mixed proteasomes, as inferred from their sedimentation characteristics (Fig. 3). It is possible that other types of mixed proteasomes exist in these mice as well. Thus, LMP2−/− mice provide a unique opportunity to study why vertebrates evolved stringent mechanisms to coordinate the assembly of immunoproteasomes.
FIGURE 1.
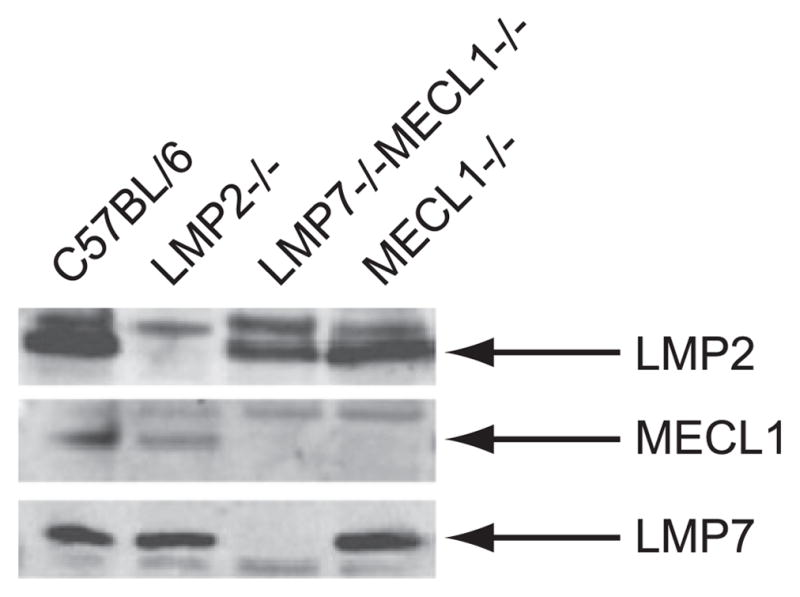
B cells express immunoproteasomes. Immunoproteasome subunit expression was determined in C57BL/6 purified B cells. LMP2−/−, LMP7−/−MECL1−/−, and MECL1−/− purified B cells were used as negative controls.
FIGURE 2.
LMP2−/− mice have reduced amount of B cells. A, Flow cytometric analysis was completed on splenocytes isolated from C57BL/6 and immunoproteasome deficient mice. Cell numbers of CD4+ T cells (CD4+, TCRβ+), CD8+ T cells (CD8+, TCRβ+), and B cells (CD19+, B220+) were determined by multiplying the splenic frequencies by the total number of cells from each spleen. Cells isolated from bone marrow (B) or spleens (C) were phenotyped using markers of B cell development (B220, IgD, and CD43). For all experiments, n = 3 mice per group. Mean ± SEM is shown. Data are representative of two independent experiments. *p < 0.05.
FIGURE 3.
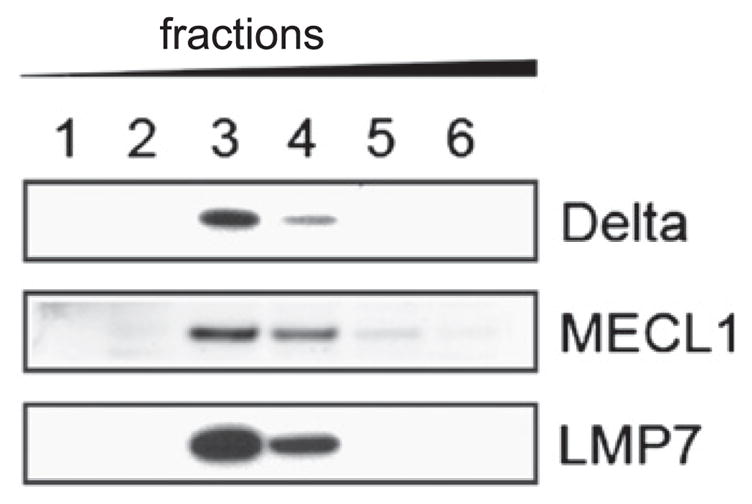
LMP2−/− B cells contain mixed proteasomes. Fractions of LMP2−/− B cell lysate were isolated using a sucrose gradient and immunoblotting was performed. The first fraction contained the lightest material and the sixth fraction contained the heaviest.
Overexpression of mixed proteasomes leads to impaired anti-IAV Ab responses
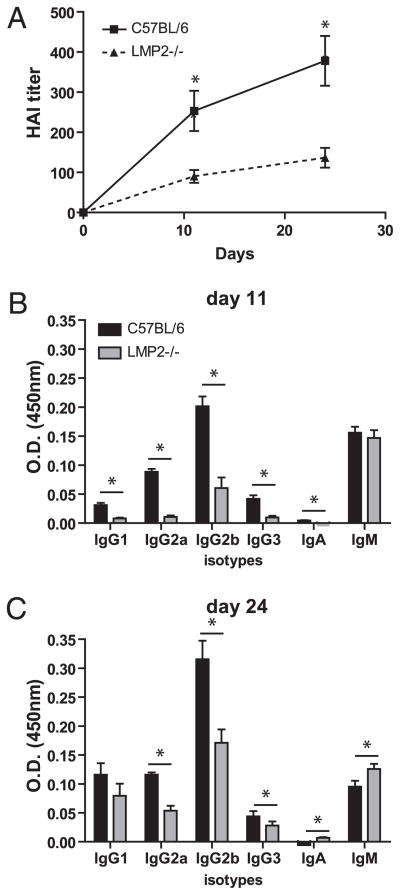
To determine whether mixed proteasomes alter the generation of primary antiviral Ab responses, we immunized LMP2−/− mice with inactivated IAV and determined anti-IAV serum titers by HAI. HAI titers were reduced more than 2-fold in LMP2−/− mice at early and late time points after vaccination (Fig. 4A). A closer examination of the anti-IAV Ab response by ELISA revealed that LMP2−/− mice were able to mount normal IgM responses but failed to isotype switch (Fig. 4B, 4C).
FIGURE 4.
Mixed proteasomes negatively affect antiviral Ab responses. Following vaccination with IAV, Ab responses were determined by HAI titers (A) and by ELISA (B, C; n = 10–12 mice per group). Data are representative of four independent experiments. Shown is mean ± SEM. *p < 0.05.
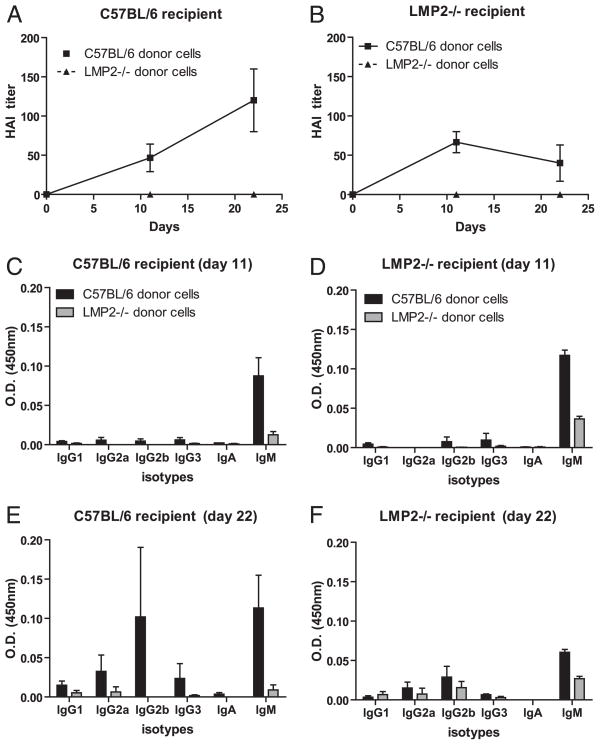
A logical explanation of why LMP2−/− mice mount defective IAVAb responses is that these mice possess reduced numbers of B cells. It is also possible that mixed proteasome expression alters the functions of mature B cells (and/or helper CD4+ T cells). To characterize the contribution of B cells, CD4+ T cells, and APCs to diminished Ab responses in LMP2−/− mice, we transferred equal numbers of WT or LMP2−/− splenic lymphocytes (i.e., splenocytes depleted of APCs) into irradiated WT recipients before immunization with inactivated IAV. Transfer of WT lymphocytes into WT mice resulted in strong Ab responses that involved delayed but robust Ig class switching (Fig. 5A, 5C, 5E). Strikingly, mice receiving LMP2−/− lymphocytes mounted undetectable (by HAI) to barely detectable (by ELISA) Ab responses.
FIGURE 5.
LMP2−/− lymphocytes are defective. CD11c/CD11b depleted C57BL/6 or LMP2−/− splenocytes were transferred into sublethally irradiated C57BL/6 or LMP2−/− recipient mice. The mice were then vaccinated with IAV and Ab titers were determined by HAI (A, B) and ELISA (C–F; n = 3 mice per group). Data are mean ± SEM.
In the same experiment, we performed adoptive transfers in LMP2−/− mice. Mice receiving LMP2−/− lymphocytes again generated weak Ab responses, whereas mice receiving WT lymphocytes generated Ab responses that failed to properly class switch (Fig. 5B, 5D, 5F). In each homologous transfer condition (wt → wt, LMP2−/− → LMP2−/−), responses were weaker than in nonmanipulated mice. This is likely due to limitations in lymphocytes homing to the proper anatomical regions as well as radiation-induced damage in lymphatic organs that interferes with the immune response.
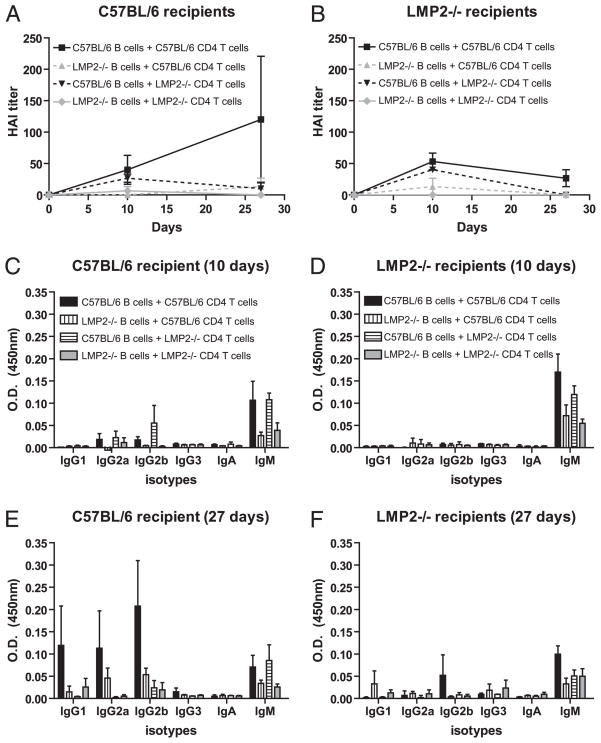
CD4+ T cells are required for the generation of fully functional IAV Ab responses in mice (26, 27), although weak responses (with H chain class switching) can occur in the absence of CD4+ T cells (28, 29). To determine the extent to which intact immunoproteasome expression in B cells versus CD4+ T cells is required for optimal Ab responses, we repeated the adoptive transfer experiment using purified B and CD4+ T cells from WT and LMP2−/− mice in all permutations (Fig. 6). Transfer of B and CD4+ T cells from the same donor completely recapitulated the observations obtained with unfractionated lymphocyte transfers. WT B cells transferred with LMP2−/− CD4+ T cells were able to generate early IAV Ab responses in both WT and LMP2−/− recipients, but these responses were transient. By contrast, all other permutations resulted in weak responses.
FIGURE 6.
Immunoproteasome expression in B and CD4+ T cells is required for the generation of IAV Ab responses. Purified B and CD4+ T cells from C57BL/6 or LMP2−/− mice were transferred into sublethally irradiated C57BL/6 or LMP2−/− recipient mice. The mice were vaccinated with IAV and Ab titers were determined by HAI (A, B) and ELISA (C–F; n = 3 mice per group; n = 2 at some late time points). Data are mean ± SEM and are representative of two independent experiments.
These data indicate that expression of mixed proteasomes in multiple cell types leads to the reduction of anti-IAV responses in LMP2−/− mice. Notably, mixed proteasome expression in B cells appears to greatly affect Ab responses, because miniscule Ab responses were generated in WT mice when LMP2−/− B cells were cotransferred with WT CD4+ T cells.
Mixed proteasome expression affects lymphocyte function
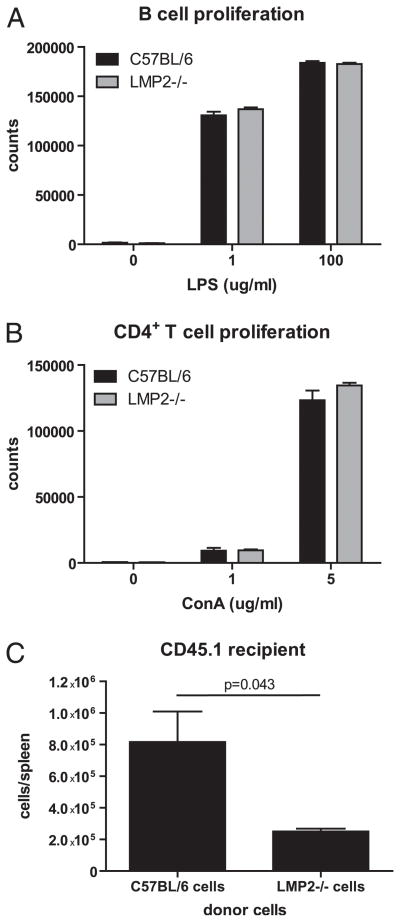
To further characterize the deleterious effects of mixed proteasomes on B and CD4+ T cell function, we measured the ability of LMP2−/− cells to proliferate after activation with their respective mitogens, LPS or Con A (Fig. 7A, 7B). LMP2 knockout did not affect proliferation of either cell type, as measured by incorporation of [3H]thymidine. We previously reported that LMP2−/− CD8+ T cells are immunodominated in anti-IAV responses by WT CD8+ T cells in adoptive transfer experiments (17, 24). Based on this finding and our observation that naive LMP2−/− mice have dramatically decreased numbers of B cells (Fig. 2), we hypothesized that LMP2−/− cells have a survival disadvantage compared with WT cells. To test this idea, we transferred WT or LMP2−/− lymphocytes into irradiated CD45.1+ congenic mice and enumerated WT or LMP2−/− lymphocytes, which express CD45.2, present in spleens 7 d later. Approximately 3-fold as many WT lymphocytes were recovered under these conditions (Fig. 7C). These findings indicate that mixed proteasome expression does not have a gross effect on lymphocyte proliferative capacity, but may affect the ability of lymphocytes to home to, survive, or homeostatically proliferate in the spleen.
FIGURE 7.
LMP2−/− lymphocytes proliferate normally but may have defects in survival. Purified B cells (A) and CD4+ T cells (B) were stimulated with LPS or Con A for a total of 3 d, and proliferation was determined by [3H] incorporation (n = 3 samples per group). Data are representative of two independent experiments. C, C57BL/6 or LMP2−/− splenocytes were transferred into partially irradiated CD45.1+ mice; 7 d later, the number of surviving lymphocytes was determined by flow cytometry (n = 3 mice per group). Splenic frequencies were multiplied by total cell numbers to determine how many transferred cells were present in each spleen. Data are mean ± SEM and are representative of two independent experiments.
Mixed proteasomes expression affects DC activation
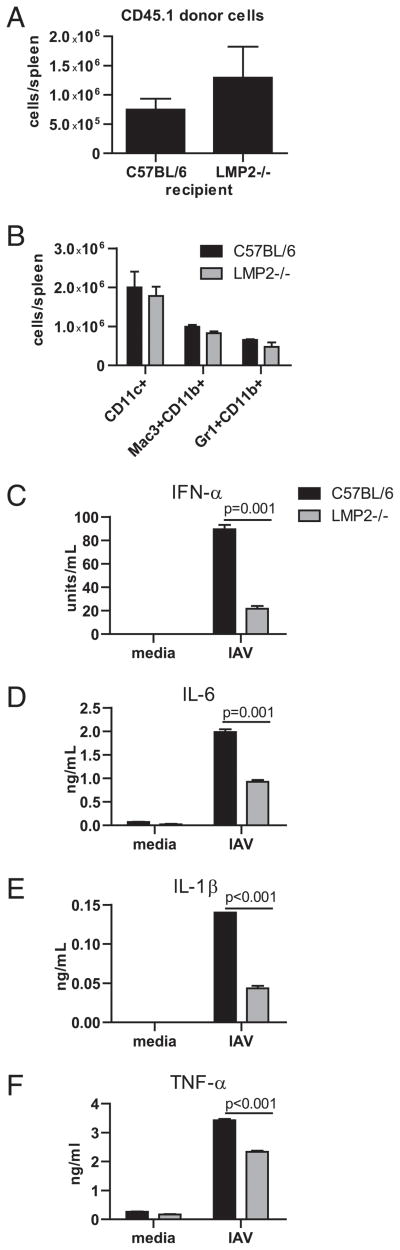
The adoptive transfer experiments described above implicated that mixed proteasomes expressed in radiation resistant cells contribute to impaired Ab responses, because after transfer of WT lymphocytes, LMP2−/− recipients generated weaker Ab responses compared with WT recipients. A trivial explanation for this finding is that WT lymphocytes were rejected by LMP2−/− recipients. We eliminated this possibility by showing that 7 d after transferring congenic WT CD45.1+ lymphocytes into irradiated mice, there were more transferred cells in LMP2−/− versus WT recipients (Fig. 8A), although this difference did not reach statistical significance.
FIGURE 8.
Mixed proteasomes negatively affect innate cytokine production. A, CD45.1+ splenocytes were transferred into partially irradiated C57BL/6 or LMP2−/− mice; 7 d later, the number of CD45.1+ cells was determined by flow cytometry (n = 3 mice per group). Splenic frequencies were multiplied by total cell numbers to determine how many transferred cells were present in each spleen. Data are representative of two independent experiments. B, Numbers of cells per spleen were determined by flow cytometry as described in Fig. 2 (n = 3 mice per group). Data are representative of two independent experiments. Bone marrow-derived DCs were infected with IAV, and supernatant was tested for levels of cytokines by ELISA (C) or by Luminex technology (D–F; n = 3 samples per group). ELISA data are representative of three independent experiments, and Luminex data are representative of two independent experiments. All data are mean ± SEM.
One explanation for the poor Ab response mounted by LMP2−/− recipients is that they exhibit defective innate immune responses, because early production of cytokines, such as IL-6 and type I IFNs, enhance Ab responses and promote isotype switching (30, 31). Could LMP2−/− mice simply possess lower numbers of innate immune cells? This was not the case, as equal numbers of DCs, macrophages, and neutrophils are present in spleens of wt and LMP2−/− mice (Fig. 8B). To examine the intriguing possibility that mixed proteasome expression alters cytokine production by innate immune cells, we infected WTand LMP2−/− bone marrow-derived DCs with IAV. Each of four cytokines measured after 18 h in culture, IFN-α, IL-6, IL-1β, and TNF-α, were present at significantly lower levels in supernatants from LMP2−/− versus WT DCs (Fig. 8C–F).
Mixed proteasomes alter NF-κB activation
How might mixed proteasomes modify B cell and DC function? Proteasomes are responsible for most protein degradation, and compromising proteasome function has potent effects on cell physiology (32). Activated B cells are particularly sensitive to limitations in proteasome function, presumably owing to increased numbers of DRiPs associated with the massive increase in Ig synthesis (33, 34). To examine the effect of mixed proteasome expression on overall proteasome function, we measured levels of polyubiquitylated proteins, which are known to accumulate when proteasome degradation is limiting. As seen in Fig. 9, there was no significant difference in the levels of polyubiquitylated proteins in activated B cells in LMP2−/− versus WT mice.
FIGURE 9.
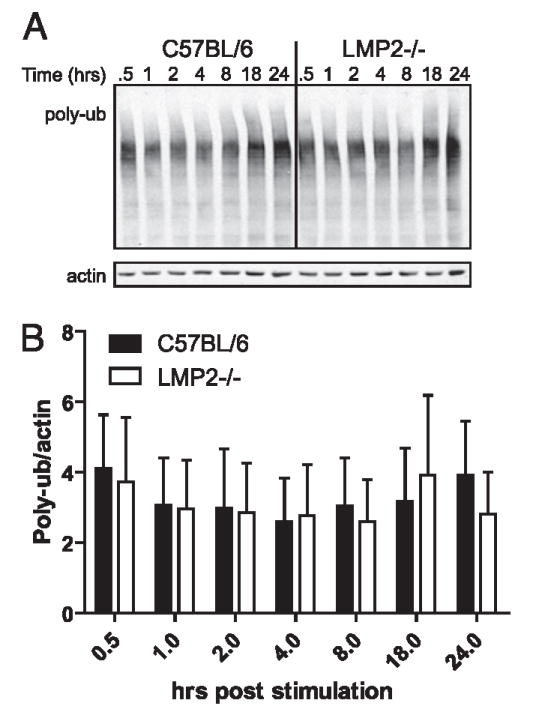
Mixed proteasomes do not alter protein degradation. A, Purified B cells were stimulated with LPS, and levels of polyubiquitinated proteins were monitored by immunoblotting over time. Levels of polyubiquitinated proteins were normalized to actin. B, The experiment was repeated in triplicate, and levels of polyubiquitinated proteins were quantified using an Odyssey imaging system.
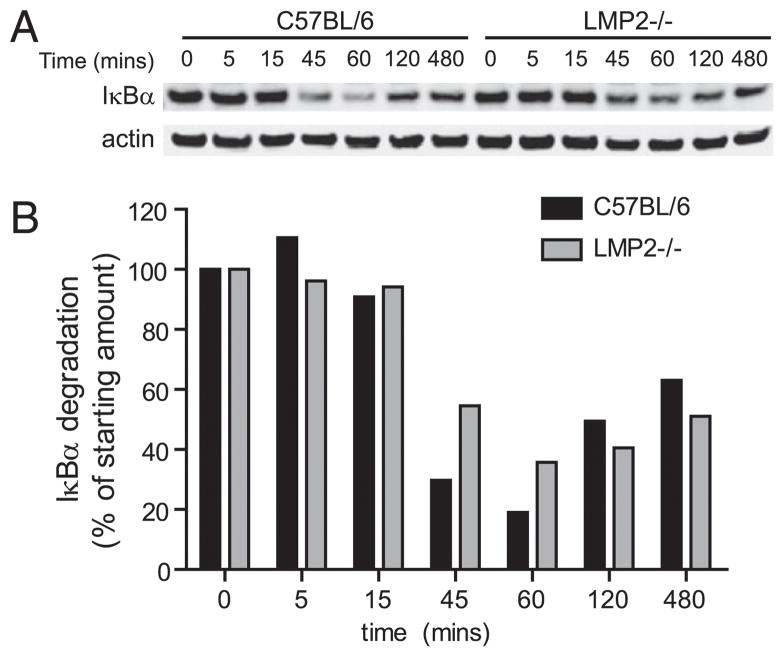
Proteasomes also play a critical role in regulating transcription factors, such as NF-κB, Spt23p, and Mga2p (3, 4, 35). Following IκB degradation by standard proteasomes, NF-κB translocates to the nucleus, rapidly activating subsets of genes, many of which are involved in immune activation. Because both lymphocyte survival and DC activation require NF-κB activation, we examined the effect of mixed proteasome expression on NF-κB activation. We stimulated purified LMP2−/− or WT B cells with LPS and measured IκBα degradation by immunoblotting. Consistent with the hypothesis that mixed proteasomes impair NF-κB activation, IκB α degradation in LMP2−/− cells was delayed and less complete than in WT cells (p < 0.05 at 60 min following LPS; Fig. 10).
FIGURE 10.
Mixed proteasomes dysregulate NF-κB signaling. Purified B cells were stimulated with LPS, and levels of IκB α were monitored by immunoblotting over time. Levels of IκB α were quantified and normalized to actin. Data are representative of three independent experiments.
Our findings indicate that mixed proteasomes present in LMP2−/− mice compromise virtually every parameter of the immune response against IAV, potentially through dysregulation of signaling pathways, such as the NF-κB pathway. The highly coordinated assembly of immunoproteasomes might be necessary to prevent immunologic chaos that can result from altered signaling pathways.
Discussion
It has long been known that splenic cells express immunoproteasomes in preference to standard proteasomes (6, 7). Inasmuch as B cells constitute a large fraction of splenocytes, it is hardly surprising that B cells—like T cells, DCs, and monocytes—preferentially express immunoproteasomes. Because B and T cells probably play limited roles in activating afferent CD8+ T cells, it is logical that the major function of immunoproteasomes in lymphocytes is unrelated to generation of class I peptide ligands.
One of the mysterious features of immunoproteasomes is that their composition is limited to a small subset of the possible permutations of immunosubunits and standard subunits in some cell types. One possibility is that certain types of mixed proteasomes compromise cellular function. In this study, we show that mixed proteasomes expressed in LMP2−/− mice decrease lymphocyte survival and reduce cytokine production by DCs. These defects are associated with impaired NF-κB signaling in LMP2−/− cells.
The involvement of immunoproteasomes in NF-κB activation is controversial (36, 37). Our studies suggest that mixed proteasomes inefficiently cleave IκB, an event required for NF-κB activation. It is entirely possible, even likely, that other signaling components are impaired in cells that express mixed proteasomes. Consistent with this possibility, it was recently reported that cardiac LMP2 expression is required for the activation of the protein kinase AKT (38).
Several recent studies suggest that immunoproteasomes are particularly adept at degrading damaged proteins (39–41). Of particular relevance is the finding that oxidized proteins are increased in the liver and brain of LMP2−/− mice (39). Resting lymphocytes are nearly inert metabolically and are unlikely to challenge proteosomal capacity. After activation, however, they divide at a rate unparalleled among vertebrate cells. Although it is possible that the defect in LMP2−/− cells is associated with a decreased ability to degrade damaged proteins, we do not find altered levels of ubiquitylated proteins in stimulated LMP2−/− cells.
Our findings demonstrate that LMP2−/− mice display severe defects in B cell and DC function, limiting their use in studying the contribution of immunoproteasomes to Ag processing. The myriad dysfunctions associated with the expression of mixed proteasomes provide a ready explanation for their low abundance in some types of WT cells. It will be of great interest in future studies to delineate the unique function performed by immunoproteasomes in cells of immune lineage.
Acknowledgments
We thank the staff of the Comparative Medical Branch of the National Institute of Allergy and Infectious Diseases for assistance with animal studies, Luc Van Kaer for LMP2−/− mice, Glennys Reynoso for technical assistance, and Dietrich Conze for expert NFκB advice.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (R21 AI073584) and the Australian National Health and Medical Research Council (ID 433608).
Abbreviations in this paper
- DC
dendritic cell
- HAI
hemagglutination inhibition
- IAV
influenza A virus
- WT
wild type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 2.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J Biol Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 3.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 4.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 5.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the Archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka K. Role of proteasomes modified by interferon-gamma in antigen processing. J Leukoc Biol. 1994;56:571–575. doi: 10.1002/jlb.56.5.571. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka K, Kasahara M. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol Rev. 1998;163:161–176. doi: 10.1111/j.1600-065x.1998.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 8.Robek MD, Garcia ML, Boyd BS, Chisari FV. Role of immunoproteasome catalytic subunits in the immune response to hepatitis B virus. J Virol. 2007;81:483–491. doi: 10.1128/JVI.01779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wieland SF, Vega RG, Müller R, Evans CF, Hilbush B, Guidotti LG, Sutcliffe JG, Schultz PG, Chisari FV. Searching for interferon-induced genes that inhibit hepatitis B virus replication in transgenic mouse hepatocytes. J Virol. 2003;77:1227–1236. doi: 10.1128/JVI.77.2.1227-1236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan S, van den Broek M, Schwarz K, de Giuli R, Diener PA, Groettrup M. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J Immunol. 2001;167:6859–6868. doi: 10.4049/jimmunol.167.12.6859. [DOI] [PubMed] [Google Scholar]
- 11.Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J Exp Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groettrup M, Standera S, Stohwasser R, Kloetzel PM. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc Natl Acad Sci USA. 1997;94:8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlmann B, Ruppert T, Kuehn L, Merforth S, Kloetzel PM. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J Mol Biol. 2000;303:643–653. doi: 10.1006/jmbi.2000.4185. [DOI] [PubMed] [Google Scholar]
- 14.Drews O, Wildgruber R, Zong C, Sukop U, Nissum M, Weber G, Gomes AV, Ping P. Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol Cell Proteomics. 2007;6:2021–2031. doi: 10.1074/mcp.M700187-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloetzel PM, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol. 2004;16:76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Norbury CC, Cho Y, Yewdell JW, Bennink JR. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J Exp Med. 2001;193:1319–1326. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caudill CM, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. T cells lacking immunoproteasome subunits MECL-1 and LMP7 hyper-proliferate in response to polyclonal mitogens. J Immunol. 2006;176:4075–4082. doi: 10.4049/jimmunol.176.7.4075. [DOI] [PubMed] [Google Scholar]
- 19.Zaiss DM, de Graaf N, Sijts AJ. The proteasome immunosubunit multicatalytic endopeptidase complex-like 1 is a T-cell-intrinsic factor influencing homeostatic expansion. Infect Immun. 2008;76:1207–1213. doi: 10.1128/IAI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yewdell JW. The seven dirty little secrets of major histocompatibility complex class I antigen processing. Immunol Rev. 2005;207:8–18. doi: 10.1111/j.0105-2896.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- 21.Nussbaum AK, Rodriguez-Carreno MP, Benning N, Botten J, Whitton JL. Immunoproteasome-deficient mice mount largely normal CD8+ T cell responses to lymphocytic choriomeningitis virus infection and DNA vaccination. J Immunol. 2005;175:1153–1160. doi: 10.4049/jimmunol.175.2.1153. [DOI] [PubMed] [Google Scholar]
- 22.Asselin-Paturel C, Boonstra A, Dalod M, Durand I, Yessaad N, Dezutter-Dambuyant C, Vicari A, O’Garra A, Biron C, Brière F, Trinchieri G. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat Immunol. 2001;2:1144–1150. doi: 10.1038/ni736. [DOI] [PubMed] [Google Scholar]
- 23.Macagno A, Gilliet M, Sallusto F, Lanzavecchia A, Nestle FO, Groettrup M. Dendritic cells up-regulate immunoproteasomes and the proteasome regulator PA28 during maturation. Eur J Immunol. 1999;29:4037–4042. doi: 10.1002/(SICI)1521-4141(199912)29:12<4037::AID-IMMU4037>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 24.Pang KC, Sanders MT, Monaco JJ, Doherty PC, Turner SJ, Chen W. Immunoproteasome subunit deficiencies impact differentially on two immunodominant influenza virus-specific CD8+ T cell responses. J Immunol. 2006;177:7680–7688. doi: 10.4049/jimmunol.177.11.7680. [DOI] [PubMed] [Google Scholar]
- 25.Van Kaer L, Ashton-Rickardt PG, Eichelberger M, Gaczynska M, Nagashima K, Rock KL, Goldberg AL, Doherty PC, Tonegawa S. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 26.Scherle PA, Gerhard W. Functional analysis of influenza-specific helper T cell clones in vivo. T cells specific for internal viral proteins provide cognate help for B cell responses to hemagglutinin. J Exp Med. 1986;164:1114–1128. doi: 10.1084/jem.164.4.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sangster MY, Riberdy JM, Gonzalez M, Topham DJ, Baumgarth N, Doherty PC. An early CD4+ T cell-dependent immunoglobulin A response to influenza infection in the absence of key cognate T-B interactions. J Exp Med. 2003;198:1011–1021. doi: 10.1084/jem.20021745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sha Z, Compans RW. Induction of CD4(+) T-cell-independent immunoglobulin responses by inactivated influenza virus. J Virol. 2000;74:4999–5005. doi: 10.1128/jvi.74.11.4999-5005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee BO, Rangel-Moreno J, Moyron-Quiroz JE, Hartson L, Makris M, Sprague F, Lund FE, Randall TD. CD4 T cell-independent antibody response promotes resolution of primary influenza infection and helps to prevent reinfection. J Immunol. 2005;175:5827–5838. doi: 10.4049/jimmunol.175.9.5827. [DOI] [PubMed] [Google Scholar]
- 30.Le Bon A, Schiavoni G, D’Agostino G, Gresser I, Belardelli F, Tough DF. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14:461–470. doi: 10.1016/s1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 31.Jego G, Palucka AK, Blanck JP, Chalouni C, Pascual V, Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–234. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 32.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 33.Cenci S, Sitia R. Managing and exploiting stress in the antibody factory. FEBS Lett. 2007;581:3652–3657. doi: 10.1016/j.febslet.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Cascio P, Oliva L, Cerruti F, Mariani E, Pasqualetto E, Cenci S, Sitia R. Dampening Ab responses using proteasome inhibitors following in vivo B cell activation. Eur J Immunol. 2008;38:658–667. doi: 10.1002/eji.200737743. [DOI] [PubMed] [Google Scholar]
- 35.Fan CM, Maniatis T. Generation of p50 subunit of NF-kappa B by processing of p105 through an ATP-dependent pathway. Nature. 1991;354:395–398. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi T, Faustman D. NOD mice are defective in proteasome production and activation of NF-kappaB. Mol Cell Biol. 1999;19:8646–8659. doi: 10.1128/mcb.19.12.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Runnels HA, Watkins WA, Monaco JJ. LMP2 expression and proteasome activity in NOD mice. Nat Med. 2000;6:1064–1065. doi: 10.1038/80349. author reply 1065–1066. [DOI] [PubMed] [Google Scholar]
- 38.Cai ZP, Shen Z, Van Kaer L, Becker LC. Ischemic preconditioning-induced cardioprotection is lost in mice with immunoproteasome subunit low molecular mass polypeptide-2 deficiency. FASEB J. 2008;22:4248–4257. doi: 10.1096/fj.08-105940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Q, Martin S, Dimayuga E, Bruce-Keller AJ, Keller JN. LMP2 knock-out mice have reduced proteasome activities and increased levels of oxidatively damaged proteins. Antioxid Redox Signal. 2006;8:130–135. doi: 10.1089/ars.2006.8.130. [DOI] [PubMed] [Google Scholar]
- 40.Khan MA, Oubrahim H, Stadtman ER. Inhibition of apoptosis in acute promyelocytic leukemia cells leads to increases in levels of oxidized protein and LMP2 immunoproteasome. Proc Natl Acad Sci USA. 2004;101:11560–11565. doi: 10.1073/pnas.0404101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Díaz-Hernàndez M, Hernández F, Martín-Aparicio E, Gómez-Ramos P, Morán MA, Castaño JG, Ferrer I, Avila J, Lucas JJ. Neuronal induction of the immunoproteasome in Huntington’s disease. J Neurosci. 2003;23:11653–11661. doi: 10.1523/JNEUROSCI.23-37-11653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]