Abstract
To study positioning of the polypeptide release factor eRF1 toward a stop signal in the ribosomal decoding site, we applied photoactivatable mRNA analogs, derivatives of oligoribonucleotides. The human eRF1 peptides cross-linked to these short mRNAs were identified. Cross-linkers on the guanines at the second, third, and fourth stop signal positions modified fragment 31–33, and to lesser extent amino acids within region 121–131 (the “YxCxxxF loop”) in the N domain. Hence, both regions are involved in the recognition of the purines. A cross-linker at the first uridine of the stop codon modifies Val66 near the NIKS loop (positions 61–64), and this region is important for recognition of the first uridine of stop codons. Since the N domain distinct regions of eRF1 are involved in a stop-codon decoding, the eRF1 decoding site is discontinuous and is not of “protein anticodon” type. By molecular modeling, the eRF1 molecule can be fitted to the A site proximal to the P-site-bound tRNA and to a stop codon in mRNA via a large conformational change to one of its three domains. In the simulated eRF1 conformation, the YxCxxxF motif and positions 31–33 are very close to a stop codon, which becomes also proximal to several parts of the C domain. Thus, in the A-site-bound state, the eRF1 conformation significantly differs from those in crystals and solution. The model suggested for eRF1 conformation in the ribosomal A site and cross-linking data are compatible.
Keywords: eukaryotes, translation termination, stop-codon recognition, ribosome-bound eRF1, cross-linking
INTRODUCTION
Translation termination ensures the formation of normal-sized proteins with high fidelity and takes place when one of the three stop codons, UAA, UAG, or UGA, is translocated to the ribosomal A site where it is recognized by class-1 polypeptide release factors (RFs) that trigger hydrolysis of the ester bond between the peptidyl and tRNA moieties of peptidyl-tRNA bound to the P site (for review, see Kisselev et al. 2003; Nakamura and Ito 2003; Poole et al. 2003). Class-2 RFs are GTPases that stimulate in vitro activity of the respective class-1 RFs and promote their release from the ribosome after peptidyl-tRNA hydrolysis (see Buckingham et al. 1997; Kisselev and Buckingham 2000; Zavialov et al. 2001; Alkalaeva et al. 2006). In prokaryotes, there are two class-1 RFs, RF1 and RF2, decoding UAA/UAG and UAA/UGA, respectively. In contrast, in eukaryotes, all three stop codons are recognized by a single protein, eRF1, which has almost no similarities with RF1/RF2 both in the sequences and spatial structures (Frolova et al. 1994; Kisselev et al. 2000; Song et al. 2000; Vestergaard et al. 2001; Shin et al. 2004). The Y-shaped crystal structure of eRF1 consists of the N-terminal (N), middle (M), and C-terminal (C) domains. The C domain is responsible for the interaction with eRF3 (Ebihara and Nakamura 1999; Eurwilaichitr et al. 1999; Merkulova et al. 1999; Cheng et al. 2009). The M domain mimics the tRNA acceptor stem and contains a universal GGQ motif common to all class-1 RFs, which is located at the tip of the M domain of eRF1 and is essential for peptidyl-tRNA hydrolysis at the peptidyl transferase center (Frolova et al. 1999; Song et al. 2000; Seit-Nebi et al. 2001; Klaholz et al. 2003; Mora et al. 2003; Rawat et al. 2003; Scarlett et al. 2003; Petry et al. 2005). The N domain mimics the tRNA anticodon arm and contains two loops with the highly conserved YxCxxxF (positions 125–131) and NIKS (positions 61–64) motifs that play a critical role in stop-codon recognition (Frolova et al. 2002; Ito et al. 2002; Seit-Nebi et al. 2002; Kolosov et al. 2005; Fan-Minogue et al. 2008; Cheng et al. 2009). Evidence that the first nucleotide of a stop codon at the A site contacts K63 in the NIKS motif of human eRF1 has been obtained (Chavatte et al. 2002) by cross-linking experiments with an mRNA analog containing a 4-thiouridine (s4U) residue at the first position of the stop codon phased on the ribosome by a tRNAAsp cognate to the Asp codon, which is located 5′ to the stop codon (Chavatte et al. 2001).
Despite numerous studies on amino acid residues of eRF1 involved in stop-codon recognition, there is still no direct experimental data on positioning of the second, third, and fourth nucleotides of a stop signal with respect to eRF1. X-ray crystallography, the powerful approach for studying the structure of ribosomal functional sites, is not applicable for eukaryotic ribosomes since they have not been crystallized so far. Cryo-electron microscopy (cryo-EM) is now actively applied to eukaryotic ribosomes (e.g., see Spahn et al. 2001; Siridechadilok et al. 2005; Chandramouli et al. 2008). However, complexes, which model various states of eukaryotic ribosomes during termination and are suitable for cryo-EM, are not yet available.
To date, site-specific cross-linking of eRF1 to mRNA analogs bearing stop signals with photoactivatable nucleotides remains for eukaryotes one of the most appropriate approaches able to provide information on the positioning of the stop signal toward other components of the translation machinery. However, only mRNA analogs with either s4U (Chavatte et al. 2001, 2002, 2003) or perfluorophenylazide-modified uridine at the first stop-codon position or with modified 3′ terminal phosphate (Bulygin et al. 2002) have been applied so far. These mRNA analogs were able to cross-link to human eRF1 when the modified codon was bound at the A site, and the cross-linking was specific for the mRNA analogs containing a stop or UGG codon (Bulygin et al. 2002; Chavatte et al. 2002). Since uridine is located in the first position of a stop signal, s4U is applicable only for studying surroundings of the first nucleotide of a stop codon.
In this study, we have applied a cross-linking approach to obtain information on the position of each nucleotide of a stop signal with respect to eRF1 in the “phased” ribosome containing the P-site-bound tRNA. We have used a set of photoactivatable mRNA analogs bearing the cross-linkers at mRNA positions +4 to +7 with respect to the first nucleotide of the P-site-bound codon. Identification of the peptides cross-linked to nucleotides of the stop signal is evidence that three distinct N-domain peptides not close in the spatial structure of free human eRF1 surround the stop signal within the “phased” ribosome. The cross-linking results are in good agreement with our data on modeling of eRF1 structure in the 80S ribosomal termination complex by using a three-dimensional structure similarity method.
RESULTS
mRNA analogs
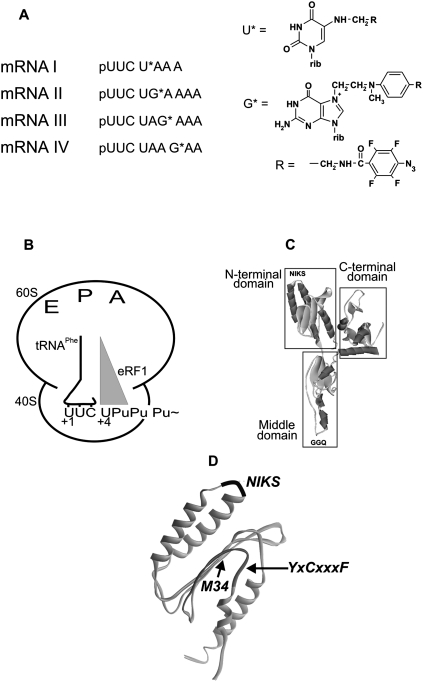
The mRNA analogs contained the Phe codon UUC followed by the UPuPuPu(p) tetraplet that carried a perfluorophenyl azide group on the C5 atom of the uridine or at the N7 atom of the guanosine (Fig. 1A). The modified nucleotide varied in position from first to fourth in the termination tetraplet. mRNA analogs containing sense codons (pUUCUG*CAAA, pUUCUCG*AAA, and pUUCUCAG*AA) were used in control experiments. The UUC codon of the mRNA analogs in the presence of tRNAPhe was targeted to the ribosomal P site, and the adjacent modified stop codon (or sense codons in controls) was bound to the A site (Fig. 1B). This complex was referred to as a “phased” ribosome. The irradiated complexes were purified to remove eRF1 cross-linked to mRNA analogs from the ribosome (Bulygin et al. 2002).
FIGURE 1.
(A) mRNA analogs used in this work. (B) 80S ribosome complexed with mRNA analog (Pu, purine), tRNA, and translation termination factors. (C) Crystal structure of human eRF1. (D) Location of the NIKS and YxCxxxF loops within the N-domain (Song et al. 2000). The distance separating modified base and the first nitrogen of the azidogroup is ≤11 Å for U* and 14 Å for G*.
Cross-linking of mRNA analogs to the components of the ribosomal complexes
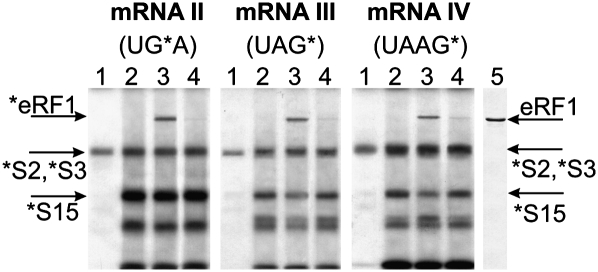
The results of cross-linking mRNA analogs II–IV (Fig. 1A) to ribosomal proteins and eRF1 are presented in Figure 2. Cross-linking patterns for analog I have been published earlier (Bulygin et al. 2002). Addition of tRNAPhe to nonphased ribosomes with mRNA analogs results in significant enhancement of cross-linking to some ribosomal proteins (Fig. 2, cf. lanes 1 and other lanes). Most likely, the protein bands in the upper and middle parts of the gels correspond to cross-linked proteins S2/S3 and S15, respectively, since the locations of these bands conform well to those of the proteins cross-linked to oligoribonucleotides (Bulygin et al. 2002; Graifer et al. 2004). Strong tRNA-dependent cross-linking to this protein has been shown to be characteristic for perfluorophenyl azide-modified nucleotides in mRNA positions from +4 to +7 (Graifer et al. 2004). The bands above and below S15 probably correspond to proteins S2/S3 and S30, respectively (Molotkov et al. 2006).
FIGURE 2.
Electrophoretic analyses of irradiated ribosomal complexes. Autoradiogram of SDS-PAGE. Upper parts of the gels with rRNA bands are not shown. Complexes are numbered according to the lane numbers (indicated above the autoradiograms). (Lane 1) 80S•mRNA, (lane 2) 80S•mRNA•tRNAPhe, (lane 3) 80S•mRNA•tRNAPhe•eRF1, (lane 4) 80S•mRNA (sense)•tRNAPhe•eRF1. Lane 5 is a stained electrophoregram of eRF1. To obtain complex 4, mRNAs were used with a single A-to-C substitution transforming stop codon to the respective sense codon. (Arrows) Bands corresponding to cross-linked eRF1 and ribosomal proteins.
Addition of eRF1 to the phased ribosomes with subsequent irradiation causes the appearance of a new radioactive band in the upper part of the gels (Fig. 2, lanes 3). This cross-linking to eRF1 is specific for the mRNA analogs II–IV containing stop signals. mRNA analogs bearing sense codons (UGC, UCG, or UCA) with modified guanosines at the same positions cross-link to eRF1 negligibly (Fig. 2, lanes 4). The specificity of cross-linking of mRNA I to eRF1 has been shown earlier (Bulygin et al. 2002). The presence of eRF1 partially quenches cross-linking to protein S15 with mRNAs III and IV (Fig. 2, cf. lanes 2 and 3) as observed earlier for a 42-nucleotide (nt)-long mRNA with s4U in the first position of the stop codon at the A site (Chavatte et al. 2001, 2002). Similarly, with all mRNA analogs, eRF1 quenches cross-linking to the 18S rRNA, as observed earlier with s4U-containing mRNAs (Bulygin et al. 2005), although cross-linked 18S nucleotides are the same as had been found with these mRNA analogs without eRF1 (Styazhkina et al. 2003) and with the sense codon-containing mRNA analogs used in this study as controls (Graifer et al. 2004; data not shown).
Initial analysis of the cross-linking sites on the eRF1 protein by CNBr-induced cleavage
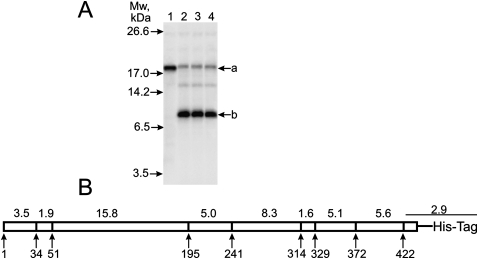
We have mapped eRF1 regions cross-linked to mRNA analogs using specific CNBr-induced cleavage of eRF1 after Met residues. There are eight internal Met residues in the wild-type human eRF1 (wt-eRF1). Therefore, nine fragments should be obtained after complete hydrolysis (Fig. 3B) if none of these methionines is cross-linked since modification of Met residues is well known to make them resistant to CNBr cleavage (e.g., see Megli et al. 1985).
FIGURE 3.
Mapping of the cross-linking site(s) within human eRF1. (A) Autoradiograms. SDS-PAGE analyses of the fragments resulted from the CNBr-induced cleavage of eRF1 cross-linked to end-labeled mRNA analogs. Lane numbers correspond to the numbers of mRNA analogs (Fig. 1). Positions of bands corresponding to molecular mass markers are given on the left. (B) Schematic representation of the CNBr-induced cleavage sites of human eRF1; calculated masses of the fragments in kilodaltons are indicated above the diagram.
After SDS-PAGE of the fragments resulting from CNBr cleavage of the eRF1 cross-linked to mRNA analogs, all lanes share a band labeled a, whose mass after subtracting the masses of the cross-linked mRNA analogs (ranging from 2.2 to 3.2 kDa) is ∼16 kDa (Fig. 3A). Evidently, this band corresponds to the eRF1 52–195 fragment (Fig. 3). For mRNA I, there is only a labeled fragment after cleavage of the cross-linked eRF1 (Fig. 3A, lane 1). However, mRNAs II–IV cross-link mainly to another site, as is evident from the appearance of the faster migrating b band.
Mapping of the cross-linking sites corresponding to the “a” band
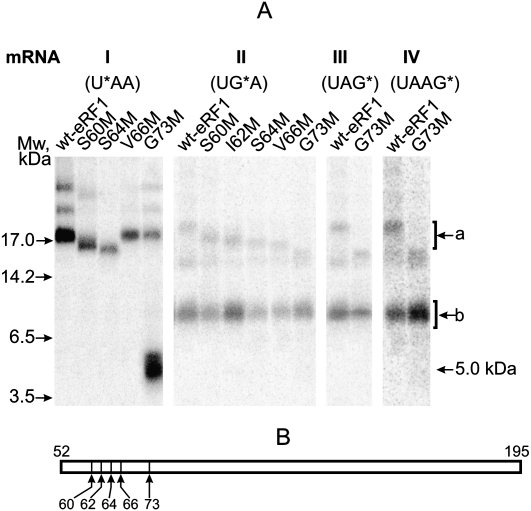
To refine the cross-link positions within the 52–195 fragment, CNBr-induced cleavage of various eRF1 mutants with single amino acid substitutions for Met in the region 60–73 (Fig. 4B) is used. The choice of this region is based on cross-linking of s4U in the first stop-codon position to tripeptide KSR (positions 63–65 of human eRF1) and indication that K63 is the most probable cross-linking site (Chavatte et al. 2002). With mRNA I, CNBr cleavage of the cross-linked S60M and S64M mutants results in formation of fragments slightly shorter than the fragment 52–195 of the wt-eRF1 (Fig. 4A). This implies that the cross-linking site is on the C-terminal side from the mutated positions. In contrast, with the G73M mutant, the major part of the label migrates as a 5-kDa band, which corresponds to the cross-linked fragment 52–73, whose calculated mass is 4.97 kDa (2.37 kDa of the fragment and 2.6 kDa of mRNA I). Thus, the modified uridine at position +4 cross-links to the 65–73 fragment, namely, to position 66 as the V66M mutant being CNBr resistant. Consequently, in the wt-eRF1, V66 might be a target for cross-linking with mRNA I.
FIGURE 4.
Mapping of the cross-linking site within the 60–73 region using eRF1 mutants. (A) Patterns of CNBr-induced cleavage fragments obtained with S60M, I62M, S64M, V66M, and G73M mutants cross-linked to mRNA analogs. Positions of bands corresponding to molecular mass markers are given on the left. (Upper bands) a; (lower bands) b. Bands above a are most likely products of incomplete CNBr-induced cleavage of cross-linked eRF1 by analogy with the results obtained with wt-eRF1 (Fig. 3); the identity of the minor band between a and b whose intensity varied from experiment to experiment is unknown. (B) Schematic representation of the cleavage sites of a 52–195 fragment corresponding to a band for the cross-linked wt-eRF1.
With mRNA II, the closer the mutation point is to the C terminus of eRF1, the faster the product corresponding to band a migrates (Fig. 4A). Therefore, the respective cross-linking site is located on the C-terminal side of all mutation points within the fragment 74–195. The mobility of the b band is not affected, indicating that the mutation points are outside of this sequence. The results with mRNAs III and IV are very similar to those obtained with mRNA II (Fig. 4A).
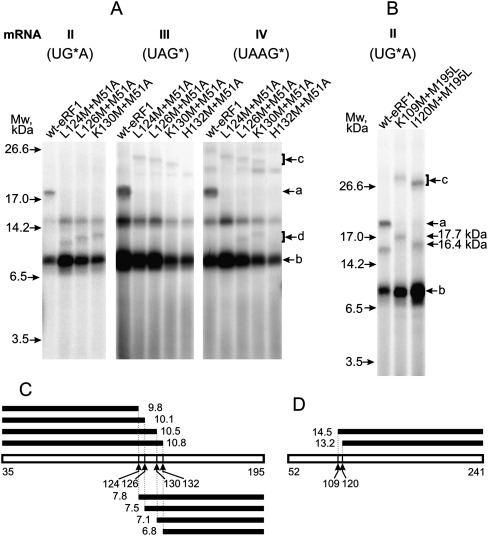
To map the cross-linking site inside the 74–195 fragment, four double mutants have been constructed. The L124M+M51A, L126M+M51A, and H132M+M51A mutants are active in an RF assay in the presence of all three stop codons, whereas the K130M+M51A mutant exhibits reduced RF activity toward UAG (data not shown). In all cases, the M51A substitution leads to abolishment of the labeled a fragment observed with the wt-eRF1 (Fig. 5A). With mRNA II, the d fragment appears instead of a, and the closer the mutation point is to the C terminus of eRF1, the slower do the products corresponding to the d bands migrate. The masses of these products, after subtracting the masses of the cross-linked mRNA analogs, range from 10 to 11 kDa. Therefore, the respective cross-linking site is on the N-terminal side of the mutation points at positions 124, 126, and 130 inside the 36–124 fragment of the eRF1 (Fig. 5C). Taking into account the data discussed in the preceding paragraph, the cross-linking site for mRNA II should be within positions 74–124.
FIGURE 5.
Mapping of the cross-linking sites within the regions 74–195 and 74–124 using double eRF1 mutants. Autoradiograms. (A,B) Patterns of CNBr-induced cleavage fragments are obtained with double mutants cross-linked to mRNA analogs. Positions of bands corresponding to molecular mass markers are given on the left. Bands a and b are marked by analogy with Figures 3A and 4. The identity of a band between a and d whose intensity varied from experiment to experiment (e.g., cf. data of Figs. 3–5) is unknown; bands c in A may be assigned to the product of incomplete CNBr-induced cleavage of cross-linked eRF1. Molecular masses of labeled products resulting from the CNBr-induced cleavage at 109 and 120 positions are given on the right. (C,D) Schematic representations of the cleavage sites of 35–195 and 52–241 fragments, respectively, together with calculated molecular masses (in kilodaltons) of the fragments resulting from the CNBr-induced cleavage.
With mRNA III, no bands are detectable that correspond to the expected products of the complete CNBr-induced cleavage of the mutants in the region 36–195 (Fig. 5A); they could be masked by the strong b band. At the same time, the c bands might correspond to products of incomplete cleavage of the cross-linked eRF1 mutants (except mutant H132M+M51A), in particular, to fragments undercleaved at M195 and M241 (in such cases, fragments about 190 amino acids long would be formed). With the wt-eRF1, longer undercleaved fragments were also detected in the upper part of the gel (not shown in Fig. 5A); in the case of undercleavage at M195 and M241, the fragment (52–314) would be longer than corresponding fragments observed with the mutants. The closer to the C-terminal side of eRF1 the mutation point is, the faster do products corresponding to the c bands migrate. Therefore, the cross-linking site is located on the C-terminal side of all mutation points within the 131–195 fragment of eRF1. The cross-linked H132M+M51A mutant is resistant to CNBr cleavage at position 132; therefore, Met132 is cross-linked. However, in wt-eRF1, cross-linking probably occurs with Phe131 since His (in position 132) is unable to cross-link with perfluorophenyl azides, whereas Phe is a good target (T Godovikova, pers. comm.).
Results obtained with mRNA IV (Fig. 5A) indicate the existence of two sites of cross-linking. One site is most likely Phe131 (by analogy with mRNA III), while existence of another site is apparent from the appearance of band d in the lanes L126M+M51A and K130M+M51A not observed with mRNA III (Fig. 5A). However, lane L124M+M51A lacks this band (Fig. 5A), probably due to the absence of a cross-linking site in the fragment 1–124. The presence of a d band in lane L126M+M51A implies that the cross-linking site is located in fragment 1–126, namely, at the positions 125 and/or 126. Cross-linking to position 126 is unlikely (in this case, eRF1 should be resistant to CNBr cleavage at this position); consequently, it should be Tyr125. However, Tyr is unable to cross-link with perfluorophenyl azides (T Godovikova, pers. comm.) and cannot be a target for mRNA IV. Therefore, the lack of band d in lane L124M+M51A might be explained by cross-linking to Met124 making the peptide resistant to CNBr cleavage after Met in this position. Thus, the most likely cross-linking involves Leu124 in the wt-eRF1.
To narrow the cross-linking site within the fragment 74–124 for mRNA II, double mutants M195L+K109M and M195L+I120M have been used (Fig. 5B). For these mutants, CNBr cleavage of the cross-linked eRF1 leads to formation of an 18-kDa or 16.5-kDa fragment instead of fragment a for wt-eRF1 (bands c at the very top of lanes with cross-linked mutants correspond to products of incomplete CNBr-induced cleavage). The 18-kDa and 16.5-kDa fragments, after subtracting the masses of the cross-linked mRNAs, fit the expected masses well if the cross-linking site is located on the C-terminal side of mutation points 109 and 120 (Fig. 5D). Consequently, the cross-linking site is inside fragment 121–241 and overlaps with fragment 74–124 by tetrapeptide 121–124. Thus, the cross-linking site is located within this tetrapeptide.
Mapping of the cross-linking sites corresponding to “b” bands
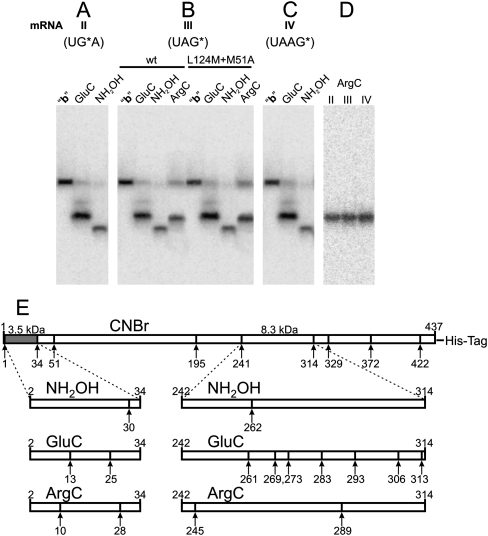
The masses of oligopeptides corresponding to bands b after subtracting the masses of the cross-linked mRNA analogs ranged from 5 to 6 kDa. However, one should take into account that the molecular mass of the major cross-linking product migrating as band b could be not exactly the same as expected from its electrophoretic mobility due to the contribution of the cross-linked oligonucleotide moiety to the rate of movement of the modified oligopeptide in the gel. This contribution could differ from that of an oligopeptide of the same mass as the oligonucleotide; the shorter the oligopeptide is, the more an effect of the cross-linked oligonucleotide is expected. Therefore, the product corresponding to bands b could be assigned in principle to any fragment resulting from CNBr-induced cleavage of the cross-linked eRF1 except for the long 52–195 fragment (Fig. 6E). Cross-linking within fragment 196–241 is excluded according to the results obtained with mutants K109M+M195L and I120M+M195L (see above) since CNBr-induced cleavage of these mutants produced the same bands b as wt-eRF1 (Fig. 5B). From other candidate fragments, only two, namely, 2–34 and 241–314, contain sites of cleavage with hydroxylamine. The results presented in Figure 6, A–C, show that with mRNAs II–IV, products corresponding to band b can be selectively cleaved with hydroxylamine; the same is observed with mutant forms of eRF1 (in Fig. 6B, the data with mutant L124M+M51A are presented as an example). Hence, the cross-linking site is located within one of two fragments, 242–314 or 2–34. The preferable candidate is the 2–34 fragment, whose electrophoretic mobility could be lower than expected because of the Y-like shape of the modified product, in which the oligonucleotide was cross-linked to the oligopeptide via its central part. To examine cross-linking in positions 2–34, we hydrolyzed the product corresponding to band b with hydroxylamine and endoproteases GluC and ArgC in parallel experiments. The results obtained with mRNA III (Fig. 6B) allow unambiguous assignment of band b to the abovementioned fragment. If the cross-linking site were within the 242–314 fragment, then GluC would produce a labeled fragment of a similar size (in the case of cross-linking in positions 242–262) to or much shorter (if the cross-link were in positions 263–314) than that obtained with hydroxylamine, which is inconsistent with the results presented in Figure 6B. Comparison of the results upon hydrolysis with endoprotease ArgC with the data obtained with GluC and hydroxylamine gives additional evidence against location of the cross-link in the 242–314 fragment (Fig. 6B).
FIGURE 6.
Mapping of the cross-linking site(s) within fragment b obtained as a result of the CNBr-induced cleavage of human eRF1 cross-linked to end-labeled mRNAs II–IV and of mutant L124M+M51A cross-linked to mRNA III. Autoradiograms. (A–D) SDS-PAGE analyses of the fragments resulting from the cleavage of the cross-linked fragment b with endoprotease GluC or ArgC or with hydroxylamine. (E) Schematic representation of the cleavage sites of human eRF1 with CNBr and the schemes of hydrolysis of fragments 2–34 and 242–314 with hydroxylamine and the endoproteases. Calculated masses of the fragments 2–34 (dark color) and 242–314 are indicated.
Our data make it possible to map the cross-linking site in the 2–34 fragment. Mutual arrangements of the bands obtained with hydroxylamine, GluC, and ArgC conform well only to a cross-link within positions 31–34 because if the cross-links were in positions 2–30, the fragment produced by hydroxylamine would be the longest, but that is not the case (Fig. 6B). Comparison of the data discussed with the results given in Figure 6A, C, and D leads to the same conclusion concerning cross-linking sites with mRNAs II and IV. Thus, with mRNAs II–IV, the major cross-linking site is located in eRF1 fragment 31–34. Moreover, our results make it possible to narrow this region to 31–33 since if M34 were modified, CNBr would not cleave at this residue, leading to formation of the 2–51 fragment. But in this case, with eRF1 mutants containing substitution M51A, the major cross-linking site would get into a large fragment (more than 120 amino acid residues long) in contrast to the short fragments corresponding to band b obtained with wt-eRF1. Taking into account that the same band b is observed with both wild-type and mutant eRF1s (Figs. 5A, 6B), one can conclude that M34 was not modified.
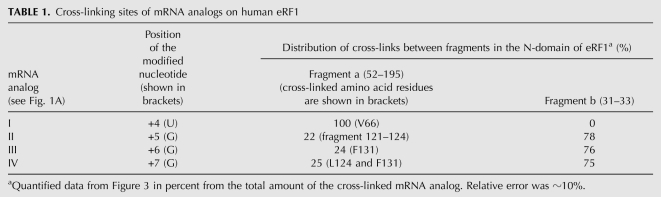
The data on cross-linking of mRNA analogs to eRF1 are summarized in Table 1. It is evident that the stop signal cross-reacts with three distinct sites in the N domain of eRF1, two of which—oligopeptide 121–131 (including the YxCxxxF motif) and the 31–33 fragment—are remote from the third one, V66 (in the region of the NIKS motif), in spatial (both in crystal and in solution) structure (Fig. 1C,D; Song et al. 2000; Kononenko et al. 2004). The major cross-linking site for all mRNAs with the exception of mRNA I was in eRF1 positions 31–33.
TABLE 1.
Cross-linking sites of mRNA analogs on human eRF1
A model for structure and location of eRF1 within the ribosome
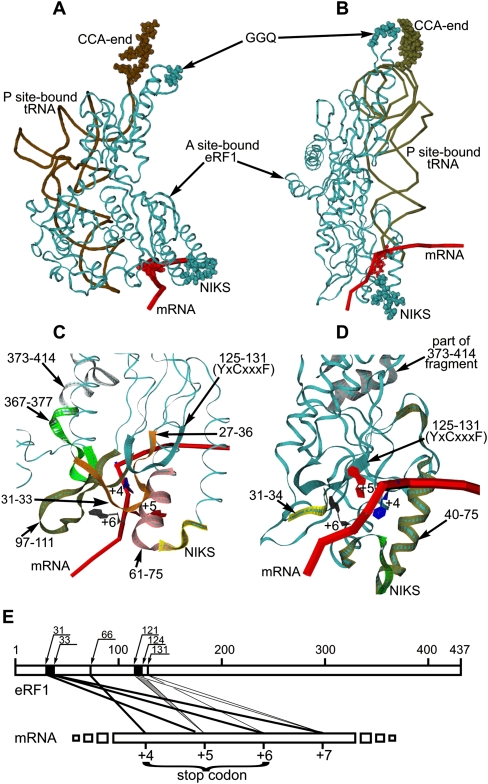
Although RF1/RF2 and eRF1 are generally considered as a tRNA mimic (see Fig. 1C; Nakamura et al. 2000), significant conformational changes in the eRF1 crystal structure are required to conform eRF1 to the shape of the tRNA inside the ribosomal A site. To model a termination complex containing mRNA, tRNA at the P site, and eRF1 at the A site, we have applied a multistage modeling procedure using a three-dimensional (3D) structure similarity method (Baker and Sali 2001), including three main stages: (1) selection of an optimal 3D template with known 3D structure, (2) adjusting the target eRF1 molecule to the template, and (3) structural optimization of eRF1 on the template taking into account the available structural data. As a template, we have used a 70S ternary complex derived from X-ray analysis comprising mRNA and two tRNAs at the P and A sites (PDB entry 2HGP) (Yusupova et al. 2006). We have assumed that arrangements of eRF1 and P-site tRNA on the 80S ribosome are structurally similar to those of the RF1 and P-site tRNA on the Thermus thermophilus ribosome, whose X-ray structure has been solved to 5.9 Å (PDB ID 2B64) (Petry et al. 2005). Using the P-site tRNA from the prokaryotic complex (2B64) mentioned as a reference molecule, we defined Cα positions of the GGQ tripeptide of RF1 relative to the CCA end of the P-site tRNA. Then, to build a ternary complex with eRF1, we replaced the A-site-bound tRNAPhe by eRF1 in the model 2HGP using these Cα positions as references on the placement of the corresponding atoms of eRF1 with respect to the P-site tRNA, keeping the 3D atomic coordinates of the mRNA and the P-site-bound tRNAPhe fixed. For this purpose, we created a model of eRF1_t0 (data not shown) whose conformation optimally fitted the A-site tRNA and was compatible with the ribosome binding pocket, but differed in its overall shape from the crystal eRF1 structure. As a result, the Y-shaped form of the eRF1 was transformed into a structure with a distance between the NIKS and GGQ motifs close to the distance between the anticodon triplet and the CCA end of the tRNA.
After that, docking of eRF1_t0 on the A-site codon was done. The main constraints for the docking were the Cα positions of the GGQ tripeptide of the eRF1 derived from the structure 2B64 (see above) toward the CCA end of the P-site-bound tRNA and the position of the NIKS motif with respect to the nucleotide in mRNA position +4 known from the cross-linking data (Chavatte et al. 2002). Two alternative models, M1 and M2, of the ternary complex of eRF1 with mRNA and the P-site-bound tRNA were generated (Fig. 7A–D). The M1 and M2 models differed in eRF1 positioning relative to the mRNA and, in particular, to the stop codon. In the M1 model (Fig. 7A,C), the mRNA passes through the cavity at the surface of the N-domain in a vicinity of the contact between the N and C domains, while in the M2 model (Fig. 7B,D), the mRNA passes through the YxCxxxF cavity. The amino acid residues of eRF1 proximal to the mRNA stop signal at the A site in these models are presented in Table 2. In the M2 model, in contrast to the M1 model, none of the amino acids of the C domain are close to the stop codon. Moreover, in the M1 model, the distance between the NIKS loop and the GGQ triplet is equal to 76 Å, while in the M2 model it corresponds to 87 Å. The distances between the stop codon and the GGQ triplet are similar in both models and are equal to 74 Å (exactly as between the 3′-terminal A and the anticodon triplet in tRNA) and to 78 Å in the M1 and M2 models, respectively (Fig. 7B,D).
FIGURE 7.
Models M1 (A) and M2 (B) of mutual arrangement of mRNA (red tube corresponding to phosphate backbone; positions +4 are in red beads), the P-site-bound tRNAPhe (brown tube; CCA-ends are in brown beads), and the A-site-bound eRF1_t0 (cyan ribbon; the NIKS and GGQ motifs are in cyan beads). Detailed views of the ribosomal decoding site in models M1 (C) and M2 (D) (mRNA positions +4, +5, and +6 are indicated; the eRF1 motifs given as colored ribbons are indicated by arrows). (E) Schematic representation of the cross-linking sites on eRF1 (numbers on the upper line indicate positions of eRF1 amino acids residues). Major sites are labeled with thick lines.
TABLE 2.
Mutual positioning of stop signal and amino acid residues of eRF1 in the ribosomal termination complex revealed from the molecular modeling
DISCUSSION
Stop signal at the ribosomal A site is located near three highly conserved GTx, NIKS, and YxCxxxF motifs of the N domain of eRF1
It is known from biochemical in vitro studies, genetic in vivo studies, and bioinformatics studies that the N domain of eRF1 is implicated in the decoding of stop codons within the ribosome (see the Introduction, and references therein), while its C domain interacts with the C domain of eRF3 (for review, see Kisselev et al. 2003; Cheng et al. 2009). However, the eRF1 lacking the C domain remains active in stop-codon recognition and peptidyl-tRNA hydrolysis in an in vitro termination assay (Frolova et al. 2000). This implies that the C domain of eRF1 is not essential for stop-codon recognition and explains why in our cross-linking experiments eRF3 has not been added to the ribosomal complex.
Earlier, it had been shown that after photoactivation, the analog of the first U of all stop codons, s4U, cross-reacts with the KSR tripeptide (positions 63–65 in human eRF1) (Chavatte et al. 2002). In this study, we have used another photoreactive group (Fig. 1A) that differs from s4U in having a wider radius of action (roughly 11–14 Å). Due to this difference, the region for cross-linking to eRF1 may be enlarged, expanding the cross-linking to V66 rather than to the KSR tripeptide (see Table 1). Summarizing the previous data (Chavatte et al. 2002; Frolova et al. 2002) and those obtained in this study (Table 1), we have estimated the human eRF1 area that is proximal to the first U of the stop codon as pentapeptide IKSRV (positions 62–66). From structural considerations, it has been suggested that to distinguish U from C, at least two amino acids are required (Saenger 1984). Therefore, probably two or three residues of this peptide may be involved in recognition of the first U.
The other cross-linking sites, oligopeptides in positions 31–33 and 121–132, are remote from the NIKS loop (positions 61–64) in the primary structure. The former represents a highly conserved GTx motif, and the latter includes the conserved YxCxxxF loop. These sites are close to each other in the spatial structure of free eRF1 (Fig. 1D; Song et al. 2000), and both of them turn out to be near the second and third stop-codon bases and even extend to the 3′-neighboring base, which affects the stop-codon decoding and is often considered as a part of the stop signal (Poole and Tate 2000). Our cross-linking data indicate that the YxCxxxF loop of eRF1 is involved in recognition of stop-codon purines and are consistent with the in vivo genetic data (Bertram et al. 2000) and the point mutagenesis data (Seit-Nebi et al. 2002; Kolosov et al. 2005). Region 31–33 of eRF1 had been earlier suggested to be involved in stop-codon recognition on the basis of various indirect and direct data (Bertram et al. 2000; Liang et al. 2005).
The NIKS loop is remote from the GTx and YxCxxxF motifs in the 3D structure of eRF1 (Fig. 1D), but all these fragments are able to cross-link to the stop codon within the ribosome. To make this possible, it has been assumed that stop-codon recognition is a two-step process (Chavatte et al. 2003). Possibly, the NIKS loop binds first to uridine of the stop codon and this binding induces a conformational change that brings the GTx motif and the YxCxxxF loop proximal to purines of the stop codon. As follows from the cross-linking patterns, the recognition requires a significant change in orientation of the NIKS toward both GTx and YxCxxxF fragments within the ribosome.
Due to two important peculiarities of the given methodology, one should avoid a misinterpretation of the cross-linking data since cross-linking of the mRNA analog to an amino acid residue does not necessarily imply that this residue is the closest and the single one in the vicinity of the respective nucleotide of the stop codon. First, some amino acid residues are highly reactive with the given photoactivatable group, while others are much less reactive or even nonreactive. For this reason, a nonreactive amino acid residue, although proximal to one of the nucleotides of the stop codon, will not be fixed by cross-linking and remains “invisible.” Second, due to the presence of the cross-linker, the most reactive amino acid residues are not necessarily the closest to mRNA analogs because the chemical reactivity can dominate over topography. But, in any case, cross-linked amino acid residues of the protein should be at the distance within 11–14 Å (radius of the reagent action in our case) of the stop codon or even contact it. Thus, there is no room for doubt that the oligopeptides in positions 31–33 and 121–131 are parts of eRF1 sites interacting with purines of the stop signals or are within 11–14 Å from them. The precise contributions of individual amino acid residues will be known when X-ray crystallography and cryo-EM techniques are successfully applied to study the eukaryotic ribosomal pre-termination and termination complexes.
Thus, it is evident that the recognition site of eRF1 is composed of three distinct parts proximal to the stop signal and to each other within the ribosomal A-site. This is in contrast to the sense-codon recognition mediated by interaction with the anticodon of the cognate tRNA. For this reason, the eRF1 possesses not a “protein anticodon,” but a 3D composite stop-codon recognition site, which, in principle, is compatible with a cavity-binding model (Bertram et al. 2000; Inagaki et al. 2002).
Concerning the decoding of the stop signal in prokaryotes, it was initially suggested that the second and third positions in UAG and UGA stop codons are recognized by PxT and SPF tripeptides in RF1 and RF2, respectively (Ito et al. 2000). But later X-ray studies on RFs bound to the 70S ribosome showed that the recognition is more complex and also involves other parts of the factors, for example, the backbone of α5 of RF2 that interacts with the second position of the stop codon (Korostelev et al. 2008; Weixlbaumer et al. 2008) and several other amino acid residues (whose identities depend on whether it is RF1 or RF2) that recognize the third position (Korostelev et al. 2008; Laurberg et al. 2008; Weixlbaumer et al. 2008). So, in prokaryotes, the 3D composite stop-codon recognition site is a well-documented feature, suggesting that the modes of stop-codon recognition in prokaryotes and eukaryotes are in general similar. But molecular details of interactions between the stop codon and class-1 RFs may be profoundly different, as one may anticipate from their distinct evolutionary origin (Frolova et al. 1994; Song et al. 2000) and differences in their translation termination mechanisms (Alkalaeva et al. 2006).
A significant conformational change of the A-site-bound eRF1
In solution, the eRF1 structure looks similar to the crystal structure (Fig. 1D; Song et al. 2000), as follows from small-angle X-ray scattering data (Kononenko et al. 2004). If eRF1 structure in a ribosome-bound state remains the same, the GTx and YxCxxxF motifs close to each other but remote from the NIKS loop could not interact with the same A-site-bound stop codon. However, our data (Table 1) are inconsistent with this prediction and imply that the distance between the stop codon and the abovementioned fragments of eRF1 are <14 Å. Evidently, significant conformational changes in eRF1 as a consequence of its binding to the “phased” ribosome should take place. This conclusion is consistent with the two-step model of eRF1 binding to the ribosome that is accompanied by conformational changes (Chavatte et al. 2003).
Rearrangements of the eRF1 structure if His residues are positively charged have been modeled by Ma and Nussinov (2004). Protonation of His residues modulates the domain interactions, making the “closed” conformation of eRF1 preferable to the “open” conformation typical for free eRF1 in the crystal or in solution. The transition of eRF1 to the “closed” conformation should be accompanied by a reduction of the distance between the NIKS and GGQ motifs of 20–30 Å. This “closed” conformation of eRF1 obtained by simulation in aqueous solution is similar to that in the eRF1_t0 model (the best A-site tRNA fit) created in this study for modeling the conformation of eRF1 in ribosomal termination complexes. The structure of eRF1 in the M1 and M2 models that correspond to these complexes slightly differs from that in the eRF1_t0 model, due to different relative positions of the stop codons and the requirement to keep the GGQ motif at the peptidyl transferase center. The distance between nucleotide +5 of a stop codon and the peptidyl transferase center remains similar to the value observed with an A-site-bound tRNA. Remarkably, the C domain in the M1 and M2 models is in close contact with the anticodon stem–loop of the P-site-bound tRNA. A tight eRF1•P-site-bound tRNA interaction may contribute to the conformational rearrangement of eRF1 and accelerate the peptidyl-tRNA hydrolysis triggered by eRF1.
The GTx and YxCxxxF motifs are within 14 Å of mRNA positions +5 and +6 in both M1 and M2 models, in agreement with our cross-linking data (see Fig. 7C; Table 2). In addition, it is worth mentioning that in the M1 model, the C4 and C5 atoms of uridine in position +4 are oriented in the direction of the NIKS loop. This orientation is favorable for cross-linking with the NIKS loop but not with the GTx and YxCxxxF motifs, consistent with the results of this study (Fig. 6E) and with the data of Chavatte et al. (2002). At the same time, the nitrogen atoms at N7 of the purines in positions +5 and +6 exhibit an orientation favorable for cross-linking to the GTx and YxCxxxF motifs but not to the NIKS loop. Again, these data are in complete agreement with our results on cross-linking (Table 1).
It is worth mentioning that in the M1 model (Fig. 7B,D), the second and third nucleotides become closer to several parts of the C domain in contrast to the M2 model. Remarkably, fragments 281–305 and 398–415 of the C domain of eRF1 crucial for binding to eRF3 (Merkulova et al. 1999; Cheng et al. 2009) are fully accessible in the M1 model (data not shown). This implies that eRF1 in the M1 conformation retains its ability to bind eRF3 within the ribosome, a prerequisite for the cooperative action of both factors during translation termination (e.g., see Alkalaeva et al. 2006; Cheng et al. 2009). Taking all these into account, we suggest that the M1 model better fits our cross-linking data and the conformation of eRF1 in the 80S ribosome termination complex than the M2 model.
Thus, molecular modeling strongly supports the idea that eRF1 undergoes a large conformational rearrangement (both mechanical and energetic) that leads to a significant overall molecular shape similarity between eRF1 and tRNA and brings part of the C domain closer to the NIKS, GTx, and YxCxxxF motifs near the stop codon. Why does the C domain of the ribosome-bound eRF1 move closer to the stop signal? One possibility is that the C domain interacts with a sugar-phosphate mRNA backbone. This interaction may contribute to stabilization of the mRNA•eRF1 complex essential to ensure a high fidelity of stop-codon recognition. Alternatively, the ribosome forces the eRF1 molecule to alter its domain orientation to fit the A site, and this adaptation may be achieved through binding of eRF1 to 18S rRNA and protein S15, whose cross-linking to a derivatized stop signal is quenched by eRF1.
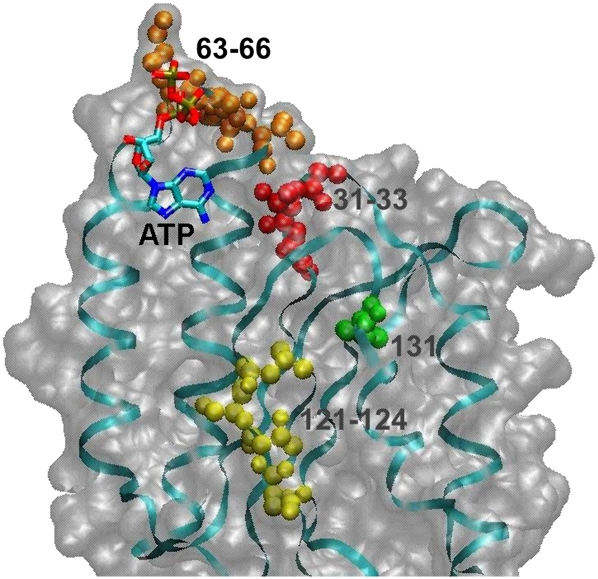
It is noteworthy to compare our cross-linking results and the data on modeling with a recent X-ray study of the crystallized eRF1•eRF3 complex containing an ATP molecule (Cheng et al. 2009). In this report it was proposed that ATP binding to the N domain of eRF1 could mimic the base interaction with the eRF1 decoding site. The adenine base was found to make contacts with hydrophobic residues Ala59, Ile62, Val71, and Ile75 from α2–loop–α3, and Ile35 from the β1 sheet, and it was suggested that the base could form hydrogen bonds with Thr32, Cys127, Gly55, and Tyr125. Further systematic mutational analysis of residues of human eRF1 involved in ATP binding revealed that mutations at Thr32, Ile35, Glu55, Val71, and Cys127 strongly changed the specificity of stop-codon recognition, suggesting that these residues are crucial for stop recognition and/or discrimination. Mapping our cross-linking results to the X-ray-derived structure of an eRF1 fragment containing the ATP binding site (Fig. 8) shows that these results conform excellently to the structure. Our major cross-linking site, the GTx fragment in positions 31–33, contains T32, which was suggested to make direct contacts with the adenine base. Residues 121–124 and 131 from the minor cross-linking site in the YxCxxxF loop are somewhat more remote than residues 31–33 from the adenine base (that could correlate with lower cross-linking extent of mRNA analogs to the YxCxxxF motif), although being within the length of the cross-linker used (14 Å). Taking all these together, one can conclude that the conserved GTx motif in positions 31–33 of human eRF1 (most probably, T32) plays a key role in the recognition of the purines of the stop codon.
FIGURE 8.
A fragment of the structure of human eRF1 bound to eRF3 (adapted from Cheng et al. 2009; PDB accession number 3E1Y) containing ATP at a putative decoding site on the N domain of eRF1. The sites of mRNA analogs cross-linking to the eRF1 found in this study are indicated.
The major conformational changes of eRF1 found in this study by modeling are similar to those observed by Cheng et al. (2009) with eRF1 bound to eRF3; in both cases, alteration of the mutual orientation of the domains of eRF1 took place, leading to formation of a bent conformation, in which the distance between the GGQ and NIKS motifs is <80 Å. Remarkably, in the case of binding to eRF3 (Cheng et al. 2009), the alteration of the eRF1 structure made its C domain closer to the N domain in analogy to that in the structure found by the modeling in this study. Thus, our results are in good agreement with the data by Cheng et al. (2009), despite the limitations of their direct comparison since the discussed X-ray data were obtained without ribosomes, tRNA, and mRNA. It is reasonable to suggest that the flexible eRF1 conformation is locked to its “tRNA-like” state by binding to eRF3 as well as to the ribosome.
MATERIALS AND METHODS
tRNAPhe (1300 pmol/A260 unit) was a kind gift from V. Katunin (Konstantinov's St. Petersburg Institute of Nuclear Physics). Isolation of the 40S and 60S ribosomal subunits from unfrozen human placenta and their association in 80S ribosomes were performed as described earlier (Matasova et al. 1991).
mRNA analogs
Oligoribonucleotides and their photoactivatable derivatives labeled with 32P at the 5′-termini were synthesized as previously described (Graifer et al. 2004; Demeshkina et al. 2005).
Cloning and mutagenesis of human eRF1
The plasmid pERF4b encoding human eRF1 (Seit-Nebi et al. 2001; Frolova et al. 2002) was used for site-directed mutagenesis. Generation of eRF1 mutants in positions 60–73 was as described earlier (Chavatte et al. 2002). Generation of M195L and M51A mutants by site-directed mutagenesis was performed according to the PCR-based “megaprimer” method as described (Frolova et al. 2002; Kolosov et al. 2005). The M51A and M195L mutants were used as templates to generate the corresponding double mutants (the primers used for generation of eRF1 mutants are available upon request at frolova@eimb.relarn.ru). The cloned DNAs were verified by sequencing, and appropriate clones were used for expression of mutant eRF1s.
Expression and purification of human eRF1
The wt-eRF1 and its mutants with the C-terminal His-tag were expressed in Escherichia coli and purified by affinity chromatography as described (Frolova et al. 2000). The eRF1 activity was measured in vitro as described (Caskey et al. 1974; Frolova et al. 1994).
Ribosomal complexes and cross-linking procedures
Complexes of 80S ribosomes (5.0 × 10−7 M) with tRNAPhe (2.5 × 10−6 M) and mRNA analogs (3.5 × 10−6 M) were obtained by incubation of these components for 40 min in buffer A (20 mM HEPES-KOH at pH 7.5, 100 mM NH4Cl, 4 mM MgCl2, 0.2 mM EDTA, 0.6 mM spermidine, 0.8 mM spermine) at room temperature. The reaction mixtures for irradiation contained 15 pmol of 80S ribosomes. Then wt-eRF1 or mutant eRF1 was added where specified in a sevenfold molar excess each over the 80S ribosomes, and the reaction mixtures were further incubated for 40 min at room temperature. Irradiation of complexes was carried out as described (Graifer et al. 2004). Reactions were stopped with 1/30 (v/v) of 5% 2-mercaptoethanol (ME) and purified by centrifugation in sucrose gradient (15%–30%) containing buffer A with 1 mM ME as described (Bulygin et al. 2002).
eRF1 mapping
CNBr-induced cleavage
Fractions of sucrose gradient containing 80S ribosomes were precipitated with trichloroacetic acid, the pellets were dissolved in 50 μL of buffer containing 2% SDS, 1% ME, 20% glycerol, 60 mM Tris-HCl (pH 7.5), and 0.05% bromphenol blue, incubated 5 min at 90°C, and resolved by 15% SDS-PAGE. The gel was dried and autoradiographed. The band corresponding to wt-eRF1 or its mutants cross-linked to labeled mRNA analog was excised. The modified factor was treated in the gel slice with 0.25 M CNBr in 70% formic acid for 3 h at room temperature (typically, 0.1 mL of the solution was applied). The solution was separated from the gel by centrifugation; the supernatant was dried in vacuum, diluted in equilibration dye buffer, and analyzed by 16.5% Tris-tricine SDS-PAGE in parallel with markers (1.1–26.6 kDa; Sigma). The dried gels were analyzed by using a PhosphorImager (Bio-Rad FX Pro Plus MultiImager). Masses of eRF1 oligopeptides cross-linked to mRNA analogs were calculated from the calibration curves obtained from mobilities of the markers in the same gel. The average values of masses of cross-linked oligopeptides were calculated based on the three to four independent cross-linking experiments.
To carry out selective cleavage of eRF1 fragments obtained as a result of CNBr-induced digestion, the respective bands were excised from the gels and the gel slices were incubated with 50–100 μL of a solution containing hydroxylamine (puriss p.a.), endoprotease GluC, or ArgC (Roche Biochemicals). Cleavage with endoproteinase GluC was carried out in 100 mM Tris-HCl buffer (pH 7.5) containing 0.02 μg/μL of the enzyme and 0.1% SDS by incubating the mixture overnight at room temperature. Hydrolysis with endoproteinase ArgC was carried out in the same buffer containing 0.005 μg/μL enzyme and 10 mM CaCl2 by incubating the mixture overnight at 37°C. The reaction with hydroxylamine was carried out as described (Changchien and Craven 1986) but with several modifications. A gel slice was incubated in a freshly prepared solution containing 2 M NH2OH•HCl, 2 M guanidinium chloride, and 0.2 M KOH (pH 9.0) for 6 h at 45°C. Then the reaction was supplied with 20 μL of 85% formic acid and 3 μL of a solution of total 60S ribosomal protein (10 μg/μL) as a carrier, diluted with 3 volumes of water, and the polypeptides were precipitated with 4 volumes of cold acetone (incubation for 16–20 h at −20°C). The eRF1 fragments resulting from the hydrolysis were resolved by 16.5% Tris-tricine SDS-PAGE.
Modeling of the optimal fit of eRF1 with the A-site-bound tRNA
Adaptation of the eRF1 conformation to the ribosomal A-site was achieved via alteration of the domain packing for optimal fit of its N and M domains with the A-site-bound tRNAPhe molecular surface, under the two constraints of positional equivalence between the invariant GGQ tripeptide and the universal CCA end, and between the anticodon loop of tRNA and the NIKS motif of eRF1. Volume exclusion interactions of eRF1 with mRNA and the P-site-bound tRNAPhe were also taken into account.
The human eRF1 structure (PDB accession number 1DT9) contained no data on side-chain atoms for 22 amino acid residues, and atomic coordinates for positions 423–437 were also absent. For this reason, only residues 1–422 were used in the structure modeling. The side-chain atom coordinates were restored and optimized using molecular dynamics. The conformational rearrangement of the eRF1 was simulated by applying a procedure of a “targeted fit” as a slow overall drift of the M domain (residues 148–294) and C domain (residues 295–422) under external “target fit” forces acting on atoms of these domains. All-atom molecular dynamics coupled with a simulated annealing protocol were used to obtain the optimal path for conformational changes of eRF1. Calculations were done using the program bison (Vorobjev 2005) with the amber94 force field (Cornell et al. 1995) and the implicit solvation model AGBNP (Gallicchio and Levy 2004). The protocol of eRF1 conformational rearrangement was the following: (1) The Cα atom of the Ser64 was targeted to the C1′ atom of A35 of tRNAPhe. (2) The Cα atom of the GGQ tripeptide was targeted to its respective target Cα atoms derived from the structure of the RF1–tRNA complex (Petry et al. 2005). (3) eRF1 was optimally fitted on the tRNA volume via forced molecular dynamics to simulate the annealing under external forces of the “target fit” potential (see Fig. 7A) to get a domain movement. (4) eRF1 conformations were equilibrated with external forces switched off and soft harmonic constraints for positions of all atoms switched on; the best fit score was found by iteration. The volume fit between eRF1 and tRNA was calculated as a best fit between the projections of atomic positions on a rectangular cell box. As a result, a tRNA-like conformation of eRF1 was achieved and designated as “eRF1_t0.” The A-site-bound tRNAPhe of the ternary complex of mRNA with the P-site- and A-site-bound tRNAs from the X-ray structure of the 70S ribosomal complex (PDB entry 2HGP) was replaced by the eRF1_t0 to get an initial “low-resolution” structure of eRF1 complexed with mRNA and the P-site-bound tRNA. The resolution (or positional uncertainty) of eRF1 relative to the stop codon at this stage was ∼25 Å.
Refinement of the structure of the eRF1•mRNA•P-site-bound tRNA complex was done by docking eRF1_t0 on the mRNA stop codon in the complex of mRNA with the P-site-bound tRNA, taking into account the constraints based on the data on direct contact of Lys63 of eRF1 with the first nucleotide of the stop codon (Chavatte et al. 2002) and the target Cα positions of the GGQ motif derived from the structure of the RF1-tRNA complex (Petry et al. 2005). Docking of eRF1_t0 on the fixed structure of mRNA bound with the P-site tRNA was done by moving the eRF1_t0 structure over the points of a rectangular grid of size 25 Å with a 5 Å cell size. For each grid position, a structure was refined via simulated annealing molecular dynamics of eRF1_t0 atoms and of the stop-codon triplet. The eRF1_t0 conformation was optimized assuming flexible N-domain, M-domain, and C-domain packing with flexible side chains and a flexible NIKS loop. The Cα atom of Gln185 in the GGQ tripeptide was positionally restrained to the position of an initial “low-resolution” eRF1_t0 structure to keep the GGQ triplet position at the peptidyl transferase center. The top codon triplet was flexible, while mRNA nucleotides (in positions from −3 to +3 and from +7 to +9) were fixed.
ACKNOWLEDGMENTS
We thank Alim Seit-Nebi for participation in experiments on the point mutagenesis of eRF1. This work was supported by the Russian Foundation for Basic Research (grants 08-04-00508 to G.G.K. and 08-04-00375a to L.Yu.F.), by grants from the Presidium of Russian Academy of Sciences (Program on Molecular and Cell Biology) to G.G.K. and to L.Yu.F., and by grant HШ-395.2008.4 for Supporting the Leading Russian Schools in Science (to L.Yu.F.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2066910.
REFERENCES
- Alkalaeva EZ, Pisarev AV, Frolova LYu, Kisselev LL, Pestova TV 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Baker D, Sali A 2001. Protein structure prediction and structural genomics. Science 294: 93–96 [DOI] [PubMed] [Google Scholar]
- Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I 2000. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6: 1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham RH, Grentzmann G, Kisselev L 1997. Polypeptide chain release factors. Mol Microbiol 24: 449–456 [DOI] [PubMed] [Google Scholar]
- Bulygin KN, Repkova MN, Ven'yaminova AG, Graifer DM, Karpova GG, Frolova LY, Kisselev LL 2002. Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes. FEBS Lett 514: 96–101 [DOI] [PubMed] [Google Scholar]
- Bulygin K, Chavatte L, Frolova L, Karpova G, Favre A 2005. The first position of a codon placed in the A site of the human 80S ribosome contacts nucleotide C1696 of the 18S rRNA as well as proteins S2, S3, S3a, S30 and S15. Biochemistry 44: 2153–2162 [DOI] [PubMed] [Google Scholar]
- Caskey C, Beaudet AL, Tate WP 1974. Mammalian release factor: In vitro assay and purification. Methods Enzymol 30: 293–303 [DOI] [PubMed] [Google Scholar]
- Chandramouli P, Topf M, Menetret J-F, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW 2008. Structure of the mammalian 80S ribosome at 8.7 Å resolution. Structure 16: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changchien LM, Craven GR 1986. The use of hydroxylamine cleavage to produce a fragment of ribosomal protein S4 which retains the capacity to specifically bind 16S ribosomal RNA. Nucleic Acids Res 14: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Frolova L, Kisselev L, Favre A 2001. The polypeptide chain release factor eRF1 specifically contacts the s4UGA stop codon located in the A-site of eukaryotic ribosomes. Eur J Biochem 268: 2896–2904 [DOI] [PubMed] [Google Scholar]
- Chavatte L, Seit-Nebi A, Dubovaya V, Favre A 2002. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J 21: 5302–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Frolova L, Laugâa P, Kisselev L, Favre A 2003. Stop codons and UGG promote efficient binding of the human polypeptide release factor eRF1 to the eukaryotic ribosomal A-site. J Mol Biol 331: 745–758 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, et al. 2009. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev 23: 1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA 1995. A second generation force field for simulation of proteins, nucleic acids and organic molecules. J Am Chem Soc 117: 5179–5197 [Google Scholar]
- Demeshkina NA, Styazhkina VA, Bulygin KN, Repkova MN, Ven'yaminova AG, Karpova GG 2005. Arrangement of mRNA on the human ribosome: Environment of mRNA nucleotide 3′ of the A-site-bound codon. Bioorg. Khem. (Mosk.) 31: 295–302 [DOI] [PubMed] [Google Scholar]
- Ebihara K, Nakamura Y 1999. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA 5: 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eurwilaichitr L, Graves FM, Stansfield I, Tuite MF 1999. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol Microbiol 32: 485–496 [DOI] [PubMed] [Google Scholar]
- Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling K, Thompson SR, Pestova T, Bedwell DM 2008. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell 30: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni A-L, Celis JE, Philippe M, et al. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703 [DOI] [PubMed] [Google Scholar]
- Frolova L, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev L 1999. Mutations in the highly conserved GGQ motif of class-1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5: 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Merkulova TI, Kisselev LL 2000. Translation termination in eukaryotes: Polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA 6: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Seit-Nebi A, Kisselev L 2002. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA 8: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicchio E, Levy RM 2004. AGBNP: An analytic implicit solvent model suitable for molecular dynamics simulations and high-resolution modeling. J Comput Chem 25: 479–499 [DOI] [PubMed] [Google Scholar]
- Graifer D, Molotkov M, Styazhkina V, Demeshkina N, Bulygin K, Eremina A, Ivanov A, Laletina E, Ven'yaminova A, Karpova G 2004. Variable and conserved elements of human ribosomes surrounding the mRNA at the decoding and upstream sites. Nucleic Acids Res 32: 3282–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y, Blouin C, Doolittle WF, Roger AJ 2002. Convergence and constraint in eukaryotic release factor (eRF1) domain 1: The evolution of stop codon specificity. Nucleic Acids Res 32: 532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y 2000. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403: 680–684 [DOI] [PubMed] [Google Scholar]
- Ito K, Frolova L, Seit-Nebi A, Karamyshev A, Kisselev L, Nakamura Y 2002. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within the domain 1. Proc Natl Acad Sci 99: 8494–8499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev LL, Buckingham RH 2000. Translation termination comes of age. Trends Biochem Sci 25: 561–566 [DOI] [PubMed] [Google Scholar]
- Kisselev LL, Oparina N Yu, Frolova L Yu 2000. Class-1 polypeptide chain release factors are structurally and functionally similar to suppressor tRNAs and comprise different structural-functional families of prokaryotic/mitochondrial and eukaryotic/archaebacterial factors. Mol Biol (Mosk) 34: 427–442 [PubMed] [Google Scholar]
- Kisselev LL, Ehrenberg M, Frolova LYu 2003. Termination of translation: Interplay of mRNA, rRNAs and release factors. EMBO J 22: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M 2003. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421: 90–94 [DOI] [PubMed] [Google Scholar]
- Kolosov P, Frolova L, Seit-Nebi A, Dubovaya V, Kononenko A, Oparina N, Justesen J, Efimov A, Kisselev L 2005. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res 33: 6418–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononenko AV, Dembo KA, Kisselev LL, Volkov VV 2004. Molecular morphology of eukaryotic class I translation termination factor eRF1 in solution. Mol Biol (Mosk) 38: 303–311 [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci 105: 19684–19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF 2008. Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857 [DOI] [PubMed] [Google Scholar]
- Liang H, Wong JY, Bao Q, Cavalcanti ARO, Landweber LF 2005. Decoding region: Analysis of eukaryotic release factor (eRF1) stop codon binding residues. J Mol Evol 60: 337–344 [DOI] [PubMed] [Google Scholar]
- Ma B, Nussinov R 2004. Release factors eRF1 and RF2: A universal mechanism controls the large conformational changes. J Biol Chem 279: 53875–53885 [DOI] [PubMed] [Google Scholar]
- Matasova NB, Myltseva SV, Zenkova MA, Graifer DM, Vladimirov SN, Karpova GG 1991. Isolation of ribosomal subunits containing intact rRNA from human placenta. Estimation of functional activity of 80S ribosomes. Anal Biochem 198: 219–223 [DOI] [PubMed] [Google Scholar]
- Megli FM, Van Loon D, Barbuti AA, Quagliarello E, Wirtz KW 1985. Chemical modification of methionine residues of the phosphatidylcholine transfer protein from bovine liver. A spin-label study. Eur J Biochem 149: 585–590 [DOI] [PubMed] [Google Scholar]
- Merkulova TI, Frolova LY, Lazar M, Camonis J, Kisselev LL 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett 443: 41–47 [DOI] [PubMed] [Google Scholar]
- Molotkov MV, Graifer DM, Popugaeva EA, Bulygin KN, Meschaninova MI, Ven'yaminova AG, Karpova GG 2006. mRNA 3′ of the A site-bound codon is located close to protein S3 on the human 80S ribosome. RNA Biol 3: 122–129 [DOI] [PubMed] [Google Scholar]
- Mora L, Heurgue-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH 2003. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol 47: 267–275 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito K 2003. Making sense of mimic in translation termination. Trends Biochem Sci 28: 99–105 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito K, Ehrenberg M 2000. Mimicry grasps reality in translation termination. Cell 101: 349–352 [DOI] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FV IV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V 2005. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Poole ES, Tate W 2000. Release factors and their role as decoding proteins: Specificity and fidelity for termination of protein synthesis. Biochim Biophys Acta 1493: 1–11 [DOI] [PubMed] [Google Scholar]
- Poole ES, Askarian-Amiri ME, Major LL, McCaughan KK, Scarlett DJ, Wilson DN, Tate WP 2003. Molecular mimicry in the decoding of translational stop signals. Prog Nucleic Acid Res Mol Biol 74: 83–121 [DOI] [PubMed] [Google Scholar]
- Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J 2003. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421: 87–90 [DOI] [PubMed] [Google Scholar]
- Saenger W 1984. Principles of nucleic acid structure (ed. Cantor CR), Chap. 18 Springer-Verlag, New York [Google Scholar]
- Scarlett DJ, McCaughan KK, Wilson DN, Tate WP 2003. Mapping functionally important motifs SPF and GGQ of the decoding release factor RF2 to the Escherichia coli ribosome by hydroxyl radical footprinting. Implications for macromolecular mimicry and structural changes in RF2. J Biol Chem 278: 15095–15104 [DOI] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, Kisselev L 2001. Class-1 translation termination factors: Invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res 29: 3982–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Kisselev L 2002. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep 3: 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim S-H 2004. Structural analyses of peptide release factor 1 from Thermogota maririma reveal domain flexibility required for its interaction with the ribosomes. J Mol Biol 341: 227–239 [DOI] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–1515 [DOI] [PubMed] [Google Scholar]
- Song H, Mugnier P, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D 2000. The crystal structure of human eukaryotic release factors eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J 2001. Structure of the 80S ribosome from Saccaromyces serevisiae—tRNA-ribosome and subunit–subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Styazhkina VA, Molotkov MV, Demeshkina NA, Bulygin KN, Graifer DM, Meshchaninova MI, Repkova MN, Ven'yaminova AG, Karpova GG 2003. Arrangement of the sense and stop codons of the template in the A site of the human ribosome as inferred from crosslinking with oligonucleotide derivatives. Mol Biol (Mosk) 37: 866–872 [PubMed] [Google Scholar]
- Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M 2001. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell 8: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Vorobjev YN 2005. Study of mechanism of interaction of oligonucleotides with the 3′-terminal region of tRNAPhe by computer modeling. Mol Biol (Mosk) 39: 777–784 [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M 2006. Structural basis for messenger RNA movement on the ribosome. Nature 444: 391–394 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M 2001. A post-termination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124 [DOI] [PubMed] [Google Scholar]