Abstract
Biogenic polyamines are found to modulate protein synthesis at different levels. This effect may be explained by the ability of polyamines to bind and influence the secondary structure of tRNA, mRNA, and rRNA. We report the interaction between tRNA and the three biogenic polyamines putrescine, spermidine, spermine, and cobalt(III)hexamine at physiological conditions, using FTIR spectroscopy, capillary electrophoresis, and molecular modeling. The results indicated that tRNA was stabilized at low biogenic polyamine concentration, as a consequence of polyamine interaction with the backbone phosphate group. The main tRNA reactive sites for biogenic polyamine at low concentration were guanine-N7/O6, uracil-O2/O4, adenine-N3, and 2′OH of the ribose. At high polyamine concentration, the interaction involves guanine-N7/O6, adenine-N7, uracil-O2 reactive sites, and the backbone phosphate group. The participation of the polycation primary amino group, in the interaction and the presence of the hydrophobic contact, are also shown. The binding affinity of biogenic polyamine to tRNA molecule was in the order of spermine > spermidine > putrescine with KSpm = 8.7 × 105 M−1, KSpd = 6.1 × 105 M−1, and KPut = 1.0 × 105 M−1, which correlates with their positively charged amino group content. Hill analysis showed positive cooperativity for the biogenic polyamines and negative cooperativity for cobalt-hexamine. Cobalt(III)hexamine contains high- and low-affinity sites in tRNA with K1 = 3.2 × 105 M−1 and K2 = 1.7 × 105 M−1, that have been attributed to the interactions with guanine-N7 sites and the backbone PO2 group, respectively. This mechanism of tRNA binding could explain the condensation phenomenon observed at high Co(III) content, as previously shown in the Co(III)–DNA complexes.
Keywords: tRNA, polyamines, preferential binding site, binding constant, hyperchromism, hypochromism, condensation, tRNA stability, FTIR spectroscopy, affinity capillary electrophoresis (ACE)
INTRODUCTION
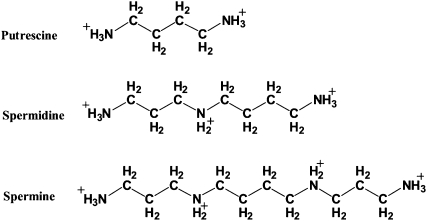
The biogenic polyamines putrescine, spermidine, and spermine (Structure 1) are small aliphatic polycationic compounds present in almost all living organisms. They are essential for normal cell growth, and their concentrations increase with cell proliferation (Tabor and Tabor 1984; Cohen 1998; Igarashi and Kashiwagi 2000; Thomas and Thomas 2001; Childs et al. 2003). In mammalian cells, they are found in millimolar concentrations (Tabor and Tabor 1984; Cohen 1998; Igarashi and Kashiwagi 2000). Of the many hypotheses advanced to explain their biological effects, the most important one concerns their interaction with nucleic acids (Tabor and Tabor 1984; Feuerstein et al. 1991; Marton and Pegg 1995; Cohen 1998; Thomas and Thomas 2001). However, it seems that a large fraction of polyamines exists in polyamine–RNA adducts, and the major part of their cellular function may be explained through structural changes of RNA by polyamines (Watanabe et al. 1991; Cohen 1998; Igarashi and Kashiwagi 2000). On the other hand, the potential effectiveness of polyamine analogs, as antiproliferative agents against many tumor cell lines and infectious diseases, provides evidence for nucleic acid interaction with the biogenic polyamines (Li et al. 1996; Frydman and Valasinas 1999; Fernandez et al. 2000; Bacchi et al. 2002; Thomas et al. 2002; Frydman et al. 2003; Thomas and Thomas 2003).
STRUCTURE 1.
Chemical structures of biogenic polyamines putrescine, spermidine, and spermine.
Interactions of polyamines with tRNA are less understood than they are with DNA, and thus, more specific theoretical and experimental studies of polyamine–tRNA bindings are required. The interactions of both spermine and spermidine with tRNA have been studied using solutions of 1H-NMR (Frydman et al. 1996, 1999), 13C-NMR (Frydman et al. 1990), and 15N-NMR (Frydman et al. 1992; Fernandez et al. 1994). The authors observed that in all polyamines, internal NH2+ groups bind to tRNA more strongly than the terminal NH3+ groups. They suggest that factors other than the electrostatic interaction are responsible for this phenomenon, since the primary amines (–NH3+) possess a higher density of positive charge than the secondary ones (–NH2+–). The 15N-NMR experiments suggest that the specificity by which the nitrogen atoms in polyamines bind to tRNA is a consequence of the various hydrogen bonds that can be established between both tRNA and the polyamines (Frydman et al. 1992; Fernandez et al. 1994). Interestingly, other researchers (Bibillo et al. 1999) found that biogenic polyamines are required for the non-enzymatic cleavage of tRNA and oligoribonucleotides at UA and CA phosphodiester bonds. This suggests that at least two linked protonated amino groups are necessary to mediate hydrolysis. They hypothesized that one ammonium group binds to the nucleotide, while the second one participates in the activation of the labile phosphodiester bond (Quigley et al. 1978). However, in spite of these extensive studies, the binding sites of the biogenic polyamines on tRNA are not still clearly established.
A spermine–tRNAPhe complex has been crystallized and its structure resolved at 2.5 Å resolution (Quigley et al. 1978; Shi and Moore 2000). The polyamine binds at two major sites—the first in the major groove at the end of the anticodon stem and the second near the variable loop around phosphate 10 at a turn, in the RNA backbone. However, 1H-NMR studies found that spermine and related polyamines bind to additional sites of tRNA, in particular at the junction of the TψC- and D-loops, that were not seen in the crystal structure (Frydman et al. 1996, 1999). Binding of biogenic polyamines to tRNA has been shown to be relevant for optimal translational accuracy and efficiency (Igarashi and Kashiwagi 2000). They are found to modulate protein synthesis (Shimogori et al. 1996; Igarashi and Kashiwagi 2006) and influence the binding of deacylated and acylated tRNA to the ribosomes (Agrawal et al. 1999; Yoshida et al. 1999). These effects may be explained by the ability of polyamines to bind and influence secondary structure of tRNA, mRNA, and rRNA (Shimogori et al. 1996; Agrawal et al. 1999; Yoshida et al. 1999; Igarashi and Kashiwagi 2000, 2006).
Recently, we have published the results of infrared spectroscopy, capillary electrophoresis, and molecular modeling on the interaction between DNA and biogenic polyamines and cobalt(III)hexamine (Ahmed Ouameur and Tajmir-Riahi 2004). In our report, several binding models were proposed for biogenic polyamines, and their biological significances have been discussed. The aim of this work was to extend our knowledge pertaining to the effects of the polyamines on nucleic acids by studying the interactions of biogenic polyamines and cobalt(III)hexamine with tRNA, using capillary electrophoresis and Fourier transform infrared spectroscopy as well as molecular modeling, in order to establish a correlation between our results and the significance of the polyamine–tRNA complex formation in vivo. Cobalt(III)hexamine was included in this study (as a control model) because it is an inorganic polyamine cation known to bind RNA at the guanine N7 and at the phosphate oxygen atoms, in the major groove of the GAAA tetraloop (Kieft and Tinoco 1997; Rüdisser and Tinoco 1999). It has been shown that cobalt(III)hexamine induces DNA condensation and conformational transitions (Ahmed Ouameur and Tajmir-Riahi 2004).
RESULTS
Interaction of polyamines with tRNA studied by FTIR spectroscopy
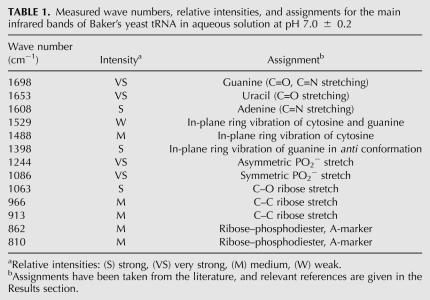
The infrared spectra of tRNA solutions (Figs. 1, 2 first spectrum) were found to be similar to the double-stranded RNA and double-stranded DNA in A-form (Tsuboi 1970; Taillandier et al. 1985; Theophanides and Tajmir-Riahi 1985; Taillandier and Liquier 1992; Arakawa et al. 2001; Ahmed Ouameur et al. 2004a; Marty et al. 2004a). Table 1 summarizes the relevant frequencies and assignments of the main infrared bands of tRNA. Bands appearing between 1700 and 1550 cm−1 were assigned to the in-plane vibration of RNA bases. The strong infrared band at 1698 cm−1 was mainly assigned to the C=O and C=N stretching vibration from guanine residues (Arakawa et al. 2001; Ahmed Ouameur et al. 2004a; Marty et al. 2004a). Both the C6=O and C8=N7 bonds are located in the major groove of double-stranded tRNA; the N7 atom is the main target for cation–RNA binding, since it is not involved in the Watson-Crick hydrogen-bond pairing. The second strong band at 1653 cm−1 was mainly assigned to the C=O stretching vibration of uracil (Arakawa et al. 2001; Ahmed Ouameur et al. 2004b; Marty et al. 2004a). This band contains both C2=O and C4=O modes, and the major contribution to its intensity comes from the C2=O stretching vibration, which is not involved in the Watson-Crick hydrogen-bond pairing. The C2=O bond is located in the minor groove, while the C4=O bond is positioned in the major groove of the double-stranded tRNA. The band at 1608 cm−1 was assigned mainly to the adenine residues (Arakawa et al. 2001; Ahmed Ouameur et al. 2004b; Marty et al. 2004a). Theoretical calculations for the adenine vibrational frequencies (Dhaoudi et al. 1993) demonstrated that the normal mode for the band at 1608 cm−1 contained major contributions from N7, N9, and N3 atoms. The N7 and N3 atoms are the main sites for cation interaction and are located in the major and the minor grooves of the tRNA duplex, respectively.
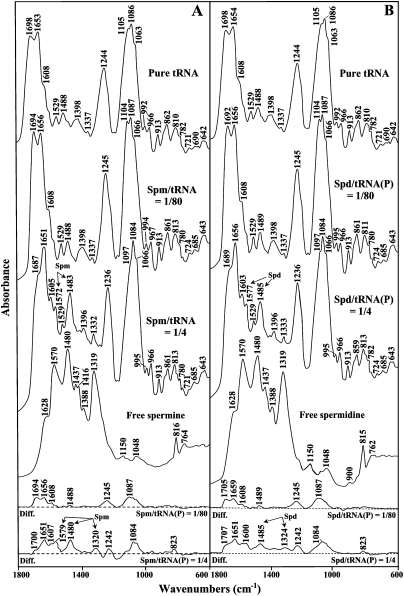
FIGURE 1.
FTIR spectra in the region of 1800–600 cm−1 for pure tRNA, free polyamine, spermine–tRNA (A), and spermidine–tRNA (B) adducts in aqueous solution at pH 7.0 ± 0.2 (top three spectra) and difference spectra for polyamine–tRNA adducts obtained at various polyamine/tRNA(P) molar ratios (bottom two spectra).
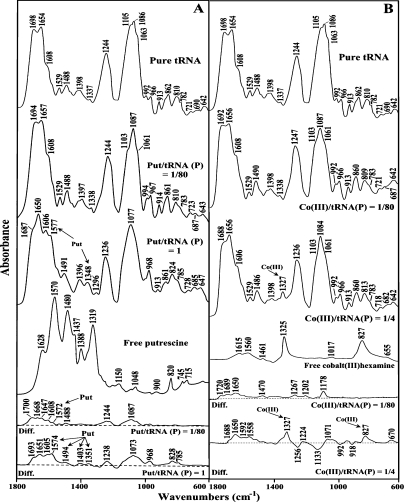
FIGURE 2.
FTIR spectra in the region of 1800–600 cm−1 for pure tRNA, free polyamine, putrescine–tRNA (A), and cobalt(III)hexamine–tRNA (B) adducts in aqueous solution at pH 7.0 ± 0.2 (top three spectra) and difference spectra for polyamine–tRNA adducts obtained at various polyamine/tRNA(P) molar ratios (bottom two spectra).
TABLE 1.
Measured wave numbers, relative intensities, and assignments for the main infrared bands of Baker's yeast tRNA in aqueous solution at pH 7.0 ± 0.2
Other infrared bands related to RNA vibrations were examined in the region between 1550 and 1250 cm−1. These absorptions were assigned to the in-plane ring vibration of the bases and contain some marker of A-form RNA (Tsuboi 1970; Arakawa et al. 2001; Ahmed Ouameur et al. 2004a; Marty et al. 2004a). The most important absorption is located at 1488 cm−1, which is attributed to cytosine in-plane ring vibration (Tsuboi 1970; Arakawa et al. 2001; Ahmed Ouameur et al. 2004a; Marty et al. 2004a). Other vibrations at 1529 cm−1 (cytosine), 1398 cm−1 (guanine in anti conformation), and 1337 cm−1 (adenine and uracil) are related to the G–C and A–U base pairs (Tsuboi 1970; Arakawa et al. 2000; Ahmed Ouameur et al. 2004b; Marty et al. 2004a).
The backbone phosphate group vibrational frequencies appeared as strong bands in the region 1250–1080 cm−1 of the tRNA spectrum (Figs. 1, 2 first spectrum). The two strong absorption bands located at 1244 and 1086 cm−1 were assigned mainly to the asymmetric and symmetric stretching vibrations of the PO2− groups, respectively (Taillandier et al. 1984a,b, 1985; Taillandier and Liquier 1992; Dhaoudi et al. 1993; Arakawa et al. 2001). The spectral region of 1080–800 cm−1 contains several infrared marker bands of tRNA in A-conformation, corresponding to the phosphodiester chain coupled with the ribose vibrational modes (Tsuboi 1970; Taillandier et al. 1985; Theophanides and Tajmir-Riahi 1985; Taillandier and Liquier 1992). Finally, several absorption bands in the region of 800–600 cm−1 were attributed to the out-of-plane ring modes of the tRNA bases: 782 cm−1 (pyrimidine), 721 cm−1 (adenine), 690 cm−1 (guanine), and 642 cm−1 (uracil) (Tsuboi 1970; Taillandier et al. 1985; Theophanides and Tajmir-Riahi 1985; Taillandier and Liquier 1992).
Infrared spectra of tRNA–polyamine complexes at low polycation concentration
The FTIR spectral features of tRNA–polyamine complexes between 1800 and 600 cm−1 are presented in Figures 1 and 2. Molar ratios from 1/80 to 1/4 were studied for spermine, spermidine, and cobalt(III)hexamine. Higher concentrations could not be investigated due to tRNA precipitation. For putrescine, molar ratios up to 1 were studied without precipitation under our experimental settings. Similar concentrations were also used for polyamine–calf-thymus DNA complexes (Ahmed Ouameur and Tajmir-Riahi 2004). These ranges are physiologically justified since millimolar concentrations of polyamines have been found in the nucleus of eukaryotic cells (Gosule and Schellman 1976).
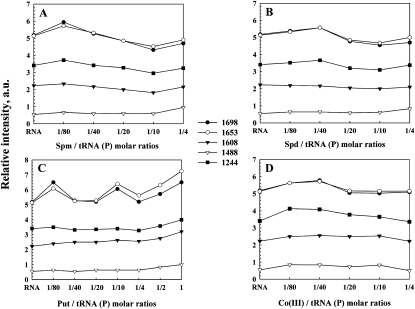
Important shifting of the absorption bands of pure tRNA was observed in the region (1700–1550 cm−1), upon polyamine complexation. The bands at 1698 cm−1 (guanine C7=N, C6=O) and 1653 cm−1 (uracil C=O) were shifted by several wave numbers (3–6 cm−1) upon the interactions with biogenic polyamines and cobalt(III)hexamine (Figs. 1, 2, r = 1/80). The shifting was associated with increase in relative intensities. The most important increase in intensity was observed mainly for a guanine band at 1698 cm−1 (10%–17%) and uracil band at 1653 cm−1 (10%–13%) (Fig. 3, r = 1/80 to 1/40). The adenine vibration at 1608 cm−1 exhibited no shifting and no major intensity changes at low polyamine concentrations (Figs. 1–3). The spectral features observed in the difference spectra of the polyamine–tRNA complexes are consistent with the mentioned spectral changes (Figs. 1, 2, diff. spectra). The positive bands at 1694 cm−1 and 1656 cm−1 (Spm-tRNA diff. spectra); 1705 cm−1 and 1659 cm−1 (Spd-tRNA diff. spectra); 1700 cm−1 and 1668 cm−1 (Put-tRNA diff. spectra); and 1720 cm−1, 1689 cm−1, and 1650 cm−1 [Co(III)-tRNA diff. spectra] are attributed to the interaction of polycation with guanine (C6=O, C=N7) and uracil (C=O) reactive sites, respectively. No major spectral differences were observed for the adenine band (1608 cm−1) in the difference spectra of the polyamine–tRNA adducts, which indicates no participation of adenine in the polycation–tRNA binding (Figs. 1, 2).
FIGURE 3.
Relative intensity variations in arbitrary units for several tRNA in-plane vibrations as a function of polyamine concentration. (A–D) Relative intensity for the tRNA bands at 1698 (guanine), 1653 (uracil), 1608 (adenine), 1488 (cytosine), and 1244 cm−1 (PO2−) for spermine, spermidine, putrescine, and cobalt(II1I)hexamine, respectively.
Other tRNA vibrational frequencies in the region of 1550–1250 cm−1 (in-plane ring vibration of the bases) showed minor spectral changes upon polyamine interaction. No major spectral shifting was observed for the bands at 1529 cm−1 (G and C), 1488 cm−1 (mainly C), 1398 cm−1 (G), and 1337 cm−1 (C) (Figs. 1, 2). In addition, the relative intensity of the cytosine band (1488 cm−1) did not change significantly at low biogenic polyamine concentrations (Fig. 3A–C). In the case of cobalt(III)hexamine, the band 1488 cm−1 was shifted upward by 2 cm−1 (Fig. 2B), which is due to the effect of the cobalt guanine N7 binding on the G–C vibrational modes. This finding was supported by the positive feature at 1470 cm−1 observed in the difference spectra of cobalt(III)hexamine–tRNA adducts (Fig. 2B).
In the phosphate region (1250–1080 cm−1), only cobalt(III)hexamine induced appreciable shifting (3 cm−1) for the band at 1244 cm−1 (asymmetric PO2− stretch), whereas the band at 1086 cm−1 (symmetric PO2− stretch) shifted by only 1 cm−1. As for the biogenic polyamines, the phosphate bands did not show considerable shifting, while intensity changes were observed. However, three new bands at 1267 cm−1, 1202 cm−1, and 1178 cm−1 appeared in the difference spectra of cobalt(III)hexamine (Fig. 2B). This indicates a weaker interaction of the biogenic polyamines with the phosphate groups than that of cobalt(III)hexamine cation.
In our previous study on polyamine–DNA interactions, we found no evidence suggesting the interaction between the polyamines and deoxyribose (Ahmed Ouameur and Tajmir-Riahi 2004). However, by examining the spectral region 1100–800 cm−1 of the polyamine–tRNA complexes, we observed some spectral perturbations, upon polyamine complexation. In fact, the band at 1063 cm−1 (C–O ribose stretch) shifts by 2–3 cm−1 in all four polyamine–tRNA complexes. In addition, the bands 992 cm−1 (C–C ribose stretch) and 862 cm−1 (ribose–phosphodiester bound) shift by 2–3 cm−1 in the biogenic polyamine–tRNA and cobalt(III)hexamine–tRNA complexes, respectively (Figs. 1, 2). The spectral changes observed can be attributed to a minor polycation interaction with the ribose moiety. Polyamine interactions with ribose 2′-OH were previously noted in a series of polycation–tRNA complexes (Bolton and Kearns 1978). However, the polycation–ribose interaction is more pronounced at high biogenic polyamine concentrations (discussed below).
Other tRNA vibrational frequencies in the region of 800–600 cm−1 showed minor spectral changes upon polyamine complexation. Bands appearing in this region were assigned to the out-of-plane vibration of the bases (Tsuboi 1970; Taillandier et al. 1984a; Theophanides and Tajmir-Riahi 1985). The band at 782 cm−1 (pyrimidine) shifted by 2 cm−1 after addition of spermine and spermidine, while it changed only by 1 cm−1 in the case of putrescine and cobalt(III)hexamine cations (Figs. 1, 2). The band at 721 cm−1 (adenine) shifted by 2–3 cm−1 upon addition of biogenic polyamines, which is indicative of other sites than N7 participation in cation interaction. The adenine-N3 site, located in the minor groove of the double helix region, could be a good target since it is not involved in the Watson-Crick network of base-pairing. Finally, the band at 690 cm−1 (guanine) shifted downward by several wave numbers (cm−1) in all four polyamine–tRNA complexes (Figs. 1, 2). Furthermore, the infrared spectra of spermine and spermidine (Fig. 3A,B) at molar ratios between 1/40 and 1/10 show a general decreasing (hypochromism) in intensity of the bands at 1698, 1653, 1608, and 1244 cm−1. This fact has been attributed to the increase in the base-stacking and base-pairing as a consequence of nucleic acid stabilization (Ahmed Ouameur and Tajmir-Riahi 2004).
Infrared spectra of tRNA–polyamine complexes at high polycation concentration
Major spectral changes were observed at high polyamine concentrations. As polycation concentration increased (r = 1/4), larger frequency shifting was observed in the double-bond region. The strong band at 1698 cm−1 (guanine, C=O6/N7) shifted downward by several wave numbers (cm−1) in the spectra of all four polycations (Figs. 1, 2). This result indicates that the preferential binding site for the polycations is the N7-guanine site, since the O6 is involved in the Watson-Crick network of base-pairing. Other bands at 1653 cm−1 (U, C=O2/O4) and 1608 cm−1 (A, C=N7) also shifted appreciably (2–5 cm−1) at high polycation concentration (Figs. 1, 2). These results are supported by the infrared difference spectra showed in the bottom of Figures 1 and 2. The positive features at 1700, 1651, and 1607 cm−1 (for spermine)—1707, 1651, and 1600 cm−1 (for spermidine); 1693, 1651, and 1605 cm−1 (for putrescine); and 1688, 1650, and 1592 cm−1 [for cobalt(III)hexamine]—are attributed to the polycation interactions with guanine-N7, uracil-O2, and adenine-N7 reactive sites, respectively (Figs. 1, 2).
Figure 3C shows the relative intensity changes of the IR marker bands in the presence of putrescine at various polycation concentrations. An important increase in intensity of the marker bands appears when the polyamine concentration increased to a maximum at high putrescine concentration (r = 1). This increase in intensity is called the hyperchromic effect (Pavia et al. 1979), and it is provoked by the base unstaking and unpairing as a consequence of the helix destabilization (Fichera et al. 2000). At high polyamine content, the helix opening contributed to putrescine binding to the single-stranded region, such as the loop of the tRNA molecule. However, the hyperchromic effect for spermine and spermidine was not observed since the maximum concentration used is at molar ratio r = 1/4 (to avoid precipitation).
The infrared spectra at higher concentrations studied in each case allowed us to detect new bands between 1600 and 1300 cm−1 for the biogenic polyamine–tRNA complexes. In fact, the bands measured at 1572, 1483, and 1332 cm−1 for spermine; 1577, 1485, and 1333 cm−1 for spermidine; and 1577, 1491, and 1348 cm−1 for putrescine (Figs. 1, 2) were assigned to the N–H bending vibration of the NH3+ groups, methylene scissoring vibrations, and symmetric bending mode of methyl groups, respectively (Pavia et al. 1979; Gunzler and Gremlich 2002). These bands shifted by 3–11 cm−1 with respect to the infrared spectra of the free polyamines (Figs. 1, 2), supporting an active participation of the primary amino groups in the interaction and the existence of the hydrophobic contact with the methyl groups of tRNA. The positive features measured in the difference spectra of each biogenic polyamine, namely, 1579, 1480, and 1320 cm−1 (spermine); 1485 and 1324 cm−1 (spermidine); and 1574, 1494, and 1351 cm−1 (putrescine) support the polycation interactions (hydrogen binding, hydrophobic contact) with the polynucleotide. The new band measured at 1327 cm−1, in the infrared spectra of the Co(III)–tRNA complex at high cobalt(III)hexamine content (Fig. 2B), is assigned to the NH3 symmetric deformation mode of cobalt(III)hexamine (Nakamoto 1997) and is not arising from tRNA vibration, which suggests that cobalt might bind tRNA by hydrogen-bonding.
The interaction between the polyamines and the backbone phosphate groups is evident by the shifting of the bands at 1244 cm−1 (νasPO2−) and 1088 cm−1 (νsPO2−) by several wave numbers (cm−1) at higher polyamine concentrations. The positive features in the difference spectra at 1242 and 1084 cm−1 (spermine and spermidine), at 1238 and 1073 cm−1 (putrescine), and 1224 and 1071 cm−1 [cobalt(III)hexamine] are due to the polycation–phosphate interactions (Figs. 1, 2). The negative bands observed in the phosphate (1256 cm−1) and ribose (1133, 992, and 918 cm−1) regions of the difference spectra of cobalt(III)hexamine indicate a minor amount of tRNA condensation. Major DNA condensation was observed upon cobalt(III)hexamine–DNA complexation at high polycation content (Ahmed Ouameur and Tajmir-Riahi 2004).
The spectral changes observed in the region of 1080–800 cm−1 exhibit polycation interaction with the ribose moiety of tRNA. The bands at 1100, 1066, 995, and 968 cm−1 assigned to the ribose C–O and C–C ribose modes showed major spectral shifting upon biogenic polyamine complexations. At high polycation content, the band at 1105 cm−1 lost its intensity and shifted toward a lower frequency at 1097 cm−1 (Figs. 1, 2). Similarly, the band at 1063 cm−1 was observed at 1066 cm−1 in the spectra of spermine–tRNA and spermidine–tRNA adducts (Fig. 1). The spectral changes were less pronounced for the cobalt(III)hexamine–tRNA complexes. This indicates some degree of polycation–ribose interaction (H-bonding) upon polyamine complexation. However, the bindings of the polycations to biopolymer did not induce tRNA conformational changes. In fact, there was no major shifting of the A-form tRNA marker bands such as 810 and 862 cm−1 (ribose–phosphodiester) upon polycation adduct formation. This indicated that tRNA remains in the A-family structure in these polyamine–tRNA complexes.
Interaction of polyamines with tRNA studied by capillary electrophoresis
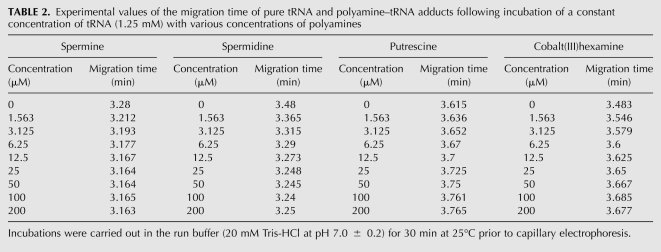
Capillary electrophoresis was used to examine further the interaction of polycations with tRNA. In Table 2, the experimental data related to the migration times of pure tRNA and its complexes with biogenic polyamines and cobalt(III)hexamine at different polycation concentrations are shown.
TABLE 2.
Experimental values of the migration time of pure tRNA and polyamine–tRNA adducts following incubation of a constant concentration of tRNA (1.25 mM) with various concentrations of polyamines
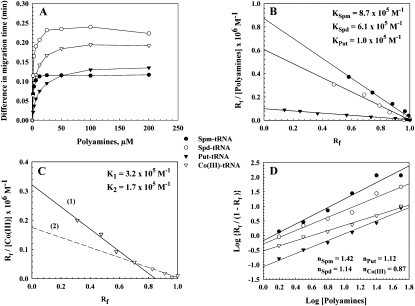
The saturation curve (Fig. 4A) shows a change in the migration time of tRNA after addition of different polycation concentrations. As polycation concentration reached 12.5 μM for spermine, 25 μM for spermidine and cobalt(III)hexamine, and 50 μM for putrescine (Table 2), no appreciable changes were measured for the polycation–tRNA complexes. Based on these results, binding constants of the polycation–tRNA complexes were determined by Scatchard analysis, as described in Materials and Methods. Scatchard analysis for the biogenic polyamine–tRNA adducts showed one overall binding constant for each polyamine, namely, KSpm = 8.7 × 105 M−1, KSpd = 6.1 × 105 M−1, and KPut = 1.0 × 105 M−1 (Fig. 4B). The stability was in the order spermine > spermidine > putrescine. This finding is in agreement with the NMR studies (Frydman et al. 1990, 1992, 1996, 1999; Fernandez et al. 1994) that found the secondary amino groups (–NH2+) of polyamine bind more strongly than the primary amino groups (–NH3+). In fact, spermine contains two -NH2+- groups, spermidine one group, while putrescine does not contain a secondary amino group. Scatchard analysis of the data for the cobalt(III)hexamine–tRNA complexes showed high- and low-affinity sites with binding constants of K1 = 3.2 × 105 M−1 and K2 = 1.7 × 105 M−1, respectively (Fig. 4C). These results suggest that cobalt(III)hexamine binds at two distinct binding sites on the tRNA molecule. Furthermore, it may be speculated that the stronger interaction is due to the guanine-N7 atom, while the weaker binding is due to the polycation-phosphate oxygen atom, which is consistent with our infrared data and other studies (Kieft and Tinoco 1997; Rüdisser and Tinoco 1999).
FIGURE 4.
(A) Plot of the difference in migration time (in minutes) of polyamine–tRNA complexes from capillary electrophoresis following incubation of a constant concentration of tRNA (1.25 mM) with various concentrations of polyamines. The difference in migration time of the polyamine–tRNA complexes was determined by subtracting the migration time of pure tRNA from that of each polyamine–tRNA adducts. (B,C) Scatchard plots for biogenic polyamine–tRNA, and cobalt(III)hexamine–tRNA complexes, respectively. (D) Hill plots for polyamine–tRNA complexes.
The analysis of the cooperativity of the binding process between the polyamines and tRNA has been done (Fig. 4D). The Hill coefficients n calculated for the biogenic polyamines were 1.42, 1.14, and 1.12, for spermine, spermidine, and putrescine, respectively. Since the values obtained were >1, this means a positive cooperativity binding of biogenic polyamines with tRNA (Jack 1995). In contrast, the Co(III)–tRNA interaction was characterized by a negative cooperativity (n < 1) (Fig. 4D). Our data on the binding parameters of the four polycations to tRNA are comparable to that measured for polyamine–DNA complexes (Ahmed Ouameur and Tajmir-Riahi 2004). This finding may reflect some common molecular mechanism for DNA and tRNA binding. Undoubtedly, one of the most important roles of these polyamines in vivo is to stabilize the nucleic acids.
Docking study
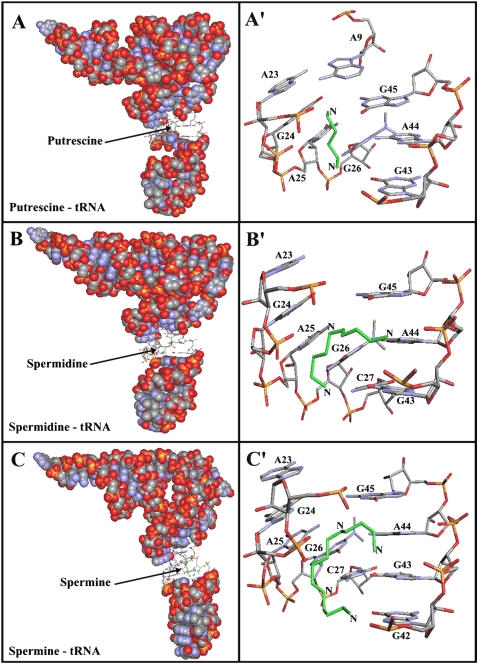
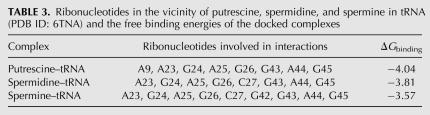
Our results from FTIR and capillary electrophoresis are accompanied by docking experiments in which the putrescine, spermidine, and spermine cations were docked to tRNA to determine the preferred binding sites on tRNA. The dockings results are shown in Figure 5 and Table 3. The models show that putrescine is surrounded by A9, A23, G24, A26, G43, A44, and G45 with a binding energy of −4.04 kcal/mol (Fig. 5A; Table 3). Spermidine is located in the vicinity of A23, G24, A25, G26, C27, G43, A44, and G45 with the binding energy of −3.81 kcal/mol (Fig. 5B; Table 3). However, spermine is positioned near A23, G4, A25, G26, C27, G43, A44, and G45 with the binding energy of −3.57 kcal/mol (Fig. 5C; Table 3). The docking results showed most of the binding sites used are similar for these biogenic polyamines (Table 3). The binding energy (ΔG) shows the stability of the complexes formed: putrescine > spermidine > spermine (Table 3).
FIGURE 5.
Best conformations for polyamines docked to tRNA (PDB entry 6TNA). (Green) The polyamines. (A) tRNA in sphere-filling model with the putrescine binding site in sticks; (A′) putrescine binding sites represented in sticks with the corresponding base residues. (B) tRNA in sphere-filling model with the spermidine binding site in sticks; (B′) spermidine binding sites represented in sticks. (C) tRNA in sphere-filling model with the spermine binding sites represented in sticks; (C′) the binding sites represented in sticks.
TABLE 3.
Ribonucleotides in the vicinity of putrescine, spermidine, and spermine in tRNA (PDB ID: 6TNA) and the free binding energies of the docked complexes
DISCUSSION
Interactions between biogenic polyamines and DNA have been extensively studied by several investigators using different methods (for reviews, see Feuerstein et al. 1991; Marton and Pegg 1995; Igarashi and Kashiwagi 2000; Thomas and Thomas 2001). Our recent report was related to the mechanism of spermine–, spermidine–, and putrescine–DNA interactions (Ahmed Ouameur and Tajmir-Riahi 2004). In this study, several binding models have been proposed, in order to support preferential binding sites of polyamines in the major and/or the minor grooves of DNA. It seems that most of the intracellular polyamines are mainly associated with RNA molecules (Igarashi and Kashiwagi 2000). In recent years, the effects of polyamines on cell growth and the dependency of drug potency on polyamines have been well investigated (Bachrach 2005; Kouvela et al. 2006). However, limited studies have been done to understand the mechanism of polyamine–tRNA binding at molecular levels. In the present study, our interest was to determine the preferential binding sites of polyamines on tRNA.
One of the main conclusions arising from the spectroscopic analyses is that at low concentrations, spermine and spermidine were capable of stabilizing the structure of tRNA. This is evidenced by a decrease in intensity of several tRNA infrared bands (hypochromic effect). This effect has been observed at higher polyamine concentrations in the case of spermine– and spermdine–DNA complexes (Ahmed Ouameur and Tajmir-Riahi 2004). Since the hypochromism effect was observed at a higher molar ratio (1/4) in the polyamine–DNA complexes (Ahmed Ouameur and Tajmir-Riahi 2004), we concluded that at low concentration, spermine and spermidine are more effective in binding tRNA than DNA. In this way, spermine and spermidine could stabilize tRNA structure at low cation content, while highly polymerized calf-thymus DNA was stabilized at high polyamine concentrations (Ahmed Ouameur and Tajmir-Riahi 2004). It has been shown that protein synthesis was stimulated at low polyamine (spermidine) concentration (Shimogori et al. 1996). This result is in accordance with our finding that tRNA molecules are stabilized at low polyamine concentration. This result also suggests that polyamine increases the binding of Met-tRNA to the ribosome subunits (Shimogori et al. 1996), which could be a consequence of polyamine binding to tRNA with an increase in RNA stability. At high polyamine concentrations, the hyperchromic effect (an increase in intensity of several infrared bands) was observed only in the putrescine–tRNA complex, due to helix destabilization. It has been found that higher concentrations of polyamine are inhibitory for the protein synthesis (Shimogori et al. 1996). The hyperchromic effect observed at high putrescine concentration could explain the inhibition of protein synthesis at high polyamine concentration. For spermine and spermidine, the hyperchromic effect was not observed since the maximum concentration used is at molar ratio 1/4, in order to ovoid precipitation.
In addition, the data from infrared spectra have allowed us to analyze the polyamine–tRNA interactions at a molecular level. The preferential binding sites are slightly different from those of polyamine–DNA complexes (Ahmed Ouameur and Tajmir-Riahi 2004). At low polyamine content, the interaction involves RNA bases and ribose moiety. The preferential binding sites are guanine-N7/O6, uracil-O2/O4, and adenine-N3 reactive sites. The interaction with the 2′-OH of the ribose is less pronounced than observed at high polyamine content. The cobalt(III)hexamine preferential binding sites are guanine-N7, uracil-O2, and cytosine-O2/N4 reactive sites and the backbone phosphate groups. At high polycation concentrations, the interaction involves guanine-N7/O6, adenine-N7, and uracil-O2 reactive sites, as well as the phosphate and the ribose backbone. In addition, the participation of the primary amino group of the polycations in polyamine binding and the presence of hydrophobic contacts were demonstrated. Capillary electrophoresis study reveals that long-chain polyamines (spermine and spermidine) bind more strongly than a small one (putrescine). This finding is in agreement with the NMR studies, which suggested that secondary amino groups bind more strongly than primary amino groups (Frydman et al. 1990, 1992, 1996, 1999; Fernandez et al. 1994). The high binding affinity of biogenic polyamines to tRNA molecule was in the order spermine > spermidine > putrescine, which correlates with their charged amino group content. Cobalt(III)hexamine contains high- and low-affinity sites in tRNA, which have been attributed to the interactions with guanine-N7 sites and the phosphate backbone. This mechanism of tRNA binding could explain the condensation phenomenon observed at high Co(III) content, as reported in the Co(III)–DNA complexes (Ahmed Ouameur and Tajmir-Riahi 2004).
In order to perform a meaningful comparison with our infrared spectroscopic results, we further studied the interactions between putrescine, spermidine, and spermine molecules and tRNA using a molecular docking approach. The results showed several models of interactions between putrescine, spermidine, spermine, and tRNA (Fig. 5; Table 3). The X-ray study (Quigley et al. 1978) showed two binding sites for spermine—one in the major groove of the double helical region in the anticodon stem and another in the deep pocket formed by bending of the D-stem nucleosides toward those of the variable loop. H-NMR analysis showed that only one spermine molecule binds tRNA (Frydman et al. 1996); the binding site was located in the TψC-loop. Using a docking approach, it would appear that spermine binds in the major groove. The docking of spermine in the TψC-loop showed a negative free binding energy (−3.19 kcal/mol, docking not shown) but still higher than the value obtained from the docking in the major groove (−3.57 kcal/mol). This would indicate that both the TψC-loop and the major groove are potential binding sites for polyamines, with the major groove being the preferred one. Other findings suggest that the major factor determining the binding of the spermine analogs to AcPhe–tRNA is the polyamine part of analogs, not the photoactivated substrate (Amarantos and Kalpaxis 2000). A recent study has suggested that polyamines are located at the bulged-out region of double-stranded RNA and affect RNA function (Higashi et al. 2008). Our infrared spectroscopic analysis of the interaction between spermine and tRNA revealed that the preferential binding sites at low spermine concentration are guanine (C=O6/N7) and uracil (C=O2). By increasing the concentration of spermine, new sites of interaction appeared [adenine (C=N3), PO2, OH2′-ribose], and the presence of hydrophobic contacts between spermine and methyl groups of tRNA are evident.
This study highlights the mechanism of interaction between biogenic polyamines and tRNA and might help to design polyamine analogs with a growth inhibitory effect. Therefore, studying the interaction between polyamine analogs and macromolecules (protein, DNA, and tRNA) is a current and future research priority.
MATERIALS AND METHODS
Materials
Biogenic polyamines (spermine, spermidine, and putrescine) and cobalt(III)-hexamine were purchased from Sigma Chemical Company. tRNA from Baker's yeast was purchased from Sigma Chemical Co. and used as supplied. The absorbance at 260 and 280 nm was used, in order to check the protein content of tRNA solution. The A260/A280 ratio was 2.2, showing that the tRNA was sufficiently free from protein (Marmur 1961). Other chemicals were of reagent grade and used without further purification.
Preparation of stock solutions
Stock tRNA solution was prepared by dissolving 10 mg of tRNA in 20 mM Tris-HCl buffer (pH 7.4). The final concentration of the stock tRNA solution was determined spectrophotometrically at 260 nm, using a molar extinction coefficient of ε260 = 9250 cm−1 M−1 (expressed as molarity of phosphate groups) (Cantor et al. 1970). The UV absorbance at 260 nm of a diluted solution (1/250) of tRNA used in our experiments was measured to be 0.925 (the path length was 1 cm), and the final concentration of the stock tRNA solution was calculated to be 25 mM in tRNA phosphate. The appropriate amount of polyamines (0.3–25 mM) was prepared in distilled water and added dropwise to tRNA solution, in order to attain the desired polyamine/tRNA(P) molar ratios (r) of 1/80, 1/40, 1/20, 1/10, 1/4, 1/2, and 1 at a final tRNA concentration of 12.5 mM (5 mg/mL) for infrared measurements. For capillary electrophoresis, the polyamine/tRNA(P) ratios were 1/800, 1/400, 1/200, 1/100, 1/50, 1/25, 1/12.5, and 1/6.25 with a final tRNA concentration of 1.25 mM. The pH of the solutions was adjusted to 7.0 ± 0.2 using NaOH solution.
FTIR spectra
Infrared spectra were recorded on a FTIR spectrometer (Impact 420 model), equipped with a DTGS (deuterated triglycine sulfate) detector and a KBr beam splitter, using AgBr windows. Spectra were collected after 2 h of incubation of polyamine with the polynucleotide solution and measured in triplicate (three individual samples with the same polynucleotide and polyamine concentrations). Interferograms were accumulated over the spectral range 4000–600 cm−1 with a nominal resolution of 2 cm−1 and a minimum of 100 scans. The difference spectra [(tRNA solution + polyamine) − (tRNA solution)] were obtained using a sharp tRNA band at 966 cm−1 as the internal reference. This band is due to ribose C–C stretching vibration, exhibits no spectral changes (shifting or intensity variation) upon polyamine–tRNA complexation, and is canceled out on spectral subtraction (Ahmed Ouameur et al. 2004a; Marty et al. 2004a).
The plots of the relative intensity (R) of several tRNA vibrations related to A–U, G–C base pairs and the PO2 stretching modes versus the polyamine concentrations were obtained after peak normalization using:
where Ii is the intensity of the absorption band for pure tRNA and tRNA in the complex with i concentration of polyamine, and I966 is the intensity of the 966 cm−1 peak (internal reference) (Ahmed Ouameur and Tajmir-Riahi 2004).
Capillary electrophoresis
A PACE System MDQ (Beckman) with photodiode array detector was used to study polyamine–tRNA interaction. An uncoated fused silica capillary of 75 μm i.d. (total length of 57 cm) and effective length of 50 cm (to the detector) was used. The capillary was conditioned each day by rinsing with 1 N sodium hydroxide for 30 min, followed by a 15-min wash with 0.1 N sodium hydroxide. Then it was rinsed with running buffer for 30 min at high pressure (50 psi), followed by a baseline run for 20 min at the voltage to be used for the experiments (25 kV). Between runs, the capillary was rinsed with 1 N NaOH for 2 min, followed by rinsing with running buffer for 3 min at high pressure. The capillary was flushed with distilled deionized water for 30 min at the end for each day and filled with deionized water overnight. Samples were injected using a voltage injection at 10 kV for 5 sec. Electrophoresis was carried out at a voltage of 25 kV for 10 min using normal polarity. All runs were carried out at 25°C. The capillary inlet and outlet vials were replenished after every five runs. The polyamine binding experiments were performed in a sample buffer containing 20 mM Tris-HCl (pH 7.0 ± 0.2), using a constant concentration of tRNA (1.25 mM) and variable concentrations of polyamines. The stock solutions of polyamines (2.5 mM) and tRNA (2.5 mM) were prepared in the sample buffer. The solutions were mixed to attain polyamine/tRNA(P) molar ratios of 1/800 to 1/6.25, in the presence of 1.25 mM tRNA. Each sample was allowed to equilibrate for 30 min and tested with two separate runs for the same stock solution. The electropherograms were monitored at 260 nm and were collected and analyzed with the Beckman P/ACE Windows controller software.
Data analysis
Affinity capillary electrophoresis (ACE) was used to detect a shift in mobility when polyamine binds to tRNA. The binding constants for the polyamine–tRNA complexes can be determined by Scatchard analysis, using mobility shift of tRNA complexes (Klotz 1982; Guszcynski and Copeland 1998). The extent of saturation (Rf) of the tRNA was determined from the change of the migration time of tRNA, in the presence of various concentrations of polyamine by the following equation:
 |
where Δm is the difference in migration time between pure tRNA and its complexes at different polyamine concentrations, while Δms corresponds to the difference in migration time between pure tRNA and tRNA-saturated complex.
The binding constant Kb, given by
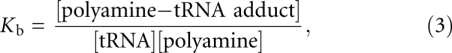 |
was determined by fitting the experimental values of Rf and polyamine concentration to the equation:
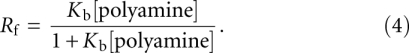 |
Rearrangement of this gives a convenient form for Scatchard analysis:
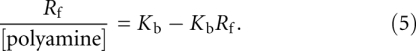 |
Capillary electrophoresis has become a useful technique for measuring binding constants (Guijt-van Duijn et al. 2000). Using the affinity capillary electrophoresis (ACE) method, many ligand–polynucleotide complexes have been successfully identified in our laboratory (Arakawa et al. 2000, 2001; Ahmed Ouameur and Tajmir-Riahi 2004; Ahmed Ouameur et al. 2004a,b; Marty et al. 2004b).
The cooperativity of the binding can be analyzed using a Hill plot. Assuming one binding site for polyamine (Equation 5), the equation shown below (Hill equation) can be established:
where n (Hill coefficient) measures the degree of cooperativity and Kd is the dissociation constant. The linear plot of 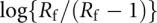 versus log[polyamine] has a slope of n and an intercept on the log[polyamine] axis of log Kd / n. The quantity n increases with the degree of cooperativity of a reaction and thereby provides a convenient and simplistic characterization of a ligand-binding reaction (Jack 1995).
versus log[polyamine] has a slope of n and an intercept on the log[polyamine] axis of log Kd / n. The quantity n increases with the degree of cooperativity of a reaction and thereby provides a convenient and simplistic characterization of a ligand-binding reaction (Jack 1995).
Molecular modeling and docking
The docking studies were performed with ArgusLab 4.0.1 software (Planaria Software LLC; http://www.arguslab.com). tRNA structure was obtained from the PDB (ID: 6TNA) (Sussman et al. 1978), and the polyamine three-dimensional structures were generated from PM3 semi-empirical calculations using Chem3D Ultra 6.0. According to X-ray (Quigley et al. 1978) and 1H-NMR (Frydman et al. 1996) data, three possible binding sites (major groove, variable loop, and TψC-loop) were searched. The docking runs were performed on the ArgusDock docking engine using high precision with a maximum of 150 candidate poses. The conformations were ranked using the Ascore scoring function, which estimates the free binding energy. Upon docking of polyamines to tRNA, the current configurations were optimized using a steepest decent algorithm until convergence, within 40 iterations, and nucleobase residues within a distance of 3.5 Å relative to polyamines were involved in the complexation.
ACKNOWLEDGMENTS
We highly appreciate the financial support of the Natural Sciences and Engineering Research Council of Canada (NSERC).
Footnotes
Abbreviations: Spm, spermine; Spd, spermidine; Put, putrescine; Co(III), cobalt(III)hexamine; CE, capillary electrophoresis; ACE, affinity capillary electrophoresis; FTIR, Fourier transform infrared; tRNA, transfer RNA; A, adenine; G, guanine; C, cytosine; U, uracil; r, polyamine/tRNA(P) molar ratio.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1994310.
REFERENCES
- Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J 1999. Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J Biol Chem 274: 8723–8729 [DOI] [PubMed] [Google Scholar]
- Ahmed Ouameur A, Tajmir-Riahi HA 2004. Structural analysis of DNA interactions with biogenic polyamines and cobalt(III)hexamine studied by Fourier transform infrared and capillary electrophoresis. J Biol Chem 279: 42041–42054 [DOI] [PubMed] [Google Scholar]
- Ahmed Ouameur A, Malonga H, Neault J-F, Diamantoglou S, Tajmir-Riahi HA 2004a. Taxol interaction with DNA and RNA—stability and structural features. Can J Chem 82: 1112–1118 [Google Scholar]
- Ahmed Ouameur A, Marty R, Neault J-F, Tajmir-Riahi HA 2004b. AZT binds RNA at multiple sites. DNA Cell Biol 23: 783–788 [DOI] [PubMed] [Google Scholar]
- Amarantos I, Kalpaxis DL 2000. Photoaffinity polyamines: Interactions with AcPhe-tRNA free in solution or bound at the P-site of Escherichia coli ribosomes. Nucleic Acids Res 28: 3733–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H, Ahmad R, Naoui M, Tajmir-Riahi HA 2000. A comparative study of calf-thymus DNA binding to Cr(III) and Cr(VI) ions: Evidence for the guanine N-7 chromium-phosphate chelate formation. J Biol Chem 275: 10150–10153 [DOI] [PubMed] [Google Scholar]
- Arakawa H, Neault J-F, Tajmir-Riahi HA 2001. Silver(I) complexes with DNA and RNA studied by Fourier transform infrared spectroscopy and capillary electrophoresis. Biophys J 81: 1580–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchi CJ, Weiss LM, Lane S, Frydman B, Valasinas A, Reddy V, Sun JS, Marton LJ, Khan IA, Moretto M, et al. 2002. Novel synthetic polyamines are effective in the treatment of experimental microsporidiosis, an opportunistic AIDS-associated infection. Antimicrob Agents Chemother 46: 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrach U 2005. Naturally occurring polyamines: Interaction with macromolecules. Curr Protein Pept Sci 6: 559–566 [DOI] [PubMed] [Google Scholar]
- Bibillo A, Figlerowicz M, Kierzek R 1999. The non-enzymatic hydrolysis of oligoribonucleotides. VI. The role of biogenic polyamines. Nucleic Acids Res 27: 3931–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton PH, Kearns DR 1978. Hydrogen bonding interactions of polyamines with the 2′OH of RNA. Nucleic Acids Res 5: 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor CR, Warshaw MM, Shapiro H 1970. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers 9: 1059–1077 [DOI] [PubMed] [Google Scholar]
- Childs AC, Mehta DJ, Gerner EW 2003. Polyamine-dependent gene expression. Cell Mol Life Sci 60: 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SS 1998. A guide to the polyamines. Oxford University Press, New York [Google Scholar]
- Dhaouadi Z, Ghomi M, Austin JC, Girling RB, Hester RE, Mojzes P, Chinsky L, Turpin PY, Coulombeau C, Jobic H, et al. 1993. Vibrational motions of bases of nucleic acids as revealed by neutron inelastic scattering and resonance Raman spectroscopy: 1. Adenine and its deuterated species. J Phys Chem 97: 1074–1084 [Google Scholar]
- Fernandez CO, Frydman B, Samejima K 1994. Interactions between polyamine analogs with antiproliferative effects and tRNA: A 15N NMR analysis. Cell Mol Biol 40: 933–944 [PubMed] [Google Scholar]
- Fernández CO, Buldain G, Samejima K 2000. Probing the interaction between N1,N4-dibenzylputrescine and tRNA through 15N NMR: Biological implications. Biochim Biophys Acta 1476: 324–330 [PubMed] [Google Scholar]
- Feuerstein BG, Williams LD, Basu HS, Marton LJ 1991. Implications and concepts of polyamine–nucleic acid interactions. J Cell Biochem 46: 37–47 [DOI] [PubMed] [Google Scholar]
- Fichera M, Gregoli L, Musumarra G 2000. 2,6-Di(heteroarylvinyl)-pyridines as new potential antitumor agents. J Phys Org Chem 13: 344–346 [Google Scholar]
- Frydman B, Valasinas A 1999. Polyamine-based chemotherapy of cancer. Exp Opin Ther Patents 9: 1055–1068 [Google Scholar]
- Frydman B, de los Santos C, Frydman RB 1990. A 13C NMR study of [5,8-13C2]spermidine binding to tRNA and to Escherichia coli macromolecules. J Biol Chem 265: 20874–20878 [PubMed] [Google Scholar]
- Frydman L, Rossomando PC, Frydman V, Fernandez CO, Frydman B, Samejima K 1992. Interactions between natural polyamines and tRNA: An 15N-NMR analysis. Proc Natl Acad Sci 89: 9186–9190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman B, Westler WM, Samejima K 1996. Spermine binds in solution to the TψC loop of tRNAPhe: Evidence from a 750 MHz 1H-NMR analysis. J Org Chem 61: 2588–2589 [DOI] [PubMed] [Google Scholar]
- Frydman B, Westler WM, Valasinas A, Kramer DL, Porter CW 1999. Regioselective binding of spermine, N1,N12-bismethylspermine, and N1,N12-bisethylspermine to tRNAPhe as revealed by 750 MHz 1H-NMR and its possible correlation with cell cycling and cytotoxicity. J Braz Chem Soc 10: 334–340 [Google Scholar]
- Frydman B, Porter CW, Maxuitenko Y, Sarkar A, Bhattacharya S, Valasinas A, Reddy VK, Kisiel N, Marton LJ, Basu HS 2003. A novel polyamine analog (SL-11093) inhibits growth of human prostate tumor xenografts in nude mice. Cancer Chemother Pharmacol 51: 488–492 [DOI] [PubMed] [Google Scholar]
- Gosule LC, Schellman JA 1976. Compact form of DNA induced by spermidine. Nature 259: 333–335 [DOI] [PubMed] [Google Scholar]
- Guijt-van Duijn RM, Frank J, van Dedem GWK, Baltussen E 2000. Recnt advances in affinity capillary electrophoresis. Electrophoresis 21: 3905–3918 [DOI] [PubMed] [Google Scholar]
- Günzler H, Gremlich H-U 2002. IR spectroscopy, An introduction. Wiley-vch, Germany [Google Scholar]
- Guszcynski T, Copeland TD 1998. A binding shift assay for the zinc-bound and zinc-free HIV-1 nucleocapsid protein by capillary electrophoresis. Anal Biochem 260: 212–217 [DOI] [PubMed] [Google Scholar]
- Higashi K, Terui Y, Suganami A, Tamura Y, Nishimura K, Kashiwagi K, Igarashi K 2008. Selective structural change by spermidine in the bulged-out region of double-stranded RNA and its effect on RNA function. J Biol Chem 283: 32989–32994 [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K 2000. Polyamines: Mysterious modulators of cellular functions. Biochem Biophys Res Commun 271: 559–564 [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K 2006. Polyamine modulon in Escherichia coli: Genes involved in the stimulation of cell growth by polyamines. J Biochem 139: 11–16 [DOI] [PubMed] [Google Scholar]
- Jack RC 1995. Basic biochemical laboratory procedures and computing. Oxford University Press, New York [Google Scholar]
- Kieft JS, Tinoco I Jr 1997. Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt(III)hexamine. Structure 5: 713–721 [DOI] [PubMed] [Google Scholar]
- Klotz MI 1982. Numbers of receptor sites from Scatchard graphs: Facts and fantasies. Science 217: 1247–1249 [DOI] [PubMed] [Google Scholar]
- Kouvela EC, Petropoloulos AD, Kalpaxis DL 2006. Unraveling new features of clindamycin interaction with functional ribosomes and dependence of the drug potency on polyamines. J Biol Chem 281: 23103–23110 [DOI] [PubMed] [Google Scholar]
- Li Y, Eiseman JL, Sentz DL, Rogers FA, Pan SS, Hu L-T, Egorin MJ, Callery PS 1996. Synthesis and antitumor evaluation of a highly potent cytotoxic DNA cross-linking polyamine analogue, 1,12-diaziridinyl-4,9-diazadodecane. J Med Chem 39: 339–341 [DOI] [PubMed] [Google Scholar]
- Marmur J 1961. A procedure for isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol 3: 208–218 [Google Scholar]
- Marton LJ, Pegg AE 1995. Polyamines as targets for therapeutic intervention. Annu Rev Pharmacol Toxicol 35: 55–91 [DOI] [PubMed] [Google Scholar]
- Marty R, Ouameur AA, Neault JF, Tajmir-Riahi HA 2004a. RNA adducts with chlorophyll and chlorophyllin: Stability and structural features. J Biomol Struct Dyn 22: 45–50 [DOI] [PubMed] [Google Scholar]
- Marty R, Ouameur AA, Neault J-F, Tajmir-Riahi HA 2004b. AZT-DNA interaction. DNA Cell Biol 23: 135–140 [DOI] [PubMed] [Google Scholar]
- Nakamoto K 1997. Infrared and Raman spectra of inorganic and coordination compounds (Part B: applications in coordination, organometallic, and bioinorganic chemistry). Wiley, New York [Google Scholar]
- Pavia DL, Lampman GM, Kriz GS 1979. Introduction to spectroscopy: A guide for students of organic chemistry. Saunders, Toronto, Canada [Google Scholar]
- Quigley GJ, Teeter MM, Rich A 1978. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci 75: 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdisser S, Tinoco I Jr 1999. Solution structure of cobalt(III)hexammine complexed to the GAAA tetraloop, and metal-ion binding to G·A mismatches. J Mol Biol 295: 1211–1223 [DOI] [PubMed] [Google Scholar]
- Shi H, Moore PB 2000. The crystal structure of yeast phenylalanine tRNA at 1. 93 Å resolution. A classic structure revisited. RNA 6: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogori T, Kashiwagi K, Igarashi K 1996. Spermidine regulation of protein synthesis at the level of initiation complex formation of Met-tRNA, mRNA and ribosomes. Biochem Biophys Res Commun 223: 544–548 [DOI] [PubMed] [Google Scholar]
- Sussman JL, Holbrook SR, Warrant RW, Church GM, Kim S-H 1978. Crystal structure of yeast phenylalanine transfer RNA: I. Crystallographic refinement. J Mol Biol 123: 607–630 [DOI] [PubMed] [Google Scholar]
- Tabor H, Tabor CW 1984. Polyamines. Annu Rev Biochem 53: 749–790 [DOI] [PubMed] [Google Scholar]
- Taillandier E, Liquier J 1992. Infrared spectroscopy of DNA. Methods Enzymol 211: 307–335 [DOI] [PubMed] [Google Scholar]
- Taillandier E, Liquier J, Taboury JA, Ghomi M 1984a. Structural transitions in DNA (A, B, Z) studied by IR spectroscopy. In Spectroscopy of biological molecules (ed. Sandorfy C, Theophanides T), pp.171–189 Reidel Dordrecht, Amsterdam [Google Scholar]
- Taillandier E, Taboury JA, Adam S, Liquier J 1984b. Left-handed helical structure of poly[d(A-C)·poly[d(G-T)] studied by infrared spectroscopy. Biochemistry 23: 5703–5706 [DOI] [PubMed] [Google Scholar]
- Taillandier E, Liquier J, Taboury JA 1985. Infrared spectral studies of DNA conformations. Adv Infrared Raman Spectrosc 12: 65–113 [Google Scholar]
- Theophanides T, Tajmir-Riahi HA 1985. Flexibility of DNA and RNA upon binding to different metal cations. An investigation of the B to A to Z conformational transition by Fourier transform infrared spectroscopy. J Biomol Struct Dyn 2: 995–1004 [DOI] [PubMed] [Google Scholar]
- Thomas T, Thomas TJ 2001. Polyamines in cell growth and cell death: Molecular mechanisms and therapeutic applications. Cell Mol Life Sci 58: 244–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Thomas TJ 2003. Polyamine metabolism and cancer. J Cell Mol Med 7: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Balabhadrapathruni S, Gallo MA, Thomas TJ 2002. Development of polyamine analogs as cancer therapeutic agents. Oncol Res 13: 123–135 [PubMed] [Google Scholar]
- Tsuboi M 1970. Application of infrared spectroscopy to structure studies of nucleic acids. Appl Spectrosc Rev 3: 45–90 [Google Scholar]
- Watanabe S, Kusama-Eguchi K, Kobayashi H, Igarashi K 1991. Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J Biol Chem 266: 20803–20809 [PubMed] [Google Scholar]
- Yoshida M, Meksuriyen D, Kashiwagi K, Kawai G, Igarashi K 1999. Polyamine stimulation of the synthesis of oligopeptide-binding protein (OppA). Involvement of a structural change of the Shine-Dalgarno sequence and the initiation codon AUG in oppa mRNA. J Biol Chem 274: 22723–22728 [DOI] [PubMed] [Google Scholar]