Abstract
The selection of cognate tRNAs during translation is specified by a kinetic discrimination mechanism driven by distinct structural states of the ribosome. While the biochemical steps that drive the tRNA selection process have been carefully documented, it remains unclear how recognition of matched codon:anticodon helices in the small subunit facilitate global rearrangements in the ribosome complex that efficiently promote tRNA decoding. Here we use an in vitro selection approach to isolate tRNATrp miscoding variants that exhibit a globally perturbed tRNA tertiary structure. Interestingly, the most substantial distortions are positioned in the elbow region of the tRNA that closely approaches helix 69 (H69) of the large ribosomal subunit. The importance of these specific interactions to tRNA selection is underscored by our kinetic analysis of both tRNA and rRNA variants that perturb the integrity of this interaction.
Keywords: tRNA, ribosome, kinetics, miscoding
INTRODUCTION
tRNA selection is the process through which the ribosome identifies cognate tRNAs for acceptance during the elongation cycle of protein synthesis. During translation, the cognate ternary complex (composed of a cognate tRNA, GTP, and EFTu) binds to the A site of the ribosome, stabilizing an “on” state that ultimately leads to the acceleration of two forward rate constants in the pathway: GTPase activation of EFTu and tRNA accommodation (Gromadski and Rodnina 2004). In addition to these kinetic contributions to tRNA selection, thermodynamic discrimination against near- and noncognate tRNAs during both an initial selection and a proofreading phase provides additional contributions to the overall fidelity of the selection process (Hopfield 1974). As a consequence of these discriminatory steps, as well as the contributions of a post-peptidyl transfer quality control step (Zaher and Green 2009b), the overall fidelity of tRNA selection is estimated to be on the order of 1/103-4 (for review, see Zaher and Green 2009a).
While much is known about the biochemical steps that are involved in the discriminatory steps of tRNA selection, there is considerably less known about how the tRNA (or the ribosome) communicates information from the small subunit decoding center (where cognate interactions must initially be deciphered) to more remote regions of the large subunit involved in GTPase activation and accommodation. CryoEM and crystallographic studies have highlighted properties of the tRNA that are likely to be important for the process of decoding. For example, we have a rather detailed understanding of how cognate tRNA anticodon stem–loops are specifically recognized in the decoding center through specific minor groove interactions between the codon:anticodon minihelix and universally conserved elements in the 16S rRNA (Ogle et al. 2001). Cryo-EM and recent high-resolution studies of tRNA bound to the ribosome in a preaccommodated A/T state (with EFTu) reveal a “bend and twist” in the tRNA. The tRNA is effectively overwound in this structure, consistent with the idea of an active role for tRNA in decoding (Valle et al. 2002; Li et al. 2008; Schmeing et al. 2009). These structures highlight a collection of interactions between the tRNA and the ribosome that are likely important for stabilizing the contorted ternary complex. From this view, the authors proposed a series of molecular rearrangements that might occur following recognition of cognate tRNA by the ribosome; these include movements in h44, r-protein S12, position 69 of the tRNA within the small subunit, and domains II and III of EFTu.
These ideas have been broadly supported by biochemical studies demonstrating that tRNA itself plays an active role in transducing information about small subunit decoding center status to the large subunit functional centers. An initial study devised a rather dramatic experiment wherein the tRNA was cut in two pieces, separating the D and anticodon stems (ACD) from the T and acceptor stems (AST) (Piepenburg et al. 2000). While each half of the tRNA could bind to the ribosome, rapid GTP hydrolysis on EFTu was not stimulated by the ACD tRNA fragment bound to the decoding center, and paromomycin could not suppress this deficiency. These studies concluded that codon recognition was not sufficient for GTPase activation, but instead depended on tRNA connectivity between the large and small subunit functional centers.
More recent biochemical studies of a miscoding tRNA variant (the G24A tRNATrp “Hirsh suppressor”) provided further support for the notion that tRNA, and its altered structure during the A/T state, is critical to signal transduction on the ribosome (Cochella and Green 2005). In this study, it was shown that a mutation at the junction between the D and anticodon stems of the tRNA could promote GTPase activation and tRNA accommodation on the ribosome, independent of cognate pairing interactions in the decoding center. Subsequent studies reported that mutations in the tertiary core of the elbow of the tRNA also stimulated miscoding with effects on both GTPase activation and tRNA accommodation (Pan et al. 2008); these authors argued that increased flexibility for these miscoding variants was key to their mode of action. Other recent studies with Escherichia coli tRNAAla have proposed that the 32–38 base pair at the top of the anticodon stem plays a crucial role in specifying codon recognition (Ledoux et al. 2009; Murakami et al. 2009). These studies collectively highlight the importance of tRNA elements somewhat remote from the anticodon in contributing to accurate decoding.
What is missing from these studies, however, is detailed mechanistic insight into how these alterations in the tRNA body facilitate kinetic steps in the tRNA selection pathway that promote acceptance of the tRNA by the ribosome. How do specific molecular changes at the junction between the anticodon and the D stems promote these two specific measurable steps on the ribosome? And, ultimately, how does the ribosome interact with the tRNA structure during the process?
To further define the mechanisms by which the tRNA body can influence tRNA selection, we set out to isolate more potent miscoding mutations in tRNATrp that might allow us to discern physical perturbations more readily. To that end, we developed an in vitro selection scheme that takes advantage of the ability of a tRNA to catalyze a peptidyl transfer reaction with biotinylated Met-tRNAfMet. Miscoding tRNA variants isolated by this approach carry mutations at the junction of the D and anticodon stems (like the Hirsh mutant), and additionally a mutation in the T loop. We demonstrate that these mutations perturb overall tRNA tertiary structure, in particular, the flexible elbow region of the tRNA that approaches the functionally important helix 69 (H69) of the large ribosomal subunit (O'Connor and Dahlberg 1995; Valle et al. 2002). We then present a detailed characterization of the kinetic properties of these selected tRNA species and of rRNA variants in nearby H69 and propose a model to account for their coordinated effects on tRNA selection.
RESULTS
In vitro selection reveals new sites in tRNATrp that stimulate miscoding
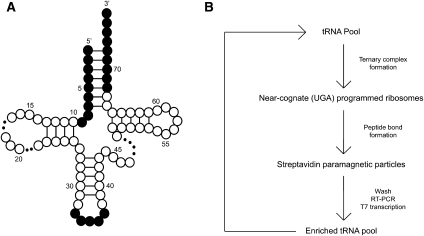
Our in vitro selection scheme relies on the reaction of a biotinylated P-site substrate (Biotin-Met-tRNAfMet, or bMet) with a provided A-site substrate (ternary complex). To discover new sites in the body of tRNATrp that promote miscoding, we carried out an in vitro selection with a mutagenized pool of tRNATrp (cognate for the UGG codon) that must recognize a near-cognate UGA codon in the A site of the programmed ribosome complex. The mutagenized tRNATrp pool was generated from a randomized DNA oligonucleotide designed to conserve as WT the anticodon region (positions 33–37 in tRNATrp) and the acceptor stem (positions 1–9 and 68–76 in tRNATrp), thus biasing mutations to the body of the tRNA (Fig. 1A). Those tRNATrp species with elevated levels of miscoding are more likely to react with the near-cognate ribosome complex, such that the tRNATrp is effectively derivatized with biotin through the peptidyl tRNA linkage. Biotinylated tRNA species were then isolated on streptavidin paramagnetic particles and the now enriched population of miscoding tRNATrp variants generated by RT-PCR and T7 transcription. This process was iterated until the miscoding activity of the population was sufficiently enriched to characterize individuals in the population (see overview of selection in Fig. 1B). We note that the selection procedure was necessarily performed with in vitro–transcribed tRNA. To address concerns about the lack of post-transcriptional modifications (given their known importance for the fidelity of tRNA selection), prior to the selection we established that the Hirsh mutation (G24A) confers a miscoding phenotype to an in vitro transcript of tRNATrp.
FIGURE 1.
In vitro selection scheme. The randomized tRNA pool (A) was constructed by mutating residues 10–32 and 38–67 at a 6% frequency (white residues). The anticodon loop and acceptor stem were left unrandomized (shaded residues). A representative round of selection (B) begins when the randomized tRNA pool is aminoacylated in the presence of EFTu and mixed with ribosome complexes. Ternary complexes able to catalyze the reaction on the UGA codon are isolated with paramagnetic streptavidin particles. The now enriched pool of tRNA variants is generated via RT-PCR and T7 transcription, and the cycle iterated again.
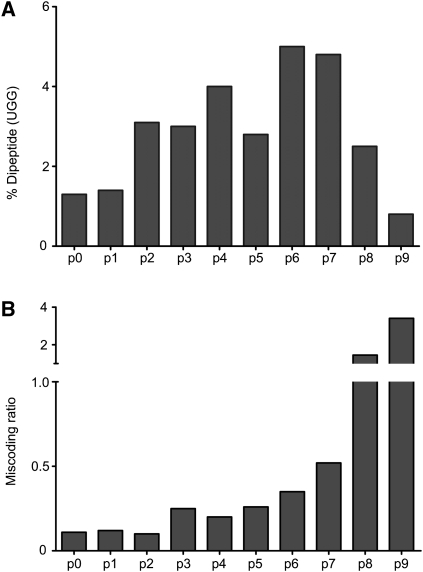
The results of nine rounds of selection are shown in Figure 2, where two parameters were evaluated: the overall extent of dipeptide formation on the UGG cognate codon (Fig. 2A) and the miscoding activity (Fig. 2B). The miscoding ratio is determined by the relative activity (amount of dipeptide formed) of the tRNA (or tRNA pool) on a UGA codon relative to the activity on a UGG codon; as anticipated, we see a clear increase in the miscoding activity from pool 0 to pool 9. However, sequencing of individual tRNAs within pool 9 revealed that most of the species that strongly “miscoded” on the near-cognate UGA ribosome complex in fact carried mutations in the anticodon of tRNATrp (CCA to UCA) that restore perfect pairing with the near-cognate codon (UGA). Indeed, analysis of the nine pools on the cognate UGG codon (Fig. 2A) revealed a decreasing ability of each pool after pool 6 to promote the cognate (UGG) peptidyl transfer reaction; this decrease correlates with an increasing number of tRNAs within these later pools carrying the undesirable anticodon mutation. As such, the best compromise between these properties was found in pool 6 where substantially increased levels of miscoding are seen, but where anticodon mutations have not yet taken over the population.
FIGURE 2.
Activity summary for nine rounds of in vitro selection (p0–p9). tRNA pools corresponding to the nine rounds of selection were evaluated (A) for the amount of dipeptide (fMet-Trp) formed on cognate (UGG)-programmed ribosome complexes and (B) for the amount of miscoding (calculated as a function of dipeptide formed on near–cognate (UGA) versus cognate (UGG)–ribosome complexes).
Identification of individual miscoding tRNA variants
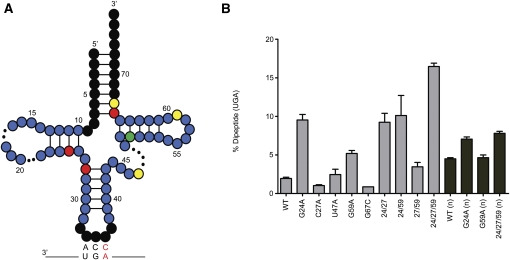
Forty individual clones from pool 6 that can be aminoacylated with Trp by the appropriate aminoacyl-tRNATrp synthetase are shown in Supplemental Figure 1. The compiled mutation frequency at each nucleotide position of tRNATrp is then represented in the secondary structure diagram of the tRNA (Fig. 3A). Nucleotides in black were conserved as WT in our pool design and, as anticipated, no mutations were found in these regions. Mutations appearing in fewer than 15% of the mutants are shaded blue and were not found to contribute significantly to miscoding. Positions in green were mutated in 15%–25% of clones, in yellow in 25%–49%, and in red in >50%. For these higher frequency positions, the mutations were always recovered in combination with one another, though no clear patterns of covariation were detected.
FIGURE 3.
Characterization of potent miscoding variants isolated by in vitro selection. The mutation frequency map (A) shows the frequency of mutation in fifty (n = 50) sequenced clones from pool 6. Nucleotides in black were not mutagenized in the experimental design. Colored nucleotides have a mutation frequency between 0% and 15% (blue), 15% and 25% (green), 25% and 49% (yellow), and >50% (red). (B) T7 transcribed tRNAs and the purified native (n) molecules were evaluated to determine the amount dipeptide formed on near-cognate (UGA)–programmed ribosome complexes after a 1-min incubation. The T7 transcripts (light gray) were assayed in HiFi buffer supplemented to 7 mM MgCl2 and the native tRNAs (dark gray) in HiFi buffer.
To determine which of the individual point mutations contribute to miscoding, individual tRNAs were transcribed from different clones and subjected to a simple miscoding assay (data not shown). These assays identified the most effective miscoders, and as such the most interesting individual mutations or combinations of mutations for further analysis. A representative selection of in vitro-transcribed tRNAs and their behavior in the miscoding assay is found in Figure 3B. Mutations C66U or G67C were present in every single tRNA sequence analyzed, although they make no apparent contribution to miscoding by tRNATrp (the data are shown for G67C). We speculate that these mutations aid in the reverse transcription step of the selection scheme, effectively lowering the melting temperature of the acceptor stem of tRNATrp and thus facilitating binding of the reverse-transcription primer. Likewise, the U47A/G individual mutations resulted in no increases in miscoding by tRNATrp in our assay; here we offer no specific explanation for their enrichment during the selection procedure.
Several of the remaining positions that emerged, U11C, G24A, and C27A, have been previously identified in screens performed in vivo (Hirsh 1971; Schultz and Yarus 1994). The mutation at position 59 (G59A/U) has not been previously isolated and was found to have an intermediate miscoding phenotype on its own. This mutation was often found in a triple mutant, G24A, C27A, G59A (24/27/59), which we report as the most efficient miscoder in our library (significantly more potent than the previously isolated Hirsh variant). Interestingly, C27A resulted in no miscoding on its own but was essential for the function of the triple mutant (Fig. 3B); neither the G24A/C27A, G24A/G59A, nor C27A/G59A mutants were able to miscode as efficiently as the triple mutant (24/27/59). All mutants were also tested on the UGG codon (Supplemental Fig. 2) and, as expected, all retained their ability to decode their cognate codon effectively.
Thermodynamic and kinetic analysis
Based on the selection results, WT tRNATrp, G24A, G59A, and the triple mutant (24/27/59) were purified from an E. coli strain (MY87) carrying a temperature-sensitive allele of tRNATrp (Eisenberg et al. 1979) so that subsequent studies could be performed with in vivo–made post-transcriptionally modified tRNA. As a first step, the natural variant transcripts were evaluated in the standard assay, thus confirming that they exhibit increased miscoding ratios like their in vitro-synthesized counterparts (Fig. 3B). Insights into the mechanism by which these variant tRNAs are promiscuously selected by the ribosome were then obtained through the determination of a number of kinetic parameters (Gromadski and Rodnina 2004). Such measurements have been used extensively to determine the role of post-transcriptional modifications or mutations in the miscoding profile of a tRNA (Cochella and Green 2005; Kothe and Rodnina 2007; Pan et al. 2008; Ledoux et al. 2009).
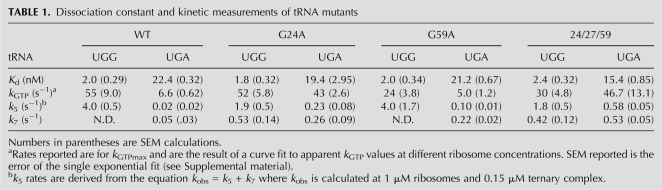
The equilibrium binding constant (Kd) was measured using an EFTu mutant (H84A) that hydrolyzes GTP at diminished rates (Daviter et al. 2003; Cochella and Green 2005; Ledoux et al. 2009). The Kd for each tRNA for the UGG cognate codon ribosome complex was ∼2 nM while it was ∼20 nM for the near-cognate UGA complex (Table 1; Supplemental Fig. 3); as anticipated, the affinity for the cognate ribosome complex is about 10-fold higher than for the near-cognate. Importantly, none of the variant tRNAs exhibit any large differences in Kd relative to WT tRNATrp that might account for the miscoding phenotype.
TABLE 1.
Dissociation constant and kinetic measurements of tRNA mutants
Rate constants for GTPase activation were determined with WT and variant tRNAs on the same ribosome complexes (programmed with mRNAs containing cognate [UGG] or near-cognate [UGA] codons in the A site). The GTPase activation rate (k3) was indirectly measured by following GTP hydrolysis, since activation is rate limiting for this event (Pape et al. 1999). Rates for single-turnover GTP hydrolysis were measured by mixing programmed ribosomes with a semipurified ternary complex (Ledoux et al. 2009) composed of EFTu:[γ-32P]GTP and either WT or variant Trp-tRNATrp and evaluating the amount of inorganic phosphate released over time (Cochella and Green 2005). GTP hydrolysis rates were measured at increasing ribosome complex concentrations, and pseudo-first-order rate constants (k3) were calculated from the extrapolation of hyperbolic fits to saturation (Table 1). We see that the measured k3 values on UGG-programmed ribosome complexes are similar for all tRNA variants tested (within twofold of one another ranging from 24 to 55 sec−1). In contrast, on the UGA-programmed ribosome complexes, the G24A and 24/27/59 variants exhibit substantially increased rates of GTPase activation relative to WT (by as much as sevenfold), such that the resulting values approach those typically observed on cognate UGG-programmed ribosome complexes. The G59A variant does not exhibit elevated levels of GTPase activation relative to WT in this assay, though we note that this native variant is a modest miscoder.
Finally, rate constants of the variant tRNAs for steps relevant to the proofreading stage of tRNA selection were determined. A rate-limiting conformational rearrangement known as “accommodation” dictates tRNA selection during the proofreading phase. As this conformational rearrangement is thought to be rate limiting for subsequent peptidyl transfer (Pape et al. 1999), we simply measure the latter event to determine the rate constant for accommodation (k5). Here, UGG- and UGA-programmed ribosomes now carrying f-[35S]-Met-tRNAfMet in the P site were mixed with the various ternary complexes (EFTu:GTP:Trp-tRNATrp), and the amount of fMet-Trp dipeptide formed over time was evaluated (Cochella and Green 2005). Since accommodation is at a branch point in the kinetic pathway, the observed rate constants obtained from ribosome titrations represent the sum (kobs = k5 + k7) of the individual rate constants for accommodation (k5) and rejection (k7). For typical cognate interactions (i.e., with UGG complexes), rejection is negligible and so the observed rate simply reflects accommodation (k5). For near-cognate interactions (i.e., with UGA complexes), the amount of dipeptide product formed is reduced, indicative of significant utilization of the discard pathway (fraction product = k5 / [k5 + k7]). These equations allow for determination of the values for k5 and k7 from direct measurements of peptide bond formation (see Table 1). For the cognate-programmed (UGG) complexes, all tRNA variants exhibited a rate of accommodation (k5) that falls within the expected range for a cognate decoding event (from 1.8 to 4.0 sec−1). On near-cognate-programmed (UGA) complexes, WT tRNATrp exhibits a low rate of 0.02 sec−1, while all three variant tRNAs substantially stimulate k5 from five- to 29-fold for G59A and 24/27/59, respectively.
We note that despite the fact that the Hirsh and triple mutants (G24A and 24/27/59) exhibit levels of GTPase activation and accommodation on a near-cognate complex nearly equivalent to those observed on a cognate one, the final extent of peptide bond formation for these variants is elevated, but still low. This apparent discrepancy is presumably explained by increased rates of rejection (k7) during the proofreading phase for each of the variant tRNAs (Table 1).
Structural analysis of tRNA variants
Structural changes in the variant tRNAs were next examined using a solution-based probing approach, in-line probing, that reports on an intramolecular “SN2-like” attack of the phosphate backbone by the adjacent 2′ OH of the ribose, a reaction that can be followed under very mildly basic conditions (pH 8.5) (Soukup and Breaker 1999). This type of chemical probing is generally useful for measuring the particular conformation of a given RNA linkage; those nucleotides involved in stable, canonical base-pairing or tertiary interactions are less likely to sample the favored “in-line” attack position while nucleotides not involved in such interactions (and that are typically more dynamic) can adopt the in-line configuration more often. While this method can sometimes identify regions of higher flexibility, it is more generally useful for defining structural differences between RNAs of interest.
We subjected the four tRNATrp species used in the kinetic studies (WT, G24A, G59A, 24/27/59) to in-line probing analysis in order to see if there might be structural correlations with their pronounced miscoding phenotypes. We note that the tRNATrp variants used for analysis are native, isolated directly from E. coli, and thus likely contain the relevant post-transcriptional modifications (Cochella and Green 2005). The tRNATrp variant species were radioactively labeled with [α-32P] ATP (Ledoux and Uhlenbeck 2008) and were incubated in a buffer at mildly elevated pH for 72 h at 25°C; products were resolved by 10% denaturing polyacrylamide gel electrophoresis (Fig. 4A). Paired regions of the tRNA including the acceptor, D, anticodon, and TΨC stems showed relatively little cleavage (as anticipated), while the most prominent cleavage sites were found within the anticodon and D loops and in the junction between the anticodon and D stems. To quantitate the cleavage pattern of the molecules and to account for any loading differences in the gel, we normalized each lane by the cleavage at position 34 (Fig. 4B). This residue is found in the anticodon loop and exhibits similar cleavage intensity for the four species being analyzed; these observations were consistent with our expectation that structural perturbations in more remote regions of the molecule (positions 24, 27, and 59) would be unlikely to directly impact the structure of the anticodon loop.
FIGURE 4.
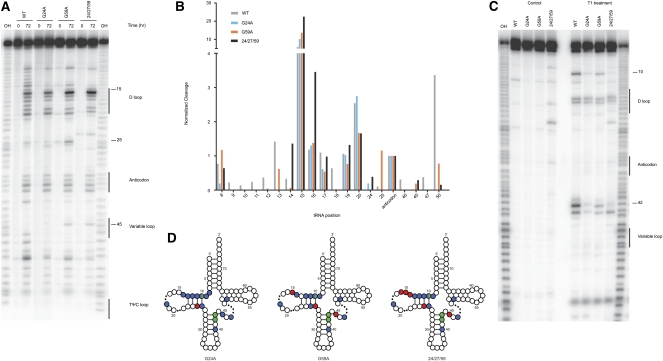
Structural analysis of variant tRNAs. (A) Ten percent PAGE in-line probing analysis where the indicated variant tRNAs were either incubated for 0 or 72 h at 25°C. Numbers highlight positions of interest in the cleavage pattern. Product identity was determined by comparison to a hydroxyl-mediated cleavage ladder (OH). (B) Quantitation of selected positions was performed using ImageQuant where the intensity of a band was normalized over the entire lane to account for loading differences and then normalized to cleavage at the same position (position 34 of the anticodon) for all variants tested. (C) Ten percent PAGE of T1 protection assay where the indicated variant tRNAs were either incubated with or without T1 nuclease. Numbers highlight positions of interest in the cleavage pattern. Product identity was determined by comparison to a hydroxyl-mediated cleavage ladder (OH). (D) Visual representation of structural data for the miscoding tRNA mutants. Sites of protection (blue circles) and enhanced cleavage (red circles) for the in-line probing assay and protection (green circles) for the T1 analysis are represented on a canonical tRNA clover-leaf structure (dots in the D and variable loops represent sites of varying length among tRNAs). Inclusion of sites was determined by comparison of individual positions to the WT molecule; the threshold for inclusion was a 2.5× difference from WT. Position 10 in all variants is protected in both assays.
What is most immediately striking in our analysis is the altered pattern of cleavage around position 15 for all three miscoding variants. While the WT pattern of cleavage in this region is diffuse, all three miscoding variants exhibit a more focused pattern of cleavage. Our most potent miscoder, 24/27/59, exhibits quantitatively stronger cleavage at positions 14, 15, and 16, suggesting that this difference may be correlated with its miscoding potency. Positions 24 and 25 show alterations in the cleavage pattern for all three variants, consistent with the fact that G24 is mutated in several variants and so this region is likely to be structurally perturbed. The G59A and 24/27/59 mutants also exhibit a slight increase in cleavage at position 45. Also striking in the analysis are the regions and residues where our miscoding variants exhibit less cleavage than WT tRNATrp. The region from residues 9–13 (as discussed above) and positions 18, 47, 40, and 50 all exhibit stronger cleavage intensity in the WT species. These data together suggest that the three miscoding variants share certain similar structural features (though there are differences in the extent to which these perturbations are observed) and are all somewhat distinct in structure from WT tRNATrp.
The idea that the isolated miscoding variants exhibit common structural perturbations was further tested using an RNase T1 cleavage assay. RNase T1 is an RNA-specific endonuclease that selectively cleaves at single-stranded guanine residues. Similar to the in-line probing experiments, the radioactively labeled tRNATrp variants were treated with RNase T1 and the digestion pattern analyzed by PAGE. As above, all three miscoding variants show a common pattern in the analysis with decreased susceptibility to cleavage at positions 10, 42, and 43 when compared with the WT tRNATrp species (Fig. 4C). Moderate protection at position 10 for the miscoding tRNA variants was also apparent in the in-line probing analysis; positions 42 and 43, found at the top of the anticodon stem, were previously inaccessible to in-line probing. Regions of protection (blue and green circles) and increased cleavage (red circles) for the miscoding tRNATrp variants, assessed by both in-line probing and T1 digestion assays, are mapped on a canonical tRNA secondary structure (Fig. 4D).
These results highlight the importance of the elbow region of the tRNA in dictating the properties of the molecule during tRNA selection. The structural changes that we document may be important only for the behavior of tRNA itself, or may also impact essential interactions with the ribosome as we explore below.
Kinetic analysis of H69 rRNA ribosome variants
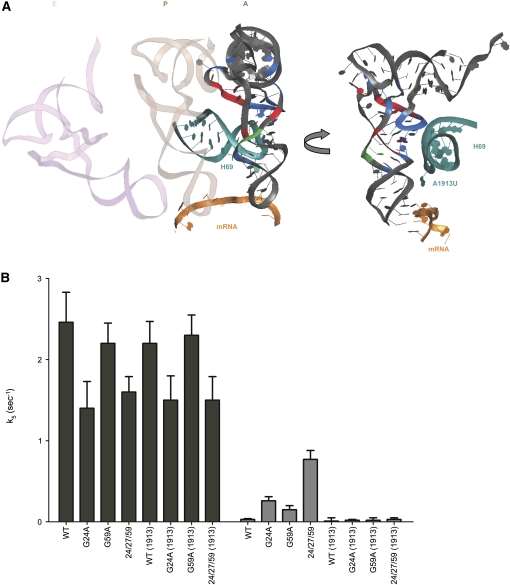
The structural changes exhibited by the tRNATrp-miscoding mutants are centered in the D stem and loop of the molecule, a region that undergoes large rearrangements upon binding to the ribosome. Based on simple analysis of the site of structural perturbation in the tRNA and the nearby sites in the ribosome (Li et al. 2008; Schmeing et al. 2009), we propose that mutations common among our miscoding variants allow for sampling of distinct conformational states during tRNA selection, establishing contacts with the intersubunit ribosomal H69 that promote promiscuous acceptance of near-cognate tRNA species. Ribosomal RNA nucleotides A1913 and C1914 at the tip of H69 (Fig. 5A) are prime candidates for playing a functionally critical role in tRNA selection (O'Connor and Dahlberg 1995; Ali et al. 2006; Schmeing et al. 2009).
FIGURE 5.
Contribution of large subunit rRNA components to tRNA selection. (A) The interactions between helix 69 (H69, teal) and a post-accomodation A-site tRNA on the ribosome are shown. The tip of H69 (residue A1913) is in close proximity to the D stem of the tRNA and the decoding center. tRNA residues highlighted (blue, green, or red) follow the same pattern as in Figure 4D. mRNA is represented in orange. E site (light pink) and P site (light brown) tRNAs are represented on the left for orientation. (B) Rates of accommodation (k5) for the formation of fMet-Trp dipeptide for WT, G24A, G59A, and 24/27/59 tRNATrp with WT and A1913U ribosomes on UGG (black bars) and UGA (gray bars) codons.
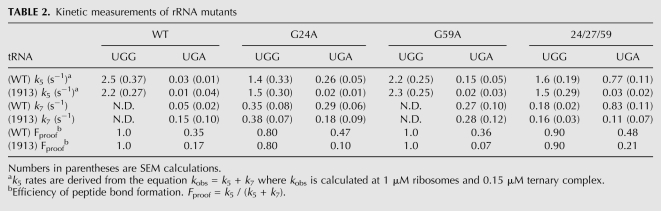
These ideas were directly tested by examining the properties of A1913U mutant ribosomes to see whether they exhibit defects in the tRNA selection process. Variant ribosomes were purified as previously described (Youngman and Green 2005) and peptidyl transfer activity was analyzed with WT, G24A, G59A, and 24/27/59 tRNATrp on cognate (UGG) and near-cognate (UGA) codons. The A1913U variant ribosomes exhibited no defects in the rate or extent of peptidyl transfer with any tRNA on the cognate UGG codon (Fig. 5B; Table 2; Supplemental Fig. 4) or with other cognate tRNA interactions (Supplemental Fig. 5). Consistent with our analysis above (Table 1), the triple mutant tRNATrp miscodes to a substantial extent on the near-cognate UGA codon (now with WT ribosomes carrying an aptamer purification tag, Youngman and Green 2005), exhibiting as much as 25-fold increases in the rate constant (k5) for peptidyl transfer. Here, however, we see striking differences as the variant A1913U ribosomes clearly fail to allow the same high level miscoding previously observed with the different miscoding tRNA variants (G24A, G59A, and 24/27/59 tRNATrp) (Fig. 5B). What is also evident is that even miscoding by the WT tRNATrp appears to be suppressed by the A1913U variant ribosomes; while the decrease in k5 (from 0.03 to 0.01 sec−1) is not above the error of the measurement, there are clearer effects on the endpoints of the reaction (from 0.35 to 0.17 for the Fproof value) (Table 2). Further evidence supporting the role of A1913 in standard tRNA selection (with natural tRNAs) is provided in Supplemental Figure 5 where miscoding by the somewhat promiscuous tRNA species, tRNALys, on AAU is suppressed in the A1913U background. These data suggest that the contacts made between tRNA and H69 of the ribosome are generally critical in promoting accommodation during the tRNA selection process.
TABLE 2.
Kinetic measurements of rRNA mutants
DISCUSSION
The isolation of robust miscoding tRNA variants through an in vitro selection approach has provided us with new insights into how the structural conformation of tRNA might be crucial to the process of tRNA selection. Earlier studies had suggested that a “sweet spot” in the tRNA structure at the junction between the D and anticodon stems might be critical for spring-like action by the tRNA important for decoding (Valle et al. 2002; Li et al. 2008). This structural insight was given some biochemical explanation in studies that followed, showing that the Hirsh variant tRNATrp (G24A) stimulated miscoding through concerted effects on the forward rate constants of GTPase activation and tRNA accommodation (Cochella and Green 2005). While more recent studies have provided additional evidence for the importance of the tRNA body in specifying the highly discriminatory process of tRNA selection (Pan et al. 2008; Ledoux et al. 2009; Murakami et al. 2009), what remains poorly understood is how the physical properties of the tRNA body make it suited for the process. Is overall tRNA flexibility generally correlated with the level of miscoding, or do the variant tRNAs that have been studied alter or establish specific contacts with the ribosome that promote steps in the tRNA selection pathway?
On the one hand, given the distortion of tRNA seen in the A/T bound ribosome structures (Valle et al. 2002; Schmeing et al. 2009), it seems reasonable to think that an increase in flexibility might facilitate tRNA loading into this state (and presumably stimulate k3). On the other hand, the accommodation step (k5) involves the release of the tRNA from EFTu and a swinging motion into the A/A state, an action that might be facilitated by a more rigid, less flexible tRNA. While these ideas are appealing and have been discussed in the literature (Schmeing and Ramakrishnan 2009), there is currently no information that addresses the question of how tRNA flexibility affects tRNA selection. How is the structure of tRNA so well suited for these different steps and their potentially different requirements? It has been argued that tRNA flexibility is increased in the Hirsh variant miscoding tRNATrp based on the biochemical observation that the miscoding activity is suppressed by a UV-induced crosslink in the tRNA between s4U8 and C13 that tethers the D- and acceptor-stems (Vacher and Buckingham 1979). While these observations are striking, their impact has been limited by the very specific nature of the experiments. What we also know is that mutations in the tRNA that increase miscoding typically stimulate both GTPase activation and accommodation, suggesting that certain key features of the tRNA are important for both steps of tRNA selection. Perhaps both key steps of tRNA selection (GTPase activation and accommodation) depend either on increased or decreased flexibility in the tRNA substrate.
As we initiated our study, we reasoned that the Hirsh variant tRNATrp (G24A) that had been isolated in an in vivo screen for stop codon read-through had relatively mild effects on miscoding because the organism would need to survive in the presence of this variant tRNA. By conducting an in vitro screen where cell viability was not demanded, we anticipated that more potent miscoding tRNA variants might be isolated, and might lead to more easily discernible physical perturbations. While our in vitro selection screen was successful in identifying novel miscoding tRNA variants, we were surprised to observe that G24A is one of the most common mutations found in the enriched population of tRNAs. Additionally, we identified positions 27 and 59 as contributors to miscoding in a triple mutant variant (with mutations at positions 24, 27, and 59) more potent than any isolated in previous in vivo screens. Position 27 is located at the top of anticodon stem of the tRNA, and on the opposite side of the coaxially stacked helix (that includes the D and anticodon stems) from position 24. Position 59 is located within the T loop in a region where in the corresponding regions of the tRNAPhe structure (Shi and Moore 2000) interactions are formed with the D loop, thus helping to specify the global L shape of the tRNA.
Our biochemical characterization of this potent miscoding tRNA variant indicated that, like the Hirsh suppressor tRNATrp, it promotes miscoding through the acceleration of forward rate constants (both GTPase activation and accommodation) in the tRNA selection process. Apparently the most effective means by which a tRNA can increase its rate of acceptance into the ribosome (in the A/A state) is through manipulation of these kinetic determinants, not through increases in overall binding affinity for the A site (Cochella and Green 2005). Interestingly, the modest miscoder G59A appears to have increased rates of tRNA accommodation, but not GTPase activation, thus potentially isolating the features required for these two steps.
Of particular interest in our analysis was the structural alteration of the variant tRNAs as measured through in-line chemical probing and T1 nuclease digestion assays. In the in-line probing analysis, we observe broadly similar patterns of cleavage in the region of the D-stem and loop for the three miscoding tRNATrp variants; overall, the pattern of cleavage is more diffuse for the wild-type tRNA than for any of the miscoding variants. Likewise, the results from our T1 protection assay show a common conformational change for all three miscoding tRNA variants, highlighted by protection at residues 42 and 43, at the top of the anticodon stem, and at residue 10, at the base of the D-stem. The observation of protected regions in the miscoding tRNA class is broadly consistent with the hypothesis that these variants have decreased flexibility (at least in these regions).
We also see enhanced cleavage for the miscoding tRNA variants at a number of positions. Position 15 is a hot spot for cleavage in the tRNA and is even more strongly targeted for cleavage in the miscoding variants. This is the precise region where the UV-induced crosslink that suppresses miscoding in the Hirsh variant is known to form. This region is quite close, in the three-dimensional structure, to position 59 where our novel mutation is found. Another site of increased cleavage for the miscoding G59A and 24/27/59 mutants is position 45, found on the other face of the tRNA molecule (in the joint region between the anticodon and T stems) where increased flexibility could impact the overall conformation of the tRNA during decoding. This site of enhanced modification is also relatively close in three dimensions to positions 24 and 27 that are mutated in this triple tRNATrp variant.
Given these data, we searched for a direct correlation between the kinetic and structural phenotypes presented by our miscoding variants. What is immediately apparent is that all three molecules exhibit both an increase in the rate of accommodation (but not GTPase activation) and an overall similar pattern of cleavage and protection during our structural analysis. If the hypothesis that decreased flexibility facilitates the transition to the A/A state (k5) is valid, then the shared cleavage protection of the D-stem (which hints at a decrease in flexibility) could reflect the behavior of a structural “motif” responsible for this kinetic phenotype.
On the other hand, if the hypothesis that increased flexibility facilitates tRNA binding and function in the A/T state (k3) is valid, then some of the pertubations in the cleavage pattern for the miscoding variants could reflect the behavior of motifs responsible for this kinetic phenotype. Reassuringly, some of the intramolecular interactions within the tRNA that are disrupted in the A/T state (27:43, 24:45, 15:48, and 16:59) are the same ones that exhibit structural differences for our miscoding variants.
The A/T state is stabilized by a number of interactions between the tRNA and the ribosome. The region of our miscoding variants where we see the greatest structural changes is the D stem which makes a contact with the tip of universally conserved H69 on the 50S subunit. H69 has been implicated through various means in initiation, elongation, termination, and recycling (O'Connor and Dahlberg 1995; Ali et al. 2006; Weixlbaumer et al. 2007), and its high level of sequence conservation further highlights its potential importance (Noller 2005). CryoEM and crystal structures of tRNA in a pre- (Valle et al. 2002; Schmeing et al. 2009) and post (Schuette et al. 2009)-accommodation state show H69 contacting tRNA around positions 25 and 26 of the D stem and position 38 in the anticodon stem, respectively. These positions that contact the tip of H69 (25/26 and 38) are relatively close to one another in the three-dimensional structure of tRNA; intriguingly, mutations in both regions of the tRNA have been shown to promote miscoding on the ribosome (Cochella and Green 2005; Ledoux et al. 2009; Murakami et al. 2009).
Genetic screens have identified a number of mutations in H69 that result in general losses in fidelity (a ram phenotype) as assessed by in vivo-based reporters for frameshifting and stop-codon read-through (O'Connor and Dahlberg 1995; Hirabayashi et al. 2006). Recently, further in vivo characterization of a number of H69 variants indicated that certain mutations in this region decrease UGA codon read-through while others increase the same event (O'Connor 2009). The authors argue that these effects can in large part be explained as defects in the termination process (mediated by class 1 RFs), thus supporting earlier characterizations of ΔH69 (Ali et al. 2006). It also seemed likely that H69 is critical to tRNA selection, an idea supported by the observed mild restrictive phenotype of ΔH69 (Ali et al. 2006). We emphasize, however, that the involvement of H69 in tRNA selection had not been previously defined in an assay that isolates individual steps in the translation cycle (a pre-steady state approach).
Here we show that ribosomes carrying a single nucleotide change (A1913U) in H69 of the large ribosomal subunit are indeed disrupted in specific steps in the tRNA selection process. While these mutant ribosomes appeared to behave indistinguishably from their WT counterparts with cognate anticodon:codon interactions, they fail to permit the more rare miscoding events on near-cognate codons (both with wild type tRNATrp and tRNALys, as well as with some miscoding variants). These observations can be classified as “synthetic effects” where the defects of the A1913U ribosomes are only visible on the already compromised reaction of near-cognate tRNA selection. These ribosomes thus have a classically restrictive phenotype, generally making fewer mistakes during the overall process of translation. It should be noted that, unlike the better characterized rpsL variants (Zaher and Green 2010), the A1913U ribosomes exhibit no apparent defects during cognate tRNA incorporation. We argue that a restrictive phenotype is the likely outcome of site-directed mutations targeting key contact points for tRNA selection, since certain steps of closure that are critical to forward steps in the process are likely made less efficient (Sharma et al. 2007).
Our in vitro selection approach allowed us to identify tRNATrp variants that more potently miscode and would thus be classified as having a ribosomal ambiguity (or ram) phenotype. These variants carry gain of function mutations that are typically more difficult to predict, even with high-resolution structures at hand (Schuette et al. 2009). Our studies highlight the importance of direct interactions between the tRNA and key bridge elements of the ribosome in promoting high-fidelity tRNA selection during elongation, as well as the critical importance of detailed biochemical assays to identify biologically meaningful molecular interactions.
EXPERIMENTAL PROCEDURES
Buffers and reagents
Ribosomes were isolated from E. coli MRE600 cells during exponential growth and purified as previously described (Moazed and Noller 1986). All experiments were carried out in HiFi buffer (50 mM Tris-HCl [pH 7.5] with 70 mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 0.5 mM spermidine, 8 mM putrescine, and 2 mM DTT) at 20°C (from Gromadski and Rodnina 2004) unless otherwise noted. mRNA for the in vitro selection and for the kinetic assays was prepared by PCR and T7 transcription as previously described and featured an epsilon sequence, a Shine-Dalgarno sequence, and a 7-nucleotide spacer followed by the coding sequence AUGUGAUAAUUGCAGAAAAAA (where AUG is the initiation codon and UGA is the stop codon). EFTu and TrpRS were purified as previously described (Cochella and Green 2005).
tRNA pool construction
A mutagenized 76-mer oligonucleotide encoding the tRNATrp sequence was obtained from IDT-DNA; the first and last 9 nucleotides of the fragment were left unmodified as were bases 33–37. These regions correspond to the acceptor stem and anticodon domains in the tRNA species. Nucleotides 10–32 and 38–67 were randomized with a frequency of 6% per nucleotide to yield a pool where individuals carried mostly one, two, or three mutations. All possible triple mutants were calculated to be included within the pool. A T7 promoter sequence was added to the population through PCR and the product was subsequently transcribed. Purification of the tRNATrp pool included nucleotide removal through a NAP-10 column (GE), DNAase I treatment, and acrylamide gel purification. tRNA was resuspended in water at a final concentration of 10 μM.
Biotinylation of tRNAfMet
E. coli tRNAfMet (Sigma) was aminoacylated with methionine and resuspended in 20 mM KOAc with 1 mM DTT. Sulfo-NHS-LC-Biotin (Pierce) was resuspended at a concentration of 3 mg/mL in 1 mL of 20 mM Na-phosphate, pH 7.0. Typically, 100 μL of tRNAfMet was added to 1 mL of biotin solution and the reaction was allowed to proceed overnight at 16°C. Biotinylated tRNAfMet was recovered by phenol/chloroform extraction and ethanol precipitation. The tRNA was resuspended in 20 mM KOAc and 1 mM DTT at a concentration of 40 μM.
In vitro selection
Ribosome initiation complexes were formed by incubating 4 μM ribosomes, 4 μM biotinylated tRNAfMet, 15 μM mRNA, and 1.5 mM GTP in HiFi buffer for 1 h at 37°C. Ternary complexes (TC) of the tRNA pool were formed by combining 4 μM tRNATrp, 10 μM EF-Tu, 2 μM TrpRS, 1.5 mM GTP, 1.5 mM ATP, 3 mM phosphoenol pyruvate, 0.1 mg/mL pyruvate kinase, and 20 mM L-Trp in HiFi buffer for 1 h at 37°C. Equal amounts (typically 25 μL) of each complex were mixed for 1 min at 25°C and the reaction was quenched with 300 μL stop buffer (0.3 M NaOAc, 12.5 mM EDTA, 0.25% SDS). The sample was incubated for 20 min at 37°C under vigorous shaking and then extracted with phenol/chloroform, precipitated with ethanol, and resuspended in 400 μL of 1× PBS, pH 5.75. To this mixture, 30 μL of Streptavidin MagneSphere Paramagnetic Particles (Promega) that had been pre-equilibrated in 1× PBS, pH 5.75, was added and incubated for 1 h at 25°C. After incubation, particles were magnetically separated and washed with 5 mL of 1× PBS with 0.1% SDS. Particles were resuspended in 3.8 μL water and 2.2 μL of 4.5× hybridization buffer (225 mM K-HEPES, pH 7.0 with 450 mM KCl). Four microliters of 20 μM 3′ primer was added and the sample was heated for 2 min at 95°C and then snap-cooled. Extension buffer (13 mM Tris-HCl, pH 8.5, 10 mM MgCl2, 100 mM DTT), 0.25 mM dNTPs, and 500 U Reverse Transcriptase (Invitrogen) were added and the reaction was incubated for 20 min at 42°C. After the reverse transcription reaction, PCR and T7 transcription were performed as described above. Peptidyl transfer analysis of pools and individual clones made during the selection were performed in HiFi buffer with 10 mM MgCl2. mRNA-programmed ribosomes (1 μM) preloaded with f-[35S]-Met-tRNAfmet were incubated with 0.15 μM TC for 1 min at 25°C and quenched with KOH. Products were resolved by electrophoretic TLC as previously described (Youngman et al. 2004).
Native tRNATrp and kinetic analysis
Native tRNATrp and mutants were isolated and purified from E. coli strain MY87 as previously described (Cochella and Green 2005). Thermodynamic and kinetic analysis of tRNA mutants was carried out largely as previously described (Cochella and Green 2005; Kothe and Rodnina 2007; Ledoux et al. 2009) with the following exceptions: The rate of peptide bond formation was determined from a time course where ribosome complexes (at 1 μM) preloaded with f-[35S]-Met-tRNAfmet and 0.15 μM ternary complex were mixed together in a Kin-Tek rapid quench apparatus. The resulting rate is presumed to be the rate at saturating conditions of initiation complex based on earlier studies (Cochella and Green 2005).
Structural analysis of tRNA variants
Native tRNAs were made and purified using standard procedures. 3′ radiolabeling was performed as previously described (Ledoux et al. 2009). For in-line probing experiments, ∼12 pmol of labeled tRNA was incubated in 20 mM Tris-HCl (pH 8.5) and 3.5 mM MgCl2 for a period of 72 h at 25°C. Reactions were quenched with an equal volume of gel loading buffer (80% formamide, 10 mM EDTA), and the products were resolved by 10% denaturing PAGE. T1 nuclease digestion was performed by incubating 12 pmol of labeled tRNA in HiFi buffer with two units of nuclease T1 for 15 min at 25°C.
Mutant ribosome analysis
Mutant ribosomes were prepared and purified as previously described (Youngman and Green 2005). The peptide bond formation assay was performed as previously described herein with the following changes: Purified 30S ribosomal subunits (3 μM) were added to 2 μM purified tagged 70S ribosomes during initiation complex formation to account for initiation defects in the A1913U mutant. All other components remain unchanged.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank L. Cochella for early contributions to the project, S. He for comments and help creating the figures, and A. Nahvi, M.E. McDonald, and H. Zaher for helpful comments on the manuscript. We also thank the U.S. National Institutes of Health for funding (GM059425) and the Howard Hughes Medical Institute for financial support (to R.G).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2232710.
REFERENCES
- Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF 2006. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell 23: 865–874 [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R 2005. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308: 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV 2003. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol 332: 689–699 [DOI] [PubMed] [Google Scholar]
- Eisenberg SP, Soll L, Yarus M 1979. The purification and sequence of a temperature-sensitive tryptophan tRNA. J Biol Chem 254: 5562–5566 [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13: 191–200 [DOI] [PubMed] [Google Scholar]
- Hirabayashi N, Sato NS, Suzuki T 2006. Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J Biol Chem 281: 17203–17211 [DOI] [PubMed] [Google Scholar]
- Hirsh D 1971. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol 58: 439–458 [DOI] [PubMed] [Google Scholar]
- Hopfield JJ 1974. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci 71: 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe U, Rodnina MV 2007. Codon reading by tRNAAla with modified uridine in the wobble position. Mol Cell 25: 167–174 [DOI] [PubMed] [Google Scholar]
- Ledoux S, Uhlenbeck OC 2008. [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S, Olejniczak M, Uhlenbeck OC 2009. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat Struct Mol Biol 16: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Agirrezabala X, Lei J, Bouakaz L, Brunelle JL, Ortiz-Meoz RF, Green R, Sanyal S, Ehrenberg M, Frank J 2008. Recognition of aminoacyl-tRNA: A common molecular mechanism revealed by cryo-EM. EMBO J 27: 3322–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF 1986. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47: 985–994 [DOI] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Suga H 2009. Bases in the anticodon loop of tRNA(Ala)(GGC) prevent misreading. Nat Struct Mol Biol 16: 353–358 [DOI] [PubMed] [Google Scholar]
- Noller HF 2005. RNA structure: Reading the ribosome. Science 309: 1508–1514 [DOI] [PubMed] [Google Scholar]
- O'Connor M 2009. Helix 69 in 23S rRNA modulates decoding by wild type and suppressor tRNAs. Mol Genet Genomics 282: 371–380 [DOI] [PubMed] [Google Scholar]
- O'Connor M, Dahlberg AE 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J Mol Biol 254: 838–847 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Pan D, Zhang CM, Kirillov S, Hou YM, Cooperman BS 2008. Perturbation of the tRNA tertiary core differentially affects specific steps of the elongation cycle. J Biol Chem 283: 18431–18440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O, Pape T, Pleiss JA, Wintermeyer W, Uhlenbeck OC, Rodnina MV 2000. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry 39: 1734–1738 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. 2009. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 28: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DW, Yarus M 1994. tRNA structure and ribosomal function. I. tRNA nucleotide 27-43 mutations enhance first position wobble. J Mol Biol 235: 1381–1394 [DOI] [PubMed] [Google Scholar]
- Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R 2007. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol 374: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moore PB 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: A classic structure revisited. RNA 6: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher J, Buckingham RH 1979. Effect of photochemical crosslink S4U(8)-C(13) on suppressor activity of su+ tRNATrp from Escherichia coli. J Mol Biol 129: 287–294 [DOI] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J 2002. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V 2007. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol 14: 733–737 [DOI] [PubMed] [Google Scholar]
- Youngman EM, Green R 2005. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods 36: 305–312 [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: 589–599 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009a. Fidelity at the molecular level: Lessons from protein synthesis. Cell 136: 746–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009b. Quality control by the ribosome following peptide bond formation. Nature 457: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2010. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol Cell 39: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]