Abstract
Modified nucleosides of ribosomal RNA are synthesized during ribosome assembly. In bacteria, each modification is made by a specialized enzyme. In vitro studies have shown that some enzymes need the presence of ribosomal proteins while other enzymes can modify only protein-free rRNA. We have analyzed the addition of modified nucleosides to rRNA during ribosome assembly. Accumulation of incompletely assembled ribosomal particles (25S, 35S, and 45S) was induced by chloramphenicol or erythromycin in an exponentially growing Escherichia coli culture. Incompletely assembled ribosomal particles were isolated from drug-treated and free 30S and 50S subunits and mature 70S ribosomes from untreated cells. Nucleosides of 16S and 23S rRNA were prepared and analyzed by reverse-phase, high-performance liquid chromatography (HPLC). Pseudouridines were identified by the chemical modification/primer extension method. Based on the results, the rRNA modifications were divided into three major groups: early, intermediate, and late assembly specific modifications. Seven out of 11 modified nucleosides of 16S rRNA were late assembly specific. In contrast, 16 out of 25 modified nucleosides of 23S rRNA were made during early steps of ribosome assembly. Free subunits of exponentially growing bacteria contain undermodified rRNA, indicating that a specific set of modifications is synthesized during very late steps of ribosome subunit assembly.
Keywords: modified nucleosides, ribosome assembly, chloramphenicol, erythromycin, rRNA
INTRODUCTION
Ribosome assembly involves several coordinated reactions. Folding, nucloelytic processing, and post-transcriptional modification of rRNA occur concomitantly with the r-protein association with the rRNA. Modification of rRNA is thereby an integral part of ribosome assembly process. Escherichia coli 16S rRNA contains 11 and 23S rRNA contains 25 modified nucleosides (Ofengand and Del Campo 2004). Eleven pseudouridines and 21 base methylations make up the bulk of modified nucleosides in E. coli rRNA. Each rRNA modification is made by a specific enzyme. In E. coli there are 32 rRNA modification enzymes in total, 25 of them are methyltransferases and seven pseudouridine synthases. All the pseudouridine synthases and most of the rRNA methyltransferases have been identified (Ofengand and Del Campo 2004; Purta et al. 2009). The substrate specificity of the rRNA modification enzymes has been studied mostly by cell-free experiments using purified enzymes. The specificity of specific rRNA modification has been shown to depend on the presence of r-proteins (Ofengand and Del Campo 2004). Substrate specificities of rRNA modification enzymes with respect to ribosome assembly are summarized in Table 1.
TABLE 1.
Modified nucleosides in Escherichia coli rRNAs and the specificities of the corresponding enzymes
Ribosome assembly in bacteria is fast and efficient. In wild-type bacteria ribosome subunits are formed within 2–3 min at 37°C (Lindahl 1975). Chloramphenicol is known to inhibit assembly of both ribosome subunits (Dagley and Sykes 1959; Kurland et al. 1962). More recently many other antibiotics were shown to cause ribosome assembly defects (for review, see Champney 2006). Chloramphenicol and erythromycin were shown to inhibit assembly of both ribosome subunits due to the unbalanced synthesis of ribosomal components (Dodd et al. 1991; Siibak et al. 2009). In the presence of these antibiotics incompletely assembled ribosome subunits accumulate in the cells (Siibak et al. 2009). Here we report the results of quantitative analysis of modified nucleosides of 16S and 23S rRNA in the antibiotic-induced ribosomal particles stalled at different assembly stages using reverse-phase, high-performance liquid chromatography (HPLC). Pseudouridines at specific positions of 23S rRNA were identified by the chemical modification/primer extension method. The results allowed dividing the rRNA modification enzymes with respect to ribosome assembly in vivo into three classes: early, intermediate, and late assembly specific enzymes.
RESULTS
Isolation of “chloramphenicol and erythromycin particles”
In order to analyze the temporal relationship between rRNA modification and ribosome subunit assembly, rRNA from incompletely assembled ribosomal subunits was analyzed with respect to the modified nucleoside composition. Accumulation of assembly defective ribosomal subunits was induced by chloramphenicol or erythromycin in an exponentially growing E. coli culture.
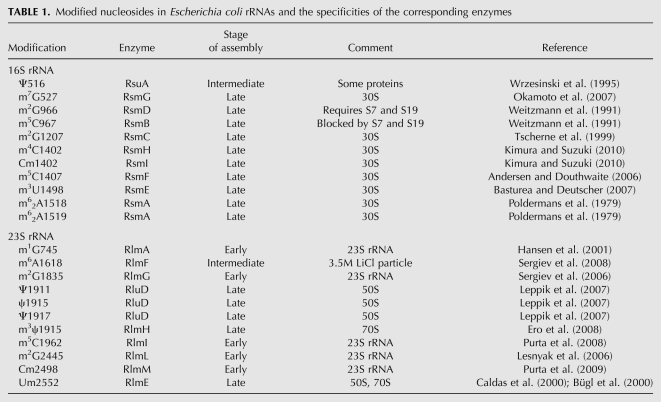
Ribosomal particles were separated by sucrose gradient centrifugation (Fig. 1). Addition of chloramphenicol or erythromycin to the growth medium leads to the appearance of three unusual ribosomal particles termed the 25S, 35S, and 45S particles. 35S and 45S particles are related to the 50S subunit and 25S particles are related to the 30S subunit (Fig. 1), in agreement with the earlier observations (Usary and Champney 2001; Siibak et al. 2009). In the absence of drugs the ribosome small subunit has one precursor (21S) and the large subunit has two precursors (34S and 43S) (Nierhaus 1991). It is possible that the 25S, 35S, and 45S assembly defective particles formed upon addition of antibiotics are related to the precursor particles found in vivo. The precursor rRNA from the “chloramphenicol particles” is converted into normal 30S and 50S ribosomes without prior degradation upon removal of the drug (Nomura and Hosokawa 1965; Adesnik and Levinthal 1969). Therefore, the subribosomal particles formed in the presence of chloramphenicol are ribosome subunit assembly intermediate particles.
FIGURE 1.
Isolation of ribosomal particles from E. coli grown in the presence of chloramphenicol or erythromycin or in the absence of a drug. Exponentially growing bacterial cells without the drug (A), or treated with chloramphenicol (B), or erythromycin (C) were lysed and centrifuged in 10%–25% sucrose. 45S, 35S, and 25S fractions of chloramphenicol and erythromycin treated cells were combined (only chloramphenicol particles are indicated by gray zones in B). Ribosomal particles were purified by a second sucrose gradient centrifugation (D–F). Indicated fractions were collected.
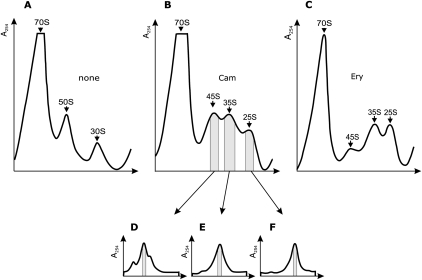
No free 30S or 50S subunits were found in the presence of chloramphenicol or erythromycin (Fig. 1), suggesting that the limiting step of ribosome subunit assembly in the presence of protein synthesis inhibitors is ribosomal protein production. Sucrose gradient fractions containing particles of interest were combined, concentrated by ultrafiltration, and repurified by a second sucrose gradient. It is worth mentioning that sedimenting the subribosomal particles leads to hardly soluble pellet. Therefore, ultrafiltration is the preferable method for concentrating the subribosomal particles. The second sucrose gradient centrifugation yielded homogenous particles. Isolation of chloramphenicol induced particles is shown in Figure 1D–F. rRNA was obtained by phenol extraction. The 25S and 45S particles contained only 16S and 23S rRNA, respectively (data not shown). The 35S particles contained a mixture of 16S and 23S rRNAs. The two rRNA species were separated from each other by sucrose gradient centrifugation (Fig. 2B).
FIGURE 2.
Purification of ribosomal RNA by sucrose gradient centrifugation. Ribosomal RNA from 70S ribosomes (A) and 35S particles (B) was deproteinized by phenol extraction and centrifuged in 5%–20% (w/w) sucrose gradient (buffer 20 mM Na-acetate, 100 mM NaCl, 1 mM EDTA) at 25,700 rpm in an SW28 rotor (Beckman) for 16 h. 16S and 23S rRNA were collected as indicated by the gray zones.
Free 30S and 50S subunits were isolated from exponentially growing E. coli cells (at 25°C) by two consecutive sucrose gradient centrifugation methods as described above for subribosomal particles. It must be noted that the free ribosome subunits in these conditions are mostly assembly intermediate particles (∼80%) with low functional activity (Peil et al. 2008). The limiting step of large ribosome subunit assembly is the final maturation at the level of the 50S particles (Lindahl 1975; Peil et al. 2008). Therefore, it was interesting to analyze the modified nucleoside composition of rRNA of the free 30S and 50S subunits. Ribosomal RNA was deproteinized by phenol extraction. Mature 16S and 23S rRNA species were isolated from 70S ribosomes by phenol extraction followed by sucrose gradient centrifugation (Fig. 2A).
Modification of 16S rRNA during 30S subunit assembly
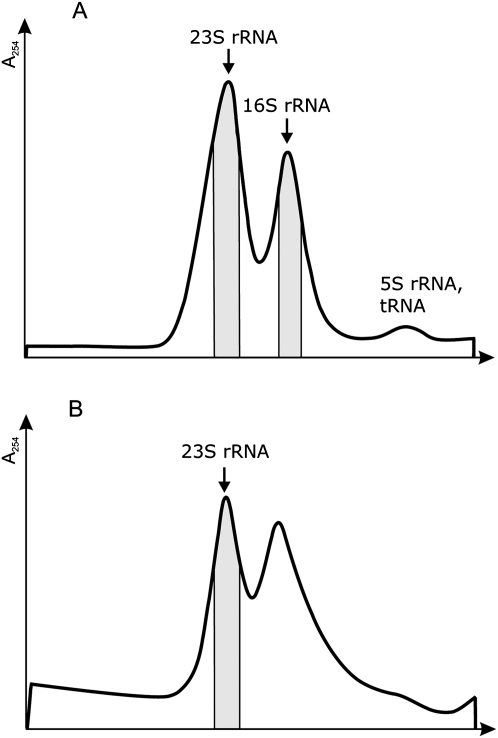
Analytical reverse-phase HPLC (RP-HPLC) allows identifying all nucleotides of E. coli 16S rRNA and nearly all nucleosides of E. coli 23S rRNA (Gehrke and Kuo 1989). Moreover, this method enables quantitative estimation of nucleosides. Therefore we have used RP-HPLC for determination of modified nucleosides in rRNA species. Nucleosides were prepared by treatment of the rRNA with nuclease P1 and bacterial alkaline phosphatase. Chromatographic peak assignments to the specific modified nucleosides were derived from relative retention times according to Gehrke and Kuo (1989). The A260/A280 ratio was used to confirm the peak identities. The RP-HPLC peak surface area corresponding to each nucleoside was calculated and compared with the respective nucleoside peak area of mature 16S rRNA of 70S ribosomes (Fig. 3; Table 2). Mature 16S rRNA isolated from 70S ribosomes has been shown to contain stochiometric amounts of modified nucleosides within 10% error limit (Gehrke and Kuo 1989). At least three independent particle preparations were analyzed with respect to modified nucleoside content.
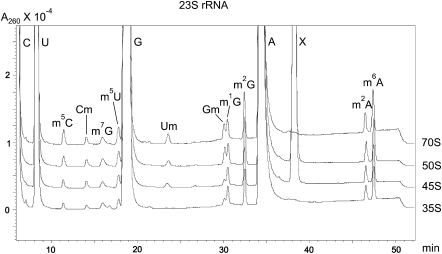
FIGURE 3.
HPLC analysis of 16S rRNA. 16S rRNA was prepared from mature 70S ribosomes, free 30S subunits, and Cam 25S particles. Nucleoside composition was determined by RP-HPLC on a Supelcosil LC-18-S. Peaks corresponding to three standard nucleosides (U, G, and A) and m5C, m7G, m3U, m4Cm, m2G, and m22A are indicated. X corresponds to an unknown compound.
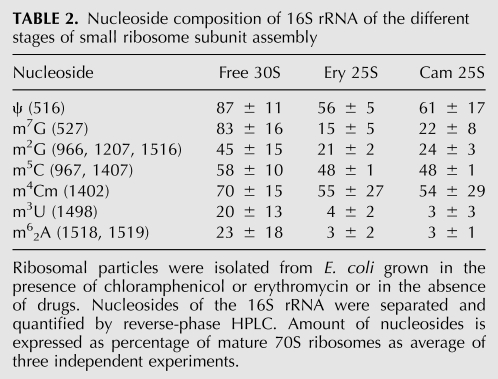
TABLE 2.
Nucleoside composition of 16S rRNA of the different stages of small ribosome subunit assembly
The 25S particles formed in the presence of Cam or Ery contain only 16S rRNA that is incompletely processed (Siibak et al. 2009). 16S rRNA of both Cam and Ery 25S particles clearly contain several modified nucleosides, albeit at a lower level compared with the mature 30S particles (Fig. 3; Table 2). Both Cam and Ery 25S particles have similar nucleoside composition (Fig. 3; Table 2). Pseudouridine, m5C, and m4Cm are present in 25S particles between 40% and 60% compared with the mature 16S rRNA. m7G and m2G are found in 25S particles in <25%. m5U and m62A are found in 25S particles only in trace amounts (Table 2). The results were well reproducible, except in m4Cm, which exhibited significant variation in different preparations. Thus, 16S rRNA is modified only at low levels during early events of 30S subunit assembly.
In the bacteria grown at 25°C, 70%–80% of the free 30S particles contain the 16S rRNA precursor with 115 extra nucleotides at the 5′ end (Siibak et al. 2009), indicating that the majority of these particles are incompletely assembled precursors of the ribosome small subunit. The modification level of 16S rRNA is still incomplete compared with the mature rRNA. Free 30S subunits contain nearly 90% of pseudouridine and 80% of m7G (Table 1). Thus the majority of these modifications are introduced into 16S rRNA during intermediate stages of small subunit assembly. Formation of m4Cm seems also to occur during intermediate assembly stages, although only 70% was found in the free 30S fraction and a large variability of results was observed (Table 2). m5U and m62A are present in the free 30S particles at ∼20% level (Table 2), which is about the same fraction as the mature 5′ end of 16S was found. Therefore, m5U and m62A can be classified as late assembly specific modifications. m2G (positions G966, G1207, and G1516) and m5C (C967 and C1407) are present at multiple positions of E. coli 16S and therefore cannot be clearly assigned to a specific ribosome assembly stage.
Modification of 23S rRNA
23S rRNA was isolated from the Ery- and Cam-induced 35S and 45S assembly intermediate particles, from the free 50S subunits, and from the mature 70S ribosomes of untreated cells (Figs. 1, 2). It must be noted that the free 50S subunits are in the majority (80%) assembly intermediate particles (Peil et al. 2008; Al Refaii and Alix 2009), The RP-HPLC method used allows identifying all 23S rRNA modifications except three. m3Ψ (present at position 1915 of 23S rRNA) and m5C (1962) have identical retention times (Kowalak et al. 1996). However, taking into account that the formation of m3Ψ occurs on the level of 70S ribosomes (Ero et al. 2008), the RP-HPLC peak at 11.4 min of 35S and 45S particles probably contain only m5C. The third nucleoside, which was not analyzed, is an incomplete modification at C2501 whose retention time is not known.
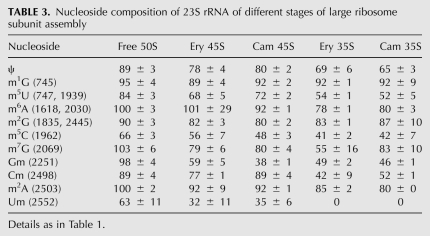
The 35S particles formed in the presence of Ery or Cam exhibited similar nucleoside composition. The particles contain several modified nucleosides in nearly stochiometric amounts. m1G, m6A, m2G, m7G, and m2A are already present in the first assembly intermediate particles by 80% or more (Table 3). This suggests that these modifications are formed during an early step of large subunit assembly. Pseudouridines, m5U, m5C, Gm, and Cm are present in the 35S particles by 40%–70% (Table 3). Um was not found in the 23S rRNA isolated from 35S particles (Fig. 4). Thus, m1G, m6A, m2G, m7G, m5C, and m2A are formed during the early steps of 50S subunit assembly.
TABLE 3.
Nucleoside composition of 23S rRNA of different stages of large ribosome subunit assembly
FIGURE 4.
HPLC analysis of 23S rRNA. 23S rRNA was isolated from mature 70S ribosomes, free 50S subunits, Cam 45S, and Cam 35S particles. Peaks corresponding to four standard nucleosides (C, U, G, and A) and m5C, Cm, m7G, m5U, Um, Gm, m1G, m2G, m2A, and m6A are indicated. Peak corresponding to the m5C contains also m3Ψ. X corresponds to an unknown compound.
A significant increase of the m5U, Gm, Cm, and Um levels was observed in the 45S particles compared with the 35S particles (Table 3). However, the Um level is still only 35% of mature 23S rRNA (Table 3). We conclude that synthesis of m5U, Gm, and Cm occurs during the intermediate steps of large ribosome subunit assembly.
Free 50S particles isolated from exponentially growing cultures represent ∼80% large subunit assembly intermediate particles according to the processing status of the 23S rRNA 5′ end and the translational activity (Peil et al. 2008). Most of the modified nucleosides are present in the free 50S particles by 90%–100% in comparison with the mature 23S rRNA (Table 3). However, Um is present in the free 50S particles by 63% of the mature 23S rRNA level (Table 3). This result is in agreement with the free 50S particles being incompletely assembled and shows that Um together with m3Ψ (see below) are added during the late steps of ribosome large subunit assembly.
Pseudouridines in 23S rRNA
Pseudouridylation is the most abundant modification in stable RNAs. There are 10 pseudouridines in E. coli 23S rRNA, one of them is methylated (m3Ψ1915). Ψ residues were quantitated by RP HPLC. According to the HPLC analysis, 35S particles contain 65%–70%, 45S particles contain 80%, and free 50S subunits contain 90% of pseudouridines compared with the 23S rRNA of 70S ribosomes (Table 2). It must be taken into account that the methylated pseudouridine (m3Ψ) co-elutes with m5C and therefore does not contribute to the pseudouridine peak. Thus, the mature 23S rRNA contains nine pseudouridines. The 23S rRNA of precursor particles lacking methylation at Ψ1915 can contain 10 pseudouridine residues.
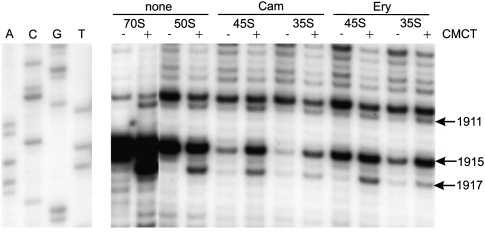
The chromatographic analysis does not answer the question of which Ψ residues are underrepresented in the subribosomal particles. To answer this question, chemical modification of 23S rRNA followed by reverse transcriptase-directed primer extension was used. The presence of pseudouridine at a particular position is indicated by the primer extension stop on the CMCT-treated RNA (+ lane) and its absence on the control RNA (− lane). We used five primers to analyze all nine pseudouridines of 23S rRNA of 35S and 45S particles, free 50S subunits, and 70S ribosomes for comparison.
All three uridines in helix 69 of E. coli 23S rRNA are isomerized to pseudouridines (Ψ1911, m3Ψ1915, and Ψ1917) by RluD (Huang et al. 1998; Raychaudhuri et al. 1998). Methylation of Ψ1915 causes a primer extension stop independent of CMCT treatment (Fig. 5). It must be noted that m3Ψ can form a Watson–Crick-like base pair in the syn conformation of glycosidic bond allowing a low level of readthrough by reverse transcriptase. Therefore, it was not possible to identify pseudouridylation at position 1915 of 50S and 70S but the upstream Ψ1911 was still detectable. In the 35S particles of both drugs, the Ψ-specific signals at 1911 and 1917 are not detectable. In the 45S particles all three pseudourindines appear to be present at low levels. The fraction of Ψ1911 and Ψ1917 is further increased in the free 50S particles but is clearly lower compared with the pseudouridylation level of 70S ribosomes (Fig. 5). The results indicate that RluD is specific to the late assembly of the large ribosome subunit. This conclusion is supported by the earlier findings that RluD-specific Ψ residues are made on the level of 50S particles in the RNA helicase DeaD deletion strain (Leppik et al. 2007) and that the RluD modifies 50S subunits and 70S ribosomes more specifically and efficiently than protein-free 23S rRNA (Vaidyanathan et al. 2007).
FIGURE 5.
Primer extension analysis of the pseudouridines in helix–loop 69 of 23S rRNA. 23S rRNA was isolated from mature 70S ribosomes, free 50S subunits, and Cam 45S, Ery 45S, Cam 35S, and Ery 35S particles and analyzed for pseudouridines by CMCT/alkali and reverse transcriptase directed primer extension. +, CMCT/alkali treatment; −, untreated RNA. Sequence of the 23S rRNA around helix–loop 69 is shown in sequencing lanes (A, C, G, T). Note that the CMCT induced stop site is one nucleotide below the actual site.
A CMCT-independent reverse transcriptase stop site at position 1915 of 23S rRNA indicates the presence of m3Ψ. 23S rRNA of 35S and 45 particles exhibit a very low CMCT-independent stop signal. In the 50S the stop is strong and in the mature 23S rRNA of 70S ribosomes the stop is very strong (Fig. 5). This shows that the enzyme RlmH responsible for methylation of Ψ1915 at C3 is specific to the very late steps of large ribosome subunit assembly. This conclusion is supported by the fact that RlmH requires the presence of the 30S subunit (Ero et al. 2008).
All other pseudouridines were present in the 35S and 45S subribosomal particles of both antibiotics. In the free 50S subunits the level of pseudouridines was similar to that of 70S ribosomes (data not shown). Taking into account the HPLC results described above, this suggests that the Ψ residues outside of helix–loop 69 are made during the early steps of large ribosome subunit assembly. Thus, the corresponding pseudouridine synthases RluA (Ψ746), RluB (Ψ2605), RluC (Ψ955, Ψ2504, Ψ2580), RluE (Ψ2457), and RluF (Ψ2604) are specific to the early assembly particles.
DISCUSSION
The nucleoside composition of subribosomal particles demonstrates that the modification pattern of rRNA is similar in both erythromycin and chloramphenicol-induced particles of the same size. Modifications are added gradually during association of r-proteins with the rRNA and thereby with the growing S-value indicating that the synthesis of rRNA modified nucleosides depends on the progression of ribosome assembly.
Two modified nucleosides are added to the rRNA at equal levels during all steps of ribosome assembly. The modifications m4Cm1402 of 16S rRNA are synthesized in an apparently stochastic way throughout the ribosome subunit assembly. Kimura and Suzuki (2010) reported recently that in the cell-free assay 30S subunit, and not free 16S rRNA, is the substrate for RsmH and RsmI in m4Cm1402 formation. Other rRNA modifications can be divided into three groups: early, intermediate, and late assembly specific modifications.
In the 16S rRNA of 25S particles the fraction of m5C is ∼50% and in the free 30S subunits ∼60% (Fig. 3; Table 1). Thus, about half of m5C is added to the 16S rRNA during the early step and the second half during the late steps of ribosome small subunit assembly. Notably, only a small fraction of m5C is added during 25S to 30S transition of the ribosome small subunit. The fact that the first half of m5C is added during the early step and the second half during the late stages of ribosome small subunit assembly suggests that the RsmB (methylates C967) and RsmF (C1407) have different specificity with respect to ribosome assembly. Cell-free studies have revealed that RsmB can methylate C967 only on the protein-free 16S rRNA and is blocked when the ribosomal proteins S7 and S19 are added (Weitzmann et al. 1991). Methylation of C1407 is thought to be a late event during ribosome assembly as the in vitro substrate of RsmF is a 30S subunit rather than naked 16S rRNA (Andersen and Douthwaite 2006). Based on the results of the nucleoside composition of assembly intermediate particles and enzyme specificity in vitro, we propose that m5C967 is made during the early step and m5C1407 during the late events of small subunit assembly.
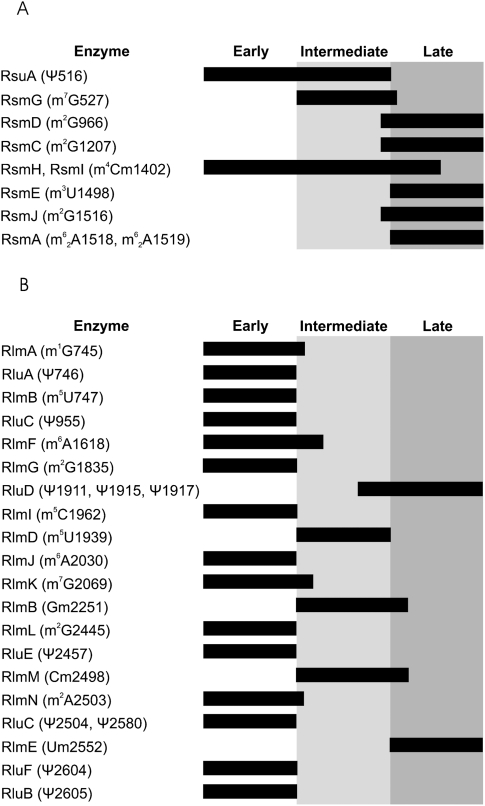
A significant fraction of Ψ516 is added during early and intermediate steps (transition 25S to 30S) of small subunit assembly. This observation agrees with the known specificity of RsuA (Wrzesinski et al. 1995). Formation of m7G527 of 16S rRNA is clearly an intermediate assembly event. Other modified nucleosides of 16S rRNA (m2G at 966, 1207, 1516; m5C1407; m3U1498; m62A at 1518 and 1519) are synthesized at the level of the 30S subunit during the late steps of ribosome assembly. Thus, seven out of 11 modified nucleosides of 16S rRNA are late assembly specific. This tendency was noted earlier by Kaczanowska and Rydén-Aulin (2007), based on a comprehensive review by Ofengand and Del Campo (2004). The results on the 16S rRNA-specific modification enzymes determined in this study are summarized in Figure 6A.
FIGURE 6.
Summary of the specificity of the rRNA modification enzymes with respect to ribosome subunit assembly. Ribosome assembly is divided into three stages (early, intermediate, and late), which are shown by white and gray zones. Activity of rRNA modification enzymes is shown by black bars. (A) Modification of 16S rRNA. (B) Modification of 23S rRNA.
The modified nucleoside composition of 23S rRNA in the early assembly particles (35S) reveals the early assembly specific modifications in the large ribosome subunit. All pseudouridines, except the RluD-specific ones in helix–loop 69 of 23S rRNA, m1G745, m6A1618, m6A2030, m7G2069, m2G1835, m2G2495, and m2A2503 are clearly early assembly specific (Fig. 6B).
Single m5C residue at position 1962 of E. coli 23S rRNA is the product of RlmI (YccW) (Purta et al. 2008). The peak at 11.4 min of the HPLC derived from 35S, 45S particles, and from free 50S subunits constitutes 40%, 50%, and 70%, respectively (Fig. 3; Table 2). This peak contains two nucleosides m5C and m3Ψ (Gehrke and Kuo 1989). The absorbance at 260 nm of m5C and m3Ψ is roughly equal. We anticipate that in 70S both nucleosides contribute to the peak at 11.4 min by 50%. Thus, the distribution of the two nucleosides is not immediately clear from the data. The intensity of the reverse transcriptase stop site at position 1915 shows that 23S rRNA of 35S particles does not contain m3Ψ, and 45S particles contain only a small fraction of m3Ψ (Fig. 5). The formation of m3Ψ was shown to occur on the level of the 70S ribosomes (Ero et al. 2008). Therefore, the 11.4-min peaks of the 35S and 45S particles probably contain mostly m5C. Moreover, ∼80% of the free 50S subunits are incompletely assembled precursors of the large ribosome subunit (Peil et al. 2008; Al Refaii and Alix 2009). Thus, the 11.4-min peak of the free 50S can contain only 20% of mature 23S rRNA and the corresponding amount of m3Ψ. Therefore, m5C is present in the free 50S particles by ∼100%. These results show that C1962 is methylated at C5 during early assembly. This is consistent with the in vitro results showing that methyltransferase RlmI is active on the naked 23S rRNA molecules. No methylation was detected on the 50S subunits or tight-couple 70S ribosomes (Purta et al. 2008).
E. coli 23S rRNA contains two m5U residues (m5U747, m5U19389). m5U is found in the 35S particles in ∼50% (Table 3). The amount of m5U methylation increases from 35S to 45S particles by 20% and reaches about 85% in the free 50S subunits (Fig. 3; Table 1). This suggests that one m5U is made during the early step and the second m5U is made during the intermediate steps of 50S maturation. The nucleotide m5U747 is located close to the peptide exit tunnel deep inside the mature 50S subunit (Schuwirth et al. 2005). Other modified nucleosides in the 750 loop of 23S rRNA (m1G745, Ψ746) are made during early assembly (Fig. 6B). Therefore it is reasonable to assume that m5U747 is an early assembly specific modification, too.
Taken together, early assembly specific modifications are m1G745, Ψ746, m5U747, Ψ955, m6A1618, m6A2030, m7G2069, m2G1835, m5C1962, Ψ2457, m2G2495, m2A2503, Ψ2504, Ψ2580, Ψ2604, and Ψ2605 (Fig. 6B). Thus, 17 out of 25 modified nucleosides of 23S rRNA are made during the early steps of ribosome assembly. The limited time window for synthesis of nucleoside modifications during the large ribosome subunit assembly requires efficient coordination of ribosome assembly events.
As discussed above, the nucleotide m5U747 is synthesized during the early stages of 50S subunit assembly. Therefore, the second m5U at position 1939 is made during the intermediate assembly events. We conclude that nucleotides m5U1939, Gm2251, and Cm2498 are synthesized during the intermediate steps of the large ribosome subunit assembly. It is evident that the corresponding regions of 23S rRNA are accessible for the modification enzymes RlmD, RlmB, and RlmM not only on the protein-free precursor-23S rRNA but also during the intermediate steps of large ribosome subunit assembly. Substrate specificity is known only for the RlmM, which is able to catalyze methylation of C2498 in the protein-free 23S rRNA but not in the tight-coupled 70S ribosomes or 50S subunits (Purta et al. 2009).
23S rRNA of free 50S subunits contains ∼60% of Um (Table 3) and low levels of U1911 and U1917 (Fig. 5). Thus, the late assembly events of the large ribosome subunit involve formation of Um2552, and isomerization of the uridines in h69.
It is important to stress that the assembly dependence of rRNA modification in vivo is in very good agreement with the published specificities of modification enzymes determined in vitro, as is evident by comparing Table 1 and Figure 6.
Coordination of the rRNA modification and the association of ribosomal protein with the subunit is important for several reasons. First, rRNA folding is a step-wise process where transient structures play an important role (Besancon and Wagner 1999; Liiv and Remme 2004; Adilakshmi et al. 2008). Therefore, the structures that are recognized by modification enzymes are formed during the progression of ehe ribosome subunit assembly. The second aspect is the role of ribosomal proteins in directing rRNA folding upon binding to the rRNA (Holmes and Culver 2004; Talkington et al. 2005). In this way, r-proteins could help to create the recognition sites for rRNA modification enzymes. On the other hand, r-proteins can inhibit rRNA modification by shielding the modification site. The dual role of r-proteins in rRNA modification is illustrated by proteins S7 and S19, which are necessary for the m2G966 formation by RsmD but block the synthesis of m5C967 by RsmB (Weitzmann et al. 1991). The modification sites that are buried deep inside the subunit must be modified during early assembly. Finally, it is noteworthy that a specific set of r-proteins are exchangeable in vivo (Pulk et al. 2010). When the ribosome bound r-proteins can be exchanged with the free r-proteins, they could be displaced by the rRNA modifications as well. In this way, the rRNA modification sites that are already masked by r-proteins, can become accessible to the modification enzymes during the late stages of ribosome assembly.
MATERIALS AND METHODS
Preparation of rRNA
Escherichia coli strain MG1655 (Blattner et al. 1997) was used in all experiments. Cells were grown at 25°C in 200 mL 2× YT medium (Sambrook and Russell 2001) until the A600 reached 0.2. At this point, either erythromycin (final concentration of 100 μg/mL) or chloramphenicol (final concentration of 7 μg/mL) was added, followed by incubation for 2 h at 25°C. The control culture was grown without antibiotics. Bacterial cells were collected by centrifugation in a Sorvall GS-3 rotor at 4000 rpm at 4°C for 10 min and were resuspended in 1 mL lysis buffer (60 mM KCl, 60 mM NH4Cl, 50 mM Tris-HCl [pH 8], 6 mM MgCl2, 6 mM β-mercaptoethanol, 16% sucrose); lysozyme and DNase I (Amresco, GE Healthcare) were added to final concentrations of 1 mg/mL and 20 U/mL, respectively. Cells were frozen for 15 min at –70°C and subsequently thawed in ice-cold water for 30 min. The freeze–thaw cycle was repeated twice, followed by centrifugation at 13,000g and 4°C for 20 min. Clear lysate was diluted twofold with buffer A (60 mM KCl, 60 mM NH4Cl, 10 mM Tris-HCl [pH 8], 12 mM MgCl2, 6 mM β-mercaptoethanol). Diluted lysates (2 mL) were loaded onto 30 mL, 10%–25% (w/w) sucrose gradient in buffer A. Ribosomal particles were separated by centrifugation at 23,000 rpm in an SW28 rotor (Beckman) at 4°C for 13.5 h. Ribosome profiles were detected by continuous monitoring of absorbance at 254 nm. 70S ribosomal particles were precipitated with 2.5 volumes of ice-cold ethanol and collected by centrifugation at 5000 rpm for 30 min in an HS4 rotor (Sorvall). 50S and 30S subunit fractions from untreated cells, and 45S, 35S, and 25S particle fractions from antibiotic-treated cells were diluted twofold with buffer A and concentrated by ultrafiltration using Amicon Ultracel-100k filters. Ribosomal particles were loaded onto sucrose gradients and separated as described above. Ribosomal particles were precipitated by addition of ethanol (2 volumes). rRNA was deproteinized by extraction with phenol and chloroform followed by ethanol precipitation. RNA was dissolved in water and stored at −80°C. 16S and 23S rRNA from 70S ribosomes and 35S particles were separated by centrifugation in 5%–20% (w/w) sucrose gradient (buffer 20 mM Na-acetate, 100 mM NaCl, 1 mM EDTA) at 25 700 rpm in an SW28 rotor (Beckman) at 4°C for 16 h. Purified 16S and 23S rRNA were precipitated with ethanol.
High-performance liquid chromatography
For HPLC analysis, 2–4 A260 Units rRNA (70–100 pmol) was digested with nuclease P1 (MP Biochemicals) and bacterial alkaline phosphatase (Fermentas Life Sciences) according to the method of Gehrke and Kuo (1989). Nucleoside composition was determined by RP-HPLC on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 μm) equipped with a precolumn (4.6 mm × 20 mm) at 30°C on a SHIMADZU Prominence HPLC system. The following buffers were used: buffer A (10 mM NH4H2PO4, 2.5% methanol at pH 5.3); buffer B (10 mM NH4H2PO4, 20% methanol at pH 5.1); and buffer C (10 mM NH4H2PO4, 35% acetonitrile at pH 4.9). RP-HPLC analysis was performed using the gradient conditions of Gehrke and Kuo (1989): flow rate 1.0 mL/min held at 0% buffer B 12 min, to 10% buffer B over 8 min, to 25% buffer B over 5 min, to 60% buffer B over 8 min, to 64% buffer B over 4 min, to 100% buffer B over 9 min, 0%–100% buffer C over 35 min, held at 100% buffer C for 10 min, and equilibration with 0% buffer B for 30 min. Nucleoside absorbance profiles were recorded at 260 and 280 nm, and peak areas were integrated. Quantitative calculations were according to the following formula: X% = (XP/NP)/[Average(X70S/N70S)]100, where X% is percentage of modified nucleoside in the subribosomal particle; XP is- modified nucleoside area of the subribosomal particle; X70S is modified nucleoside area of the 70S ribosomes; NP is area of corresponding unmodified nucleoside of the subribosomal particles; and N70S is area of corresponding unmodified nucleoside of the 70S ribosomes.
Determination of pseudouridines
Pseudouridines were determined according to the method of Ofengand et al. (2001). Fifteen micrograms rRNA was dissolved in 20 μL water, 80 μL of BEU buffer (7 M urea, 4 mM EDTA, 50 mM Bicine/NaOH [pH 8.5]), and 20 μL of CMCT/BEU buffer (1 m CMCT in BEU buffer) (CMCT; Sigma-Aldrich Chemie GmbH) were added. One hundred microliters of BEU buffer was added to 15 μg of rRNA in 20 μL of water serving as the negative control. Both samples were incubated at 37°C for 20 min for CMCT modification of U, G, and Ψ residues. Reaction was stopped by addition of 38 μL 4 M NaOAc and 600 μL of cold 96% ethanol. Samples were kept at −20°C for 10 min, and the RNA precipitate was collected by centrifugation at 6000g and 4°C. The supernatant was carefully removed and RNA was washed twice with 600 μL of 70% ethanol. The precipitate was dried at 37°C for 10 min. rRNA was dissolved in 50 μL of NPK buffer (20 mM NaHCO3, 30 mM Na2CO3, 2 mM EDTA), and the samples were incubated at 37°C for 4 h to allow removal of CMCT from U and G residues. After incubation, rRNA was precipitated and washed as described above. The precipitate was dissolved in 20 μL of water and stored at −20°C. Pseudouridine sequencing of rRNA was carried out by primer extension using primers U1 (CAGCCTGGCCATCATTACGCC), C7 (ACACCAGTGATGCGTCCAC), C17 (CCACTTTAAATGGCGAAC), C33 (GTTTGATTGGCCTTTCACCC), and T3 (GCTTTCTTTAAATGATGGCTGCTT), and AMV reverse transcriptase (Seikagaku Corp.) in the presence of [α-32P]dCTP (Amersham Biosciences). The resulting DNA fragments were resolved in 7% polyacrylamide-urea gel. Radioactivity was visualized by a Typhoon PhosphorImager (GE Healthcare).
ACKNOWLEDGMENTS
We thank Aivar Liiv, Lauri Peil, Margus Leppik, Tanel Tenson, Kai Virumäe, and Rya Ero (all from the University of Tartu) for help and advice. This work was supported by Estonian Science Foundation Grant No. 7509.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2160010.
REFERENCES
- Adesnik M, Levinthal C 1969. Synthesis and maturation of ribosomal RNA in Escherichia coli. J Mol Biol 46: 281–303 [DOI] [PubMed] [Google Scholar]
- Adilakshmi T, Bellur DL, Woodson SA 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Refaii A, Alix J 2009. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol Microbiol 71: 748–762 [DOI] [PubMed] [Google Scholar]
- Andersen N, Douthwaite S 2006. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J Mol Biol 359: 777–786 [DOI] [PubMed] [Google Scholar]
- Basturea G, Deutscher M 2007. Substrate specificity and properties of the Escherichia coli 16S rRNA methyltransferase, RsmE. RNA 13: 1969–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besancon A, Wagner R 1999. Characterization of transient RNA–RNA interactions important for the facilitated structure formation of bacterial ribosomal 16S RNA. Nucleic Acids Res 27: 4353–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 [DOI] [PubMed] [Google Scholar]
- Bügl H, Fauman E, Staker B, Zheng F, Kushner S, Saper M, Bardwell J, Jakob U 2000. RNA methylation under heat shock control. Mol Cell 6: 349–360 [DOI] [PubMed] [Google Scholar]
- Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem 275: 16414–16419 [DOI] [PubMed] [Google Scholar]
- Champney W 2006. The other target for ribosomal antibiotics: Inhibition of bacterial ribosomal subunit formation. Infect Disord Drug Targets 6: 377–390 [DOI] [PubMed] [Google Scholar]
- Dagley S, Sykes J 1959. Effect of drugs upon components of bacterial cytoplasm. Nature 183: 1608–1609 [DOI] [PubMed] [Google Scholar]
- Dodd J, Kolb J, Nomura M 1991. Lack of complete cooperativity of ribosome assembly in vitro and its possible relevance to in vivo ribosome assembly and the regulation of ribosomal gene expression. Biochimie 73: 757–767 [DOI] [PubMed] [Google Scholar]
- Ero R, Peil L, Liiv A, Remme J 2008. Identification of pseudouridine methyltransferase in Escherichia coli. RNA 14: 2223–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke C, Kuo K 1989. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J Chromatogr 471: 3–36 [DOI] [PubMed] [Google Scholar]
- Hansen L, Kirpekar F, Douthwaite S 2001. Recognition of nucleotide G745 in 23 S ribosomal RNA by the rrmA methyltransferase. J Mol Biol 310: 1001–1010 [DOI] [PubMed] [Google Scholar]
- Holmes KL, Culver GM 2004. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat Struct Mol Biol 11: 179–186 [DOI] [PubMed] [Google Scholar]
- Huang L, Ku J, Pookanjanatavip M, Gu X, Wang D, Greene P, Santi D 1998. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry 37: 15951–15957 [DOI] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén-Aulin M 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71: 477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Suzuki T 2010. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res 38: 1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak JA, Bruenger E, Hashizume T, Peltier JM, Ofengand J, McCloskey JA 1996. Structural characterization of U*-1915 in domain IV from Escherichia coli 23S ribosomal RNA as 3-methylpseudouridine. Nucleic Acids Res 24: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C, Nomura M, Watson J 1962. The physical properties of the chloromycetin particles. J Mol Biol 4: 388–394 [DOI] [PubMed] [Google Scholar]
- Leppik M, Peil L, Kipper K, Liiv A, Remme J 2007. Substrate specificity of the pseudouridine synthase RluD in Escherichia coli. FEBS J 274: 5759–5766 [DOI] [PubMed] [Google Scholar]
- Lesnyak D, Sergiev P, Bogdanov A, Dontsova O 2006. Identification of Escherichia coli m2G methyltransferases: I. the ycbY gene encodes a methyltransferase specific for G2445 of the 23 S rRNA. J Mol Biol 364: 20–25 [DOI] [PubMed] [Google Scholar]
- Liiv A, Remme J 2004. Importance of transient structures during post-transcriptional refolding of the pre-23S rRNA and ribosomal large subunit assembly. J Mol Biol 342: 725–741 [DOI] [PubMed] [Google Scholar]
- Lindahl L 1975. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol 92: 15–37 [DOI] [PubMed] [Google Scholar]
- Nierhaus KH 1991. The assembly of prokaryotic ribosomes. Biochimie 73: 739–755 [DOI] [PubMed] [Google Scholar]
- Nomura M, Hosokawa K 1965. Biosynthesis of ribosomes: Fate of chloramphenicol particles and of pulse-labeled RNA in Escherichia coli. J Mol Biol 12: 242–265 [DOI] [PubMed] [Google Scholar]
- Ofengand J, Del Campo M, Kaya Y 2001. Mapping pseudouridines in RNA molecules. Methods 25: 365–373 [DOI] [PubMed] [Google Scholar]
- Ofengand J, Del Campo M 2004. Modified nucleosides of Escherichia coli ribosomal RNA. In EcoSal—Escherichia coli and Salmonella: Cellular and molecular biology (ed. Björk G), ASM Press, Washington, DC: http://www.ecosal.org/ [DOI] [PubMed] [Google Scholar]
- Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K 2007. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol 63: 1096–1106 [DOI] [PubMed] [Google Scholar]
- Peil L, Virumäe K, Remme J 2008. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J 275: 3772–3782 [DOI] [PubMed] [Google Scholar]
- Poldermans B, Roza L, Van Knippenberg PH 1979. Studies on the function of two adjacent N6,N6-dimehyadenosines near the 3′end of 16 S ribosomal RNA of Escherichia coli. III Purification and properties of the methylating enzyme and methylase-30 S interaction. J Biol Chem 254: 9094–9100 [PubMed] [Google Scholar]
- Pulk A, Liiv A, Peil A, Maiväli Ü, Nierhaus KH, Remme J 2010. Ribosome reactivation by replacement of damaged proteins. Mol Microbiol 75: 801–814 [DOI] [PubMed] [Google Scholar]
- Purta E, O'Connor M, Bujnicki J, Douthwaite S 2008. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J Mol Biol 383: 641–651 [DOI] [PubMed] [Google Scholar]
- Purta E, O'Connor M, Bujnicki J, Douthwaite S 2009. YgdE is the 2′-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol Microbiol 72: 1147–1158 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Conrad J, Hall B, Ofengand J 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4: 1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW 2001. Molecular cloning: A laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schuwirth B, Borovinskaya M, Hau C, Zhang W, Vila-Sanjurjo A, Holton J, Cate J 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Sergiev P, Lesnyak D, Bogdanov A, Dontsova O 2006. Identification of Escherichia coli m2G methyltransferases: II. The ygjO gene encodes a methyltransferase specific for G1835 of the 23 S rRNA. J Mol Biol 364: 26–31 [DOI] [PubMed] [Google Scholar]
- Sergiev P, Serebryakova M, Bogdanov A, Dontsova O 2008. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J Mol Biol 375: 291–300 [DOI] [PubMed] [Google Scholar]
- Siibak T, Peil L, Xiong L, Mankin A, Remme J, Tenson T 2009. Erythromycin- and chloramphenicol-induced ribosomal assembly defects are secondary effects of protein synthesis inhibition. Antimicrob Agents Chemother 53: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington M, Siuzdak G, Williamson J 2005. An assembly landscape for the 30S ribosomal subunit. Nature 438: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne J, Nurse K, Popienick P, Ofengand J 1999. Purification, cloning, and characterization of the 16 S RNA m2G1207 methyltransferase from Escherichia coli. J Biol Chem 274: 924–929 [DOI] [PubMed] [Google Scholar]
- Usary J, Champney W 2001. Erythromycin inhibition of 50S ribosomal subunit formation in Escherichia coli cells. Mol Microbiol 40: 951–962 [DOI] [PubMed] [Google Scholar]
- Vaidyanathan P, Deutscher M, Malhotra A 2007. RluD, a highly conserved pseudouridine synthase, modifies 50S subunits more specifically and efficiently than free 23S rRNA. RNA 13: 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzmann C, Tumminia S, Boublik M, Ofengand J 1991. A paradigm for local conformational control of function in the ribosome: Binding of ribosomal protein S19 to Escherichia coli 16S rRNA in the presence of S7 is required for methylation of m2G966 and blocks methylation of m5C967 by their respective methyltransferases. Nucleic Acids Res 19: 7089–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzesinski J, Nurse K, Bakin A, Lane B, Ofengand J 1995. A dual-specificity pseudouridine synthase: An Escherichia coli synthase purified and cloned on the basis of its specificity for ψ 746 in 23S RNA is also specific for ψ 32 in tRNAphe. RNA 1: 437–448 [PMC free article] [PubMed] [Google Scholar]