Abstract
Pre-mRNA splicing is catalyzed by the spliceosome, and its control is essential for correct gene expression. While splicing repressors typically interfere with transcript recognition by spliceosomal components, the yeast protein L30 blocks spliceosomal rearrangements required for the engagement of U2 snRNP (small ribonucleoprotein particle) to its own transcript RPL30. Using a mutation in the RPL30 binding site that disrupts this repression, we have taken a genetic approach to reveal that regulation of splicing is restored in this mutant by deletion of the cap-binding complex (CBC) component Cbp80. Indeed, our data indicate that Cbp80 plays distinct roles in the recognition of the intron by U1 and U2 snRNP. It promotes the initial 5′ splice site recognition by U1 and, independently, facilitates U2 recruitment, depending on sequences located in the vicinity of the 5′ splice site. These results reveal a novel function for CBC in splicing and imply that these molecular events can be the target of a splicing regulator.
Keywords: regulated splicing, Cbp80, 5′SS, U1 snRNP/U2 snRNP
INTRODUCTION
Most eukaryotic pre-mRNAs need to be spliced before being translated. During pre-mRNA splicing, intervening sequences (introns) are precisely removed and the adjacent sequences (exons) are spliced together. This process takes place in the spliceosome, a large complex ribonucleoprotein particle (RNP) (for review, see Wahl et al. 2009). Assembly of the spliceosome starts cotranscriptionally with the recognition of the 5′ splice site (5′SS) by the U1 small nuclear RNP (snRNP), committing the transcript to splicing. This can be assisted by the cap-binding complex (CBC), an heterodimer of the factors Cbp20 and Cbp80 (Izaurralde et al. 1994; Fortes et al. 1999b). Subsequently, the 3′ end of the intron, including the branch site (BS) and the 3′ splice site (3′SS), is identified by the factors BBP and Mud2 in budding yeast, and SF1 and U2AF in metazoans. An interaction between these components and U1 snRNP has been shown in vitro in this complex (Abovich and Rosbash 1997; Kent and MacMillan 2002), known as commitment complex or CC (complex E in metazoans). In the next step, a poorly understood remodeling of the CC takes place leading to the association of U2 snRNP with the BS, to form the pre-spliceosome or complex A (Parker et al. 1987; Wu and Manley 1989; Zhuang and Weiner 1989). Next, the pre-spliceosome is further remodeled and the tri-snRNP U4/U6.U5 engages in the nascent particle (complex B). Subsequent rearrangements involving the displacement of U1 and U4 snRNP lead to the formation of a catalytically active spliceosome (complex C) (Wahl et al. 2009). In addition to its role in the formation of the CC, the CBC has also been shown to promote association of U4/U6.U5 to the nascent spliceosome (O'Mullane and Eperon 1998; Staley and Guthrie 1999; Gornemann et al. 2005).
Spliceosome assembly is tightly regulated at multiple levels (Graveley 2000; Dreyfuss et al. 2002; Hertel and Graveley 2005; House and Lynch 2008), and work in Saccharomyces cerevisiae has provided relevant data on splicing mechanisms and regulation (Brow 2002; Meyer and Vilardell 2009). One example is the essential RPL30 gene, encoding the ribosomal protein L30 which, when in excess, binds to its own transcript and stalls spliceosome assembly. L30 interacts with a kink-turn structure (Klein et al. 2001) that mimics the L30 rRNA binding site (Vilardell et al. 2000). This prevents association of U2 snRNP with the BS by a distinct mechanism likely to involve an interference with conformational changes that occur during spliceosome assembly (Macías et al. 2008). Here we follow a genetic approach to address the molecular mechanisms involved in this regulation. Taking advantage of silent RPL30 mutants that affect splicing regulation by L30, we have sought mutations in trans that restore control of splicing. We have identified multiple mutations in the CBC component Cbp80, indicating that this factor has a wider role in spliceosome assembly than previously assumed. This includes a function in U2 recruitment that is the target of a splicing regulator.
RESULTS
C9 in RPL30 is required by L30 to repress U2 snRNP recruitment
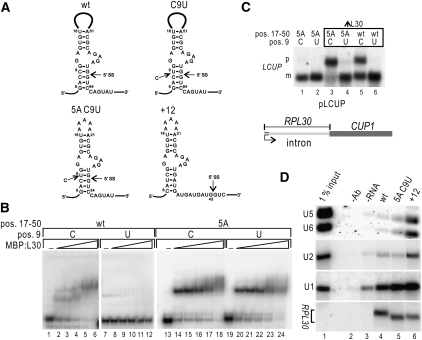
To dissect mechanisms involved in the repression of U2 snRNP recruitment by L30, we undertook a genetic approach based on RPL30 mutant transcripts known to abolish control of splicing (Vilardell and Warner 1994). Mutations in RPL30 used here, localized in the kink-turn motif recognized by L30, are shown in Figure 1A. C9U perturbs a helix involved in the kink-turn, and it is predicted to weaken it. 5A reduces the large loop (positions 17–50, Fig. 1A), not required for L30 binding (Chao and Williamson 2004), to five adenines, thus likely stabilizing the kink-turn. +12 combines 5A with moving the 5′SS 12 nucleotides (nt) downstream. We have previously shown that in this transcript L30 binding and splicing are compatible, due to the added space between L30 and the intron (Macías et al. 2008), and we use this RNA as a control for the experiments described below. As expected, C9U disrupts L30 binding in vitro (Fig. 1B, lanes 7–12; Vilardell et al. 2000), 5A shows more affinity to L30 (lanes 12–18), and, importantly, the combination 5A C9U binds L30 as well (lanes 19–24).
FIGURE 1.
L30 binds, but does not repress, the RPL30 5A C9U transcript. (A) Schematic representation of the RPL30 kink-turn bound by L30, wt, and mutant versions, as indicated (SS: splice site, the AUG is at the end of the first exon). Numbers refer to the start of transcription. (B) Effect of mutations in RPL30 on L30 binding. RPL30 transcripts (0.5 pmol, nt 1–123) were incubated with buffer (lanes 1,7,13,19) or with increasing amounts of MBP:L30 (50, 100, 200, 500, and 1000 ng; lanes 2–6, 8–12, 14–18, and 20–24, respectively). Transcripts were wt (lanes 1–6), C9U (lanes 7–12), 5A (lanes 13–18), and 5A-C9U (lanes 19–24). Reactions were analyzed in a 6% acrylamide gel. (C) Both C9U and 5A C9U transcripts fail to accumulate pre-mRNA under conditions of L30 excess. W303 (lanes 1,2) or yJV25 (producing excess L30, lanes 3–6) cells were transformed with pLCUP plasmids (schematized on the bottom) bearing the indicated mutations (top). RNA was extracted, subjected to Northern analysis, and probed with LCUP sequences (D) snRNP coimmunoprecipitation with L30. RPL30 transcripts (nt 1–347) were incubated under splicing conditions with MBP:L30. Reactions were immunoprecipitated with anti-MBP, and pelleted RNA was subjected to Northern analysis to detect RPL30, U1 and U2 snRNA, as indicated. Lane 1, RPL30 +12; lane 2, RPL30 5A C9U; lane 3, RPL30; lane 4, no transcript added; lane 5, no antibody added; lane 6, 1% of the input.
To determine the effect of 5A C9U on splicing regulation in vivo we used the LCUP1 reporter system, based on fusing RPL30 exon 1 and intron to the CUP1 ORF (Vilardell and Warner 1997). Northern analyses (Fig. 1C) indicate that splicing of wt LCUP1 transcripts is regulated by L30 (lane 5), while C9U mutants fail to be regulated (lane 6). LCUP1 5A transcripts are highly repressed under excess L30 (lane 3 vs. 1), consistent with increased affinity to L30. Interestingly, regulation of splicing by L30 is not restored in LCUP1 5A C9U transcripts (lane 4 vs. 2). We conclude that both the stability and the sequence of the RPL30 kink-turn are important for regulation, and that the C9U mutation abolishes regulation even of transcripts in which the kink-turn is stably bound by L30.
There are two alternate possibilities to explain the C9U phenotype of lack of repression. Either L30 is dislodged during intron recognition or it remains associated with the transcript but fails to repress U2 snRNP recruitment. To distinguish between them, we determined the levels of co-immunoprecipitation of L30 with U1 and U2 snRNAs in presence of 5A C9U transcripts. We used the +12 RNA as a positive control, where splicing and L30 binding are compatible (Macías et al. 2008). Figure 1D shows similar levels of U1 coimmunoprecipitation in wt (lane 4) and 5A C9U transcripts (lane 5), consistent with the initial intron recognition by U1 snRNP being compatible with L30 binding. There is lack of U2 coimmunoprecipitation in both transcripts, but not +12 (lane 6). This indicates repression of splicing in wt RNA (Macías et al. 2008); however, since RPL30 5A C9U is spliced even in presence of L30; lack of U2 coimmunoprecipitation is consistent with L30 removal during U2 association with RPL30. Accordingly, we detect U1 cross-linking with the 5A C9U 5′SS, as well as association of this intron with BBP, under repressive conditions (Supplemental Fig. S1). We conclude that a C at position 9 is required for repression of splicing by L30, and that C9U blocks repression of splicing after intron recognition by U1. Thus, we decided to search for mutations that restore regulation of splicing by L30 to transcripts with this mutation.
Screen for regulation of splicing of C9U transcripts
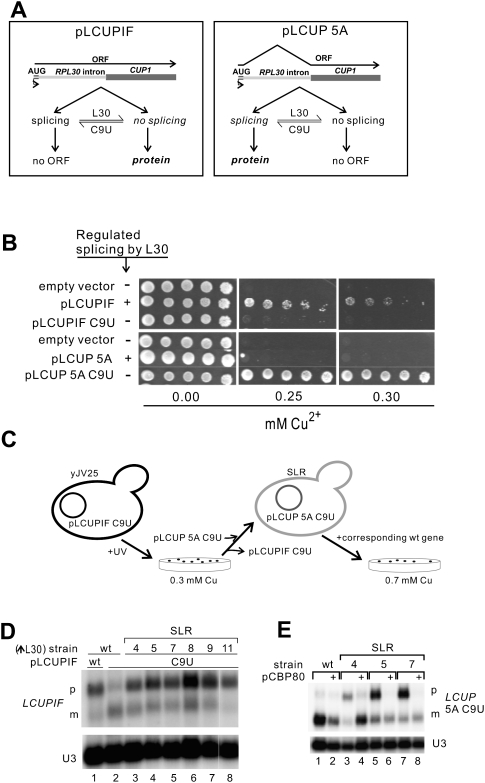
Our genetic screen is based on the CUP1-based reporter LCUPIF, in which Cup1 protein can only be expressed from unspliced RNA (Fig. 2A); thus, LCUPIF splicing repression by L30 is expected to produce increased copper tolerance. Accordingly, LCUPIF confers copper resistance (>0.2 mM) to a strain engineered to produce excess L30 (yJV25), while LCUPIF C9U does not (Fig. 2B). Therefore, we anticipated that cells with the LCUPIF C9U reporter would grow in copper (>0.2 mM) if they bore a mutation-enhancing L30 repression of splicing. To select such mutants, yJV25 cells carrying LCUPIF C9U were UV-irradiated and selected on 0.3 mM Cu2+ (see the diagram in Fig. 2C and Materials and Methods), and six mutants showed splicing repression of this reporter (Fig. 2D, lanes 3–8). These mutants, called SLR (Suppressors of Lack of Repression by L30), all exhibit similar Cu2+ tolerance with LCUPIF C9U (Supplemental Fig. S2). To test more specifically the effects on splicing repression of our mutants we used the LCUP 5A C9U reporter (Fig. 2A), not repressed in wt cells, but expected to be repressed in SLR mutants. Complementation of slr with the wt allele should restore full splicing of the 5A C9U reporter, allowing selection by increased copper tolerance (as schematized in Fig. 2C). Using this approach, a YCp50 plasmid containing the CBP80 gene was found to restore wt levels of copper tolerance to SLR4, SLR5, and SLR7 (Supplemental Fig. S3). Northern analyses further verified that in these cells the repression of splicing by L30 is similar to that in wt (Fig. 2E, even lanes). We determined that the genomic copy of CBP80 includes the mutation L82Stop in SLR4, L157P in SLR5, and D291Stop in SLR7 (Supplemental Fig. S4).
FIGURE 2.
Screen for synthetic enhancers of L30 repression of splicing. (A) Reporter plasmids based on the fusion between RPL30 exon 1 and intron with the CUP1 ORF. LCUPIF transcripts (left) produce Cup1 protein only when unspliced, while LCUP RNAs (right) need to be spliced to encode the protein. (B) Phenotype, identified as growth in medium-containing copper, of cells with constitutive excess of L30 (yJV25) and transformed with either pLCUPIF (upper panel) or pLCUP 5A (bottom panel). Under repression conditions (“+” rows), pLCUPIF confers copper resistance while pLCUP-5A does not. When L30 repression is abolished by the C9U mutation (“−” rows), pLCUP confers tolerance while pLCUPIF does not. Serial one-fifth dilutions were spotted in each case. (C) Screen strategy to select mutations that restore inhibition of splicing by L30 on a C9U transcript. Corresponding Northern analyses are shown in panels D and E. Strain yJV25 with the plasmid pLCUPIF-C9U (Cu-sensitive) was UV-irradiated and SLR mutants were selected on plates containing 0.3 mM copper. Colonies showing LCUPIF C9U pre-mRNA accumulation (panel D) were cured of the plasmid and transformed with pLCUP 5A C9U, rendering them Cu sensitive again because of increased repression, unless the slr mutation is complemented or suppressed. Thus, cells were transformed with a YCp50-based wt genomic library and the transformants selected on 0.7 mM copper, and pCBP80 was identified (panel E). (D) Northern analysis of RNA from SLR mutant cells transformed with pLCUPIF C9U (lanes 3–8). As controls, RNA extracted from yJV25 cells harboring pLCUPIF wt (lane 1) or C9U (lane 2) were loaded in the same gel. Precursor (p) and mature (m) LCUPIF transcripts are indicated. (E) Northern analysis of RNA extracted from SLR4, SLR5, and SLR7 (lanes 3–8) and yJV25 cells (lanes 1,2), transformed with pLCUP 5A C9U alone (odd lanes), or plus pCBP80 (even lanes). Precursor (p) and mature (m) LCUP transcripts are indicated. In D and E U3 was used as loading control.
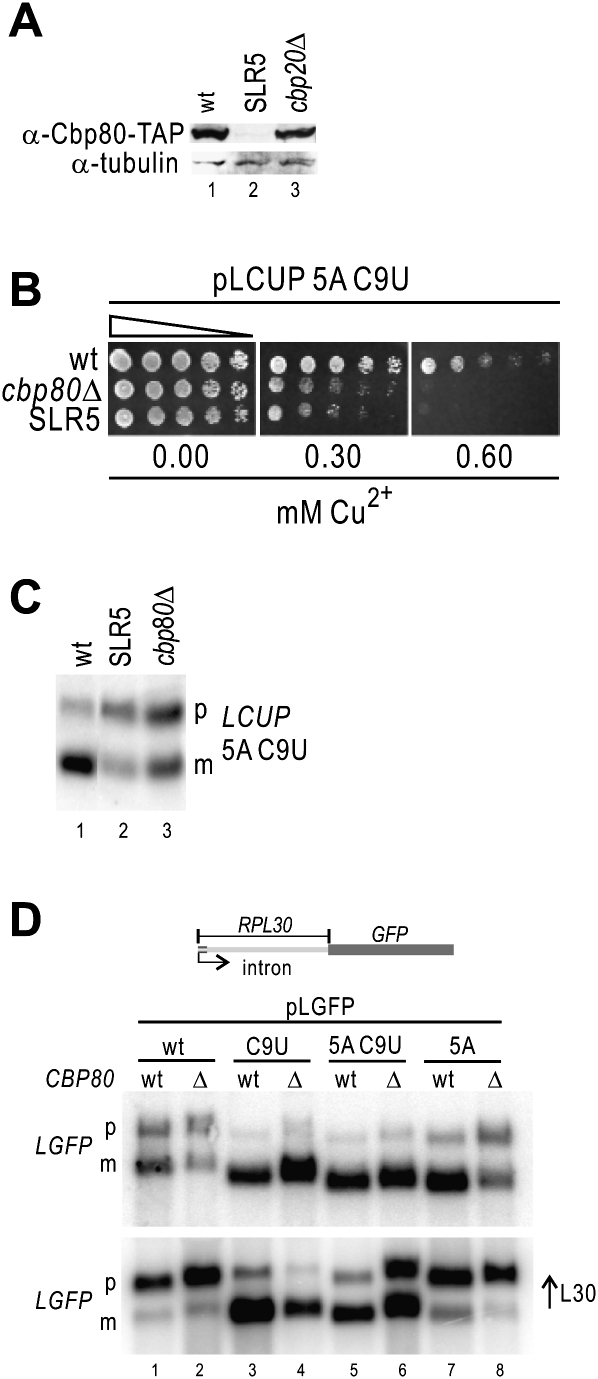
Deletion of CBP80 suppresses the requirement for C9 by L30
The isolation of two cbp80 alleles with premature Stop codons prompted us to verify the Cbp80 levels in the L157P mutant (SLR5). Western blot analyses indicate that they are drastically reduced by this mutation (Fig. 3A), suggesting that deletion of CBP80 in yJV25 could produce an SLR phenotype. To assess this possibility we determined that the Cu2+ tolerance of yJV25 cbp80Δ cells, bearing a LCUP 5A C9U reporter, was indistinguishable from that of SLR5 (Fig. 3B), and we verified by Northern analysis that in cbp80Δ cells splicing of this reporter was repressed by L30 (Fig. 3C). To verify that this effect is not due to a general decrease in splicing efficiency in this intron, we analyzed its splicing and regulation by L30 in cbp80Δ cells, using the pLGFP reporter (based on pLCUP, with the ORF of CUP1 replaced by GFP) (Macías et al. 2008). Our results show that splicing of the C9U reporter (insensitive to L30) is not affected by deletion of Cbp80 (Fig. 3D upper panel, lanes 3,4), indicating that the RPL30 intron is efficiently spliced in absence of Cbp80. In addition, splicing of the 5A C9U RNA, but not that of C9U, becomes repressed by excess L30 in cbp80Δ cells (Fig. 3D, lower panel, lane 6 vs. 4). Therefore we conclude that absence of functional Cbp80 is the cause of augmented splicing regulation by L30 on transcripts bound by L30. This is also consistent with the increased pre-mRNA levels in cbp80Δ in absence of additional L30 (endogenous L30 levels suffice to trigger some repression; Fig. 3D, upper panel, lane 2). We also verified that both splicing of C9U and splicing regulation of wt transcripts were effective in cbp80Δ, cbp20Δ, and cbp80Δ cbp20Δ (cbcΔ) strains (Supplemental Fig. S5).
FIGURE 3.
Deletion of CBP80 is synthetic with L30 repression of splicing. (A) Decreased levels of Cbp80-TAP in SLR5 mutants. Cbp80 protein was TAP-tagged at the C terminus in yJV25 (wt, lane 1), SLR5 (lane 2), and cbp20Δ (Y02074). Extracts were subjected to Western analyses, as indicated. Tubulin was used as loading control. (B) Deletion of CPB80 produces the same phenotype as that of SLR5. One-fifth serial dilutions of wt (yJV25, top), cbp80Δ (yJV35, middle), and SLR5 (bottom) cells transformed with pLCUP 5A C9T were spotted on copper-containing media, as indicated. Copper sensitivity indicates splicing repression of the LCUP 5A C9U transcript (C). Deletion of CBP80 leads to repression of LCUP 5A C9U splicing by L30. Northern analysis of RNA from wt (yJV25, lane 1), SLR5 (lane 2), and cbp80Δ (lane 3) cells. Positions of precursor (p) and mature (m) LGFP 5A C9U are indicated on the right. (D) Splicing efficiency and repression by L30 of the RPL30 intron in wt and cbp80Δ cells, with or without excess L30. Northern analysis of RNA from either wt (even lanes) or cbp80Δ cells (odd lanes) transformed with the pLGFP reporter plasmid, as indicated at the top. Samples in the bottom panel are from cells under excess L30 (pMB73). Positions of precursor (p) and mature (m) LGFP are indicated on the left. pLGFP contains the GFP ORF instead of Cup1 in pLCUP (Vilardell and Warner 1997). pMB73 encodes L30 without the autoregulatory loop (Macías et al. 2008).
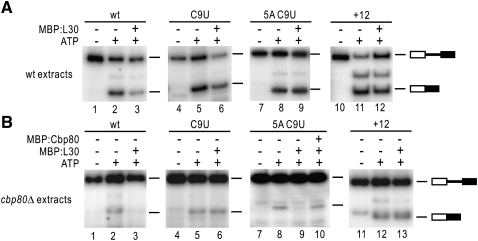
To test whether our findings from the genetic screen could be confirmed in vitro, RPL30 transcripts were incubated with extracts from either wt or cbp80Δ cells, supplemented with recombinant L30. Our results (Fig. 4) show that, while overall splicing activity is reduced in cbp80Δ extracts (Fig. 4B), 5A C9U transcripts are specifically repressed by L30 in these extracts (cf. lanes 8,9 from panels A and B). Importantly, addition of recombinant Cbp80 restores splicing of 5A C9U transcripts repressed by L30 (Fig. 4B, lane 10). These data provide further support to the interaction between Cbp80 and repression of splicing by L30 and prompted us to analyze the molecular bases for this interaction.
FIGURE 4.
In vitro regulation of splicing of RPL30 5A C9U by L30 in the absence of Cpb80. Synthetic RPL30 transcripts, indicated at the top, were incubated under splicing conditions with wt (A) and cbp80Δ (B) extracts, supplemented with MBP:L30 or MBP:Cbp80, as indicated. Upon completion of the reaction, RNA was extracted and analyzed by semiquantitative RT-PCR. Bands corresponding to substrate and spliced RNA are shown. Amounts of RNA, extracts, and recombinant protein were equivalent in all reactions.
Effects of Cbp80 deletion on cotranscriptional spliceosome assembly and L30 regulation
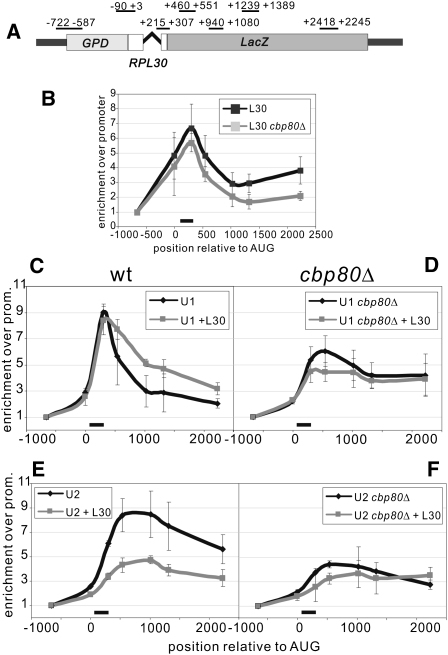
During control of splicing by L30 U2 recruitment is abolished and U1 association with the intron becomes stabilized (Macías et al. 2008). Our data, showing enhancement of L30 repression by deletion of Cbp80, raise the possibility of a novel role for CBC on U2 recruitment. Thus, we decided to monitor cotranscriptional splicing regulation by L30 in wt and cbp80Δ cells. For this we used the RPL30-LacZ reporter, based on RPL30 with the LacZ ORF as the second exon. This provides a convenient tool to assess cotranscriptional spliceosome assembly and its regulation by ChIP (Macías et al. 2008). The reporter was introduced in strains either producing or not producing excess L30 (from plasmid pMB73) (Macías et al. 2008). ChIP analyses from these cells are shown in Figure 5. We first verified that deletion of Cbp80 does not lead to an increased cotranscriptional recruitment of L30, which could otherwise be consistent with enhanced regulation in cbp80Δ cells (Fig. 5B). Next we followed cotranscriptional engagement of U1 snRNP on RPL30-LacZ (ChIP of the U1 component Snu71-HTB; Macías et al. 2008). In the absence of excess L30, the maximum U1 ChIP signal is reduced in cbp80Δ cells, but it lingers toward the 3′ end of the gene (Fig. 5C,D, black lines). Under excess L30, ChIP of U1 shows a slight reduction and a similar persistence (Fig. 5D, gray line), which can also be observed in wt cells (Fig. 5C, gray). Cotranscriptional recruitment of U2 (ChIP of the U2 component Lea1-HTB) (Macías et al. 2008) in wt cells is strongly repressed by excess L30 (Fig. 5E). In cbp80Δ cells, U2 recruitment is diminished, with a further signal reduction under excess L30 (Fig. 5F). Taking into account the persistence of the U1 ChIP profile and the low U2 recruitment observed in cbp80Δ cells, our data suggest that in the RPL30 intron cotranscriptional engagement of U2 is dependent on Cbp80. A persistence of U1 ChIP on the target gene has also been described in cbcΔ cells (Gornemann et al. 2005). However, in the same cells U2 ChIP on ECM33 resembles that of wt (Gornemann et al. 2005). We examined this apparent discrepancy by analyzing the cotranscriptional recruitment of U2 snRNP on the ACT1 gene, which is not affected by L30 overexpression, in a cbp80Δ strain. Neither deletion of CBP80 nor excess L30 have a significant effect on cotranscriptional recruitment of either U1 or U2 snRNP on the ACT1 intron (Supplemental Fig. S6). This is consistent with previously published results (Gornemann et al. 2005) and indicates that the role of Cbp80 in cotranscriptional spliceosome assembly is transcript specific.
FIGURE 5.
Cotranscriptional U1 and U2 recruitments on the RPL30-LacZ transcript in wt and cbp80Δ cells. Horizontal axes show the distance in nucleotides from the start codon. Vertical axes indicate the signal relative to that of the promoter (first primer pair, or PP1). The black bar indicates intron position. The ChIP profiles correspond to wt (panels C and E) or cbp80Δ cells (panels D and F), under normal conditions (black lines) or under L30 excess (gray lines). (A) Scheme showing the positions of the PCR primers used for the ChIP analyses of the RPL30-LacZ intron, relative to the translation start. (B) ChIP against L30 (L30-TAP). In both cases there is L30 excess (pMB73, see Materials and Methods). Black line, wt cells; gray, cbp80Δ cells. In the following panels, ChIP profiles of the indicated proteins are shown, performed on RPL30-LacZ. (C,D) ChIP against U1 snRNP (Snu71-HTB). (E,F) ChIP against U2 snRNP (Lea1-HTB).
Cbp80 and 5′ splice site sequences impact U1 and U2 snRNP cotranscriptional recruitment
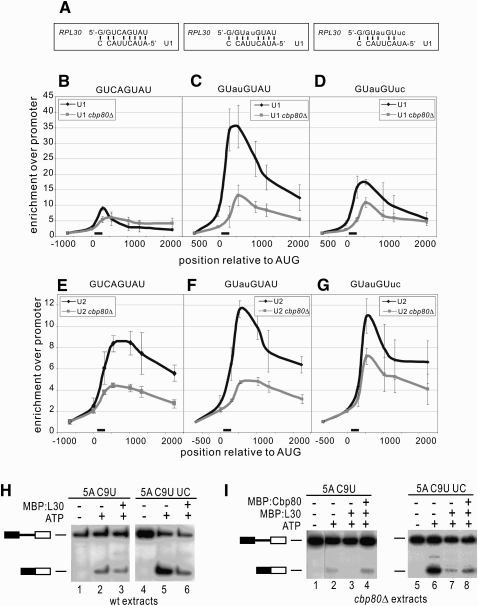
The persistence of U1 on the RPL30 transcript in cbp80Δ cells (Fig. 5D) can reflect a limited cotranscriptional recruitment of U2 (Macías et al. 2008), suggesting a role for Cbp80 in this recruitment. To further dissect the contribution of Cbp80 to U1 and U2 recruitments we investigated whether sequence features in RPL30 could play a specific role. A particular attribute of the RPL30 intron is the evolutionarily conserved 5′SS sequence, GUCAGUAU, unique in yeast introns. This sequence can form 7 base pairs with U1, while the ACT1 intron, with GUAUGUUC at its 5′ end, has the potential to form 5 (Fig. 6A) and produces U1 and U2 ChIPs more resilient to Cbp80 deletion (Supplemental Fig. S6). Thus, we investigated the role of these sequences in combination with Cbp80 on spliceosome assembly in the context of the RPL30-LacZ transcript. Mutants GUauGUuc, containing the first 8 nt of ACT1 (lowercase indicates changes in wt RPL30 to ACT1), and GUauGUAU, with positions 3 and 4 as in ACT1, were compared with the wt version GUCAGUAU. These mutations have been shown not to affect RPL30 splicing in wt cells (Supplemental Fig. S7; Macías et al. 2008). The results (Fig. 6) can be summarized as follows.
FIGURE 6.
Effect of mutations in the RPL30 intron and Cbp80 on cotranscriptional recruitment of U1 and U2 on the RPL30-LacZ gene. Horizontal axes show the distance in nucleotides from the start codon. Vertical axes indicate the signal relative to that of the promoter (first primer pair, or PP1). The black bar indicates intron position. The ChIP profiles correspond to wt (black lines) or cbp80Δ cells (gray lines). (A) Panels indicate different intronic 5′ ends, with a schematic representation of the possible base-pairing with U1 snRNA. GUCAGUAU panels are based on data from Figure 5. (B–D) ChIP profiles of U1 snRNP (Snu71-HTB). (E–G) ChIP profiles of U2 snRNP (Lea1-HTB). (H,I) In vitro splicing of the RPL30 intron, either wt or with reduced affinity for U1 snRNA, in wt or cbp80Δ extracts. Splicing reactions were set up and analyzed as in Figure 4, using wt extracts (H) or extracts from cbp80Δ cells (I), supplemented with MBP:L30 or MBP:Cbp80, as indicated. In the UC transcript, intron positions 6 and 7 (AU) have been mutated to UC, which cannot base-pair to U1. Bands corresponding to substrate and spliced RNA are shown. Amounts of RNA, extracts, and recombinant protein were equivalent in all reactions (see Materials and Methods).
In wt cells, U1 cotranscriptional recruitment is influenced not only by the potential base-pairing between the intron and U1, but also by the particular 5′SS sequence (Fig. 6B–D, black). Thus, GUauGUAU (Fig. 6C) produces a ChIP signal more than three times higher than that of wt RPL30 (Fig. 6B). This difference is striking considering that the potential to base-pair with U1 is similar in both constructs. Consistent with this, disrupting the potential base-pairing of U1 to positions 7 and 8 (GUauGUuc) produces intermediate levels of U1 ChIP (Fig. 6D), despite having less potential for base-pairing with U1 than RPL30 5′SS (Fig. 6B). This indicates that AU at positions 3 and 4 facilitates U1 recruitment. The corresponding U1 ChIPs in cbp80Δ cells (gray lines) indicate that Cbp80 is required to take full advantage of the increased recruitment afforded by the extra base-pairing of U1 to intronic positions 7 and 8. Thus, deletion of Cbp80 has a greater effect on the intron with GUauGUAU than on the one with GUauGUuc at the 5′SS, to the point that both reporters recruit similar levels of U1 in absence of Cbp80 (Fig. 6C,D, respectively, gray lines).
U2 cotranscriptional recruitment in wt cells (Fig. 6E–G, black) shows efficiencies that do not necessarily correlate with those of U1. While U2 associates better with a GUauGUAU intron than with GUCAGUAU (Fig. 6F,E, respectively), the improvement does not match that of U1. Moreover, GUauGUAU and GUauGUuc introns produce similar U2 ChIPs (Fig. 6F,G), despite having distinct U1 ChIPs. This absence of correlation between U1 and U2 recruitments persists in cbp80Δ cells (Fig. 6E–G, gray lines). Interestingly, this is more marked in the GUauGUAU and GUauGUuc introns (Fig. 6F,G). While their U1 ChIP profiles are comparable (Fig. 6C,D, gray), U2 recruitment to GUauGUuc introns remains closer to that of the wt. These data indicate that the role of Cbp80 in U2 recruitment becomes more apparent in introns with increased base-pairing to U1, consistent with the low recruitment of U2 in GUCAGUAU introns in cbp80Δ cells (Fig. 6E, gray). We conclude that Cbp80 has roles in U1 as well as U2 cotranscriptional recruitment, and these become more evident when the interaction between U1 and the intron is hyperstabilized. We verified that the introduced mutations do not induce alterations in pol II recruitment (Supplemental Fig. S8), and we ascertained that the ChIPs on ACT1 remained similar across the samples (Supplemental Fig. S9).
Our model predicts that reducing the affinity for U1 of RPL30 should improve its splicing in cbp80Δ extracts, and to assess this possibility we synthesized two transcripts: RPL30 5A C9U, corresponding to the wt intron, and RPL30 5A C9U UC, with UC at intron positions 7 and 8 abolishing the potential extra base-pairing to U1 (Fig. 6A). Our results show that in wt extracts both RNAs splice (Fig. 6H), with 5A C9U UC yielding more product (Fig. 6H, lanes 2,5). As expected, they are both only marginally sensitive to L30 (Fig. 6H, lanes 3,6). In contrast, in cbp80Δ extracts (Fig. 6I) splicing of 5A C9U is reduced to low levels (Fig. 6I, lane 2), while that of the 5A C9U UC remains active (Fig. 6I, lane 6). Both are repressed by L30 (Fig. 6I, lanes 3,7), and addition of recombinant Cbp80 restores splicing to some extent. This effect is specific to RPL30 5A C9U introns (Supplemental Fig. S10).
DISCUSSION
Data on the control of splicing by L30 support a novel strategy for regulation, based on interference with spliceosomal transitions during U2 snRNP association with the intron (Macías et al. 2008). In a genetic screen for mutations that alter this regulation we have identified Cbp80. This factor forms, in conjunction with Cbp20, the CBC, and is linked to the recognition of the 5′SS during both early and late steps in spliceosome assembly (Gornemann et al. 2005, and references therein). Yet our genetic analysis suggests an additional role for Cbp80 in U2 recruitment, and we have tested this hypothesis. Here we show that U2 snRNP recruitment is assisted by Cbp80, is influenced by the sequence next to the 5′SS, and can be targeted by splicing regulatory factors like L30.
A mutation in the exon 1 of RPL30 that restores U2 snRNP recruitment requires Cbp80
We have determined that the C9U RPL30 mutation abolishes regulation of splicing even under conditions where L30 binding is restored (Fig. 1C). This suggests that destabilizing the RPL30 kink-turn allows the spliceosome to compete with L30 binding. Consistent with this, we show that L30 remains associated with the 5A C9U transcript during initial intron recognition by U1 but it fails to coimmunoprecipitate with U2 (Fig. 1D; Supplemental Fig. S1). A spliceosomal conformational change is required for U2 binding (Abovich et al. 1994), and it is plausible that L30 fails to prevent this because it is dislodged from the C9U mutant RNAs at this step. In a genetic screen based on this model, we have found that Cbp80 is required for the spliceosome to successfully compete with L30 (Fig. 2E), but not to splice RPL30 under nonrepressive conditions (Fig. 3D). This can be reproduced in vitro (Fig. 4). These data suggest an additional function of Cbp80, beyond its known roles on recruitment of U1 and U6 (Gornemann et al. 2005). Therefore, to determine the molecular basis for this, we have analyzed in detail U1 and U2 cotranscriptional spliceosome assembly in cbp80Δ cells.
Role of Cbp80 on U1 and U2 snRNP recruitment
Our results are consistent with a role for Cbp80 in cotranscriptional U2 recruitment, which is more evident when the potential base-pairing between the intron and U1 snRNP is unusually strong, as in RPL30. This explains the genetic interaction of Cbp80 with L30 regulation, as RPL30 is an effective U1 snRNP recruiter (Fig. 5; Tardiff et al. 2006). Regarding this U1 recruitment, comparison of ChIP profiles from GUAUGUAU and GUCAGUAU introns reveals a critical role for intronic sequences in the vicinity of the 5′ splice site, even when the number of potential base pairs to U1 snRNA does not change (Fig. 6B,C). This is in agreement with previous reports indicating that initial recognition of an intron by U1 is not limited by base-pairing, and that GUAUGU could be favored over GUCAGU by the U1 snRNP factor U1-C (Du and Rosbash 2002). Interestingly, however, there is no apparent change in the splicing efficiency of both transcripts, or in their regulation (Supplemental Fig. S7; Macías et al. 2008). The effective recruitment of U1 to the RPL30 intron depends on both its sequence and Cbp80, since in cbp80Δ cells U1 ChIP on GUAUGUAU and GUAUGUUC introns are equivalent (Fig. 6B–D). Thus, our results indicate that positions 3 and 4, and 7 and 8, have different effects on U1 recruitment. They also reveal that the contribution of positions 7 and 8 is mostly dependent on Cbp80, in contrast to that of positions 3 and 4. Remarkably, our ChIP data indicate that U2 recruitment does not always correlate with that of U1, arguing that cotranscriptional association of U2 does not simply follow U1 recruitment. Thus, while mutation of GUAUGUAU to GUAUGUUC in cbp80Δ cells does not have a significant effect on U1 ChIP, there is a clear reduction in U2 ChIP on a GUAUGUAU intron (Fig. 5E). This suggests that in cbp80Δ cells hyperstabilized U1 binding interferes with U2 recruitment, and we have verified this possibility in vitro by showing that a reduced potential for base-pairing to U1 improves splicing of RPL30 substrates in cbp80Δ extracts (Fig. 6I). This was surprising, as Cbp80 promotes U1 recruitment (Colot et al. 1996; Fortes et al. 1999b), and it could be expected that a lower affinity for U1 is detrimental for splicing in cbp80Δ extracts. Yet, it does not, suggesting a different limitation, more consistent with a function for Cbp80 in U2 recruitment, antagonized by the binding of U1 to the intron. Significantly as well, L30 repression is active in all 5A C9U substrates (Fig. 6I; Supplemental Fig. S10), consistent with our finding that repression in 5A C9U RNA is not due to a general reduction in splicing activity. The interference with U2 snRNP recruitment by hyperstabilized intron binding to U1 in cbp80Δ cells evokes the blocking of U4/U6.U5 tri-snRNP recruitment in prp28 cells (Staley and Guthrie 1999), suggesting a U1 snRNP remodeling prior to U2 recruitment. There are genetic and molecular data supporting this U1 remodeling. Genetic interactions between U1 snRNP, the CBC, and the BS have been observed (Fortes et al. 1999a). In addition, mutations in U1 predicted to weaken the interaction with the 5′SS allow for some U2 recruitment in the absence of ATP (Liao et al. 1992). Furthermore, the structure of the human U1 snRNP bound to the 5′SS argue in favor of this possibility as well, with the factor U1-C stabilizing the interaction U1-5′SS while being subjected to long-range protein connections within the particle (Pomeranz Krummel et al. 2009). Perhaps remodeling of the bridging between U1 snRNP and the BS affects the binding of U1 to the 5′SS, an interaction clearly influenced by CBC (Gornemann et al. 2005). In this context, the genetic interaction between Cbp80 and L30 could be explained by their antagonistic roles on spliceosome assembly (Supplemental Fig. S11). L30 inhibits U2 recruitment by stabilizing a particular conformation of U1 on the intron, while Cbp80 promotes U2 recruitment and a likely U1 remodeling. In a C9U mutant L30 binding may not be stable enough to repress unless Cbp80 is absent. Thus, Cbp80 could facilitate the conformational change that competes with L30 binding or that L30 inhibits.
Cbp80 and regulated splicing
Given its multiple roles in RNA processing (including an indirect effect on histone ubiquitination) (Hossain et al. 2009), deletion of Cbp80 can have diverse effects on different transcripts, depending on when its function is more critical. Thus, while early spliceosome events on ACT1 are not severely affected in cbp80Δ cells (Supplemental Fig. S6), microarray analyses (Clark et al. 2002) indicate strong changes in ACT1 splicing in these cells, suggesting an effect on later stages of splicing. Consistent with this, microarray data from cbp80Δ cells correlate well with those from cells with deletions of factors involved either in U2 recruitment (Mud2/U2AF, Msl1/U2B) or U4/U6.U5 tri-snRNP function (Snu66) (Fig. 2 in Clark et al. 2002). This is also in agreement with the ChIP data showing a role for Cbp80 in early and late stages of spliceosome assembly (Gornemann et al. 2005). Furthermore, the same analyses indicate that some of the most affected transcripts in cbp80Δ cells are subjected to regulated splicing and do not have consensus branch site sequences. One example is theYRA1 transcript, with highly regulated splicing in a scheme that includes the link between splicing and RNA export (Preker and Guthrie 2006; Dong et al. 2007), processes both affected by CBC. Therefore, the involvement of Cbp80 in steps involving the recognition of the 3′ end of the intron by U2 snRNP could be targeted as well by several other strategies of regulation.
Our findings may have implications for mammalian systems, with higher variability in splicing signals and more regulated splicing than yeast. CBC is part of the first exon definition complex (Berget 1995), and as such it may play a relevant role in splicing decisions involving alternative 5′ and 3′ splicing sites in these introns, consistent with a possible role in alternative splicing (Raczynska et al. 2010). Moreover, as CBC has been shown to affect cotranscriptional events downstream from the first exon (Wong et al. 2007; Fong et al. 2009), it is not inconceivable that the described modulation of U1 and U2 cotranscriptional recruitment affects a variety of splice site choices along mammalian transcripts.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains and plasmids used in this study are described in the Supplemental material.
Genetic screen
108 yJV25 cells containing pLCUPIF C9U were UV-irradiated and selected on 0.3 mM copper. Splicing of the (refreshed) reporter construct was verified in 46 colonies by Northern analyses, and six SLR mutants were isolated. To identify the mutation, the pLCUP5A-C9U reporter was used, which confers copper resistance to wt cells but not to SLR mutants. Details can be found in the Supplemental material.
Copper assays, Northern analyses, and in vitro assays
Northern and in vitro assays were performed as in Macías et al. (2008). Gel-shifts were done as in Vilardell and Warner (1994). Copper assays were done as in Konarska et al. (2006). RT-PCRs were done as in Hossain et al. (2009), except that our primers contained an M13 sequence tag to ensure specific amplification, and products were analyzed on a 10% nondenaturing, 1× TBE polyacrylamide gel. One nanogram of RNA and 0.5 μg of recombinant protein were used in the reactions. Probes are detailed in the Supplemental material.
ChIP
Chromatin immunoprecipitation and quantitative PCR assays were performed as described in Macías et al. (2008), using the same primer pairs. Error bars are based on three independent biological replicas. RPL30-LacZ mutants described in Figure 6 were generated by in vivo gap-repair cloning, via cotransformation of pJV44 digested with Bam/Spe and a PCR fragment containing the corresponding mutations in the RPL30 intron. Positive colonies were sequenced for verification. (See the Supplemental material for a list of used primers.)
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to C. Query and J. Valcárcel for discussions and critiques to the manuscript, and to F. Azorín, F. Gebauer, R. Méndez, and J. Warner for comments on the manuscript. This research has been supported by the MEC (BFU grants), an EU-MC Contract (#510183), and Agaur. S.M. and M.M. are supported in part by an Agaur fellowship and J.V. has been supported by a Ramón y Cajal/IP3 contract (MEC).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2366310.
REFERENCES
- Abovich N, Rosbash M 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412 [DOI] [PubMed] [Google Scholar]
- Abovich N, Liao XC, Rosbash M 1994. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev 8: 843–854 [DOI] [PubMed] [Google Scholar]
- Berget SM 1995. Exon recognition in vertebrate splicing. J Biol Chem 270: 2411–2414 [DOI] [PubMed] [Google Scholar]
- Brow DA 2002. Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Chao JA, Williamson JR 2004. Joint X-ray and NMR refinement of the yeast L30e-mRNA complex. Structure 12: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M Jr 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296: 907–910 [DOI] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev 10: 1699–1708 [DOI] [PubMed] [Google Scholar]
- Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F 2007. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell 25: 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Du H, Rosbash M 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419: 86–90 [DOI] [PubMed] [Google Scholar]
- Fong N, Ohman M, Bentley DL 2009. Fast ribozyme cleavage releases transcripts from RNA polymerase II and aborts co-transcriptional pre-mRNA processing. Nat Struct Mol Biol 16: 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Bilbao-Cortes D, Fornerod M, Rigaut G, Raymond W, Seraphin B, Mattaj IW 1999a. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev 13: 2425–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW 1999b. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol 19: 6543–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Graveley BR 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ, Graveley BR 2005. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem Sci 30: 115–118 [DOI] [PubMed] [Google Scholar]
- Hossain MA, Claggett JM, Nguyen T, Johnson TL 2009. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA 15: 1515–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW 2008. Regulation of alternative splicing: More than just the ABCs. J Biol Chem 283: 1217–1221 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668 [DOI] [PubMed] [Google Scholar]
- Kent OA, MacMillan AM 2002. Early organization of pre-mRNA during spliceosome assembly. Nat Struct Biol 9: 576–581 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: A new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC 2006. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell 21: 543–553 [DOI] [PubMed] [Google Scholar]
- Liao XC, Colot HV, Wang Y, Rosbash M 1992. Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res 20: 4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macías S, Bragulat M, Tardiff DF, Vilardell J 2008. L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Mol Cell 30: 732–742 [DOI] [PubMed] [Google Scholar]
- Meyer M, Vilardell J 2009. The quest for a message: Budding yeast, a model organism to study the control of pre-mRNA splicing. Brief Funct Genomics Proteomics 8: 60–67 [DOI] [PubMed] [Google Scholar]
- O'Mullane L, Eperon IC 1998. The pre-mRNA 5′ cap determines whether U6 small nuclear RNA succeeds U1 small nuclear ribonucleoprotein particle at 5′ splice sites. Mol Cell Biol 18: 7510–7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49: 229–239 [DOI] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Guthrie C 2006. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA 12: 994–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska KD, Simpson CG, Ciesiolka A, Szewc L, Lewandowska D, McNicol J, Szweykowska-Kulinska Z, Brown JW, Jarmolowski A 2010. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res 38: 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64 [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell 24: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell J, Warner JR 1994. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev 8: 211–220 [DOI] [PubMed] [Google Scholar]
- Vilardell J, Warner JR 1997. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol 17: 1959–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell J, Yu SJ, Warner JR 2000. Multiple functions of an evolutionarily conserved RNA binding domain. Mol Cell 5: 761–766 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wong CM, Qiu H, Hu C, Dong J, Hinnebusch AG 2007. Yeast cap binding complex (CBC) impedes recruitment of cleavage factor IA to weak termination sites. Mol Cell Biol 27: 6520–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Manley JL 1989. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev 3: 1553–1561 [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM 1989. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev 3: 1545–1552 [DOI] [PubMed] [Google Scholar]